SESN2 Could Be a Potential Marker for Diagnosis and Prognosis in Glioma
Abstract
1. Introduction
2. Materials and Methods
2.1. Collecting and Processing Data
2.2. Survival and Statistical Analyses
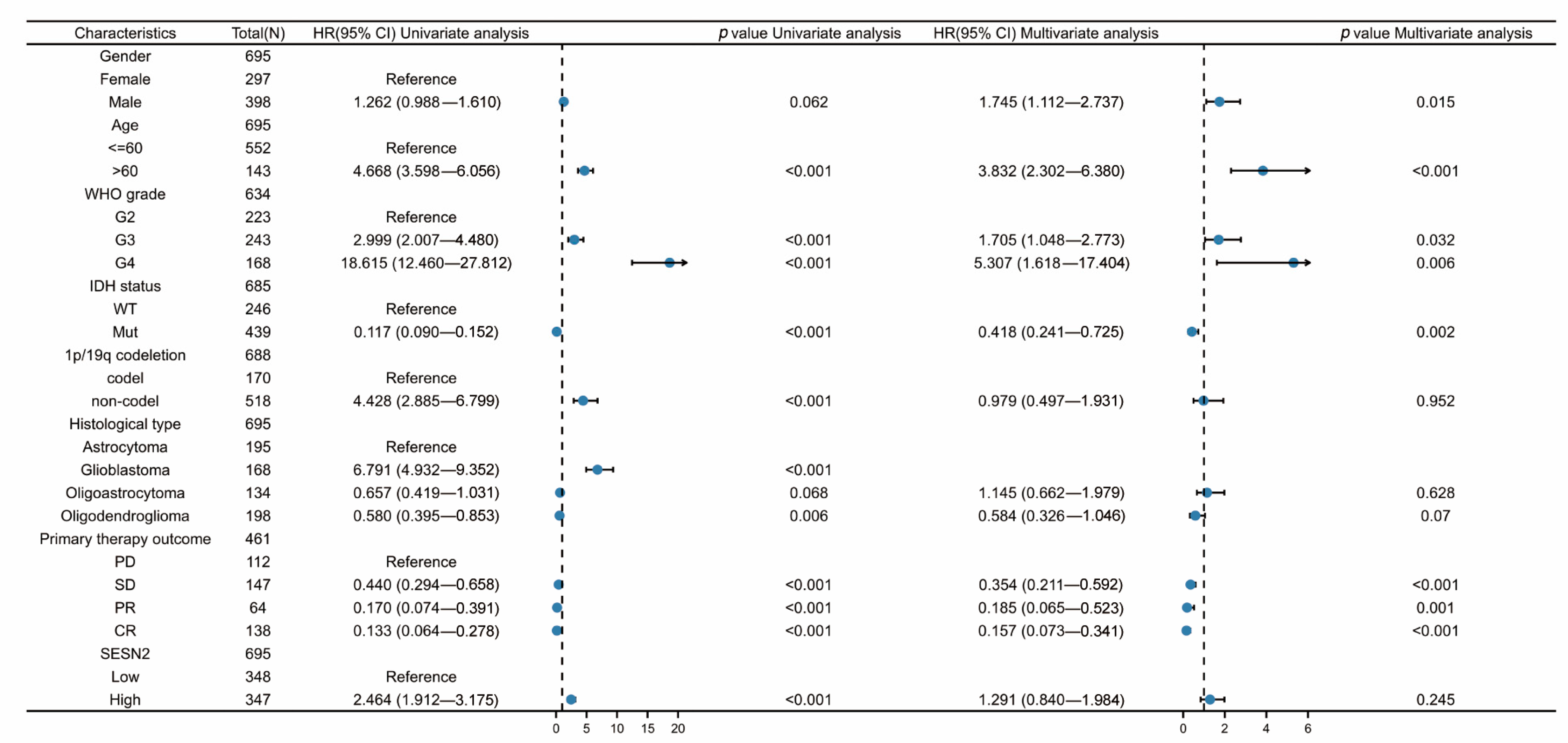
2.3. Univariate and Multivariate Cox Regression Analyses
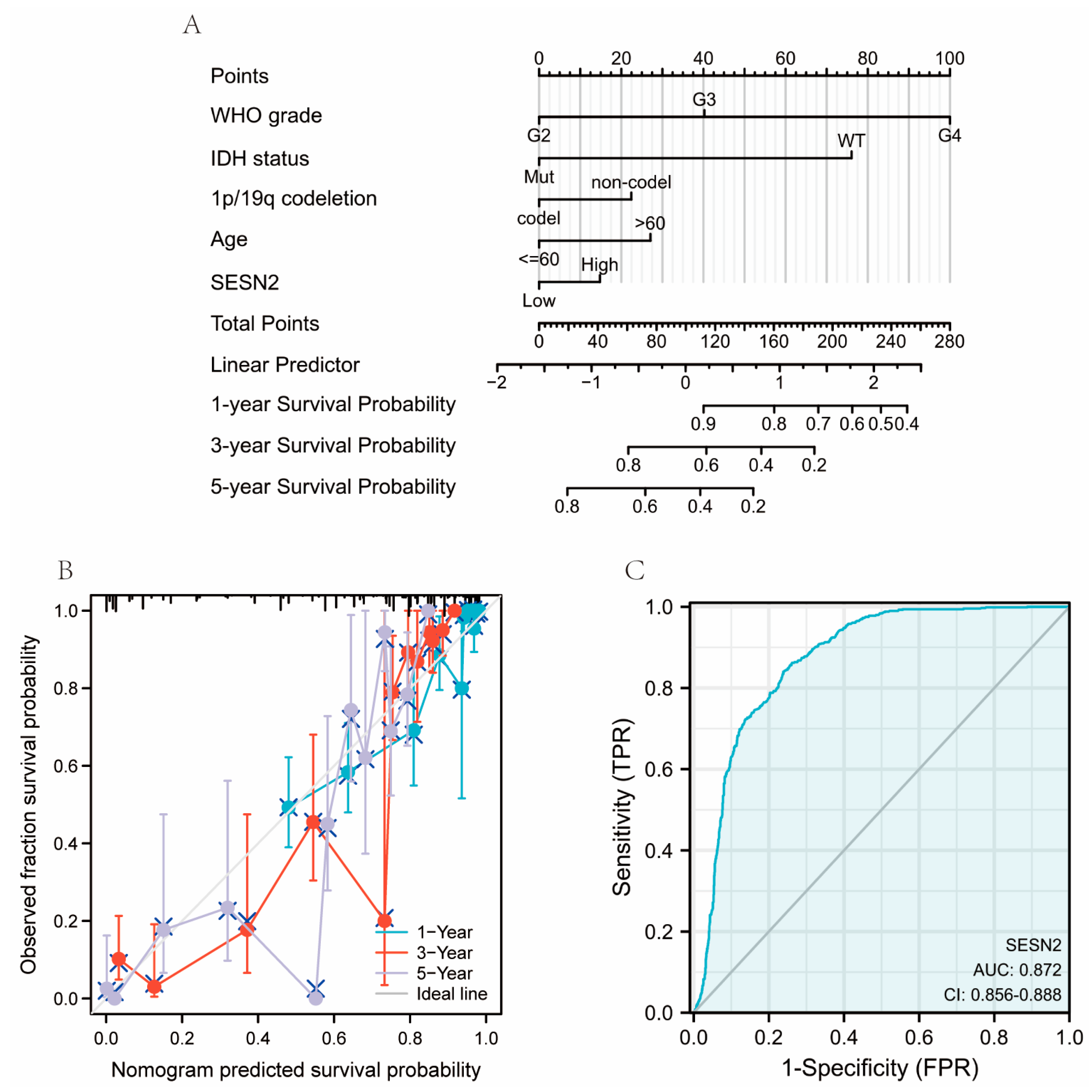
2.4. Construction of Nomograms, Calibration Plots, and ROC Curves
2.5. Analyses of SESN2-Related Gene Function Enrichment and Immune Cell Infiltration
3. Results
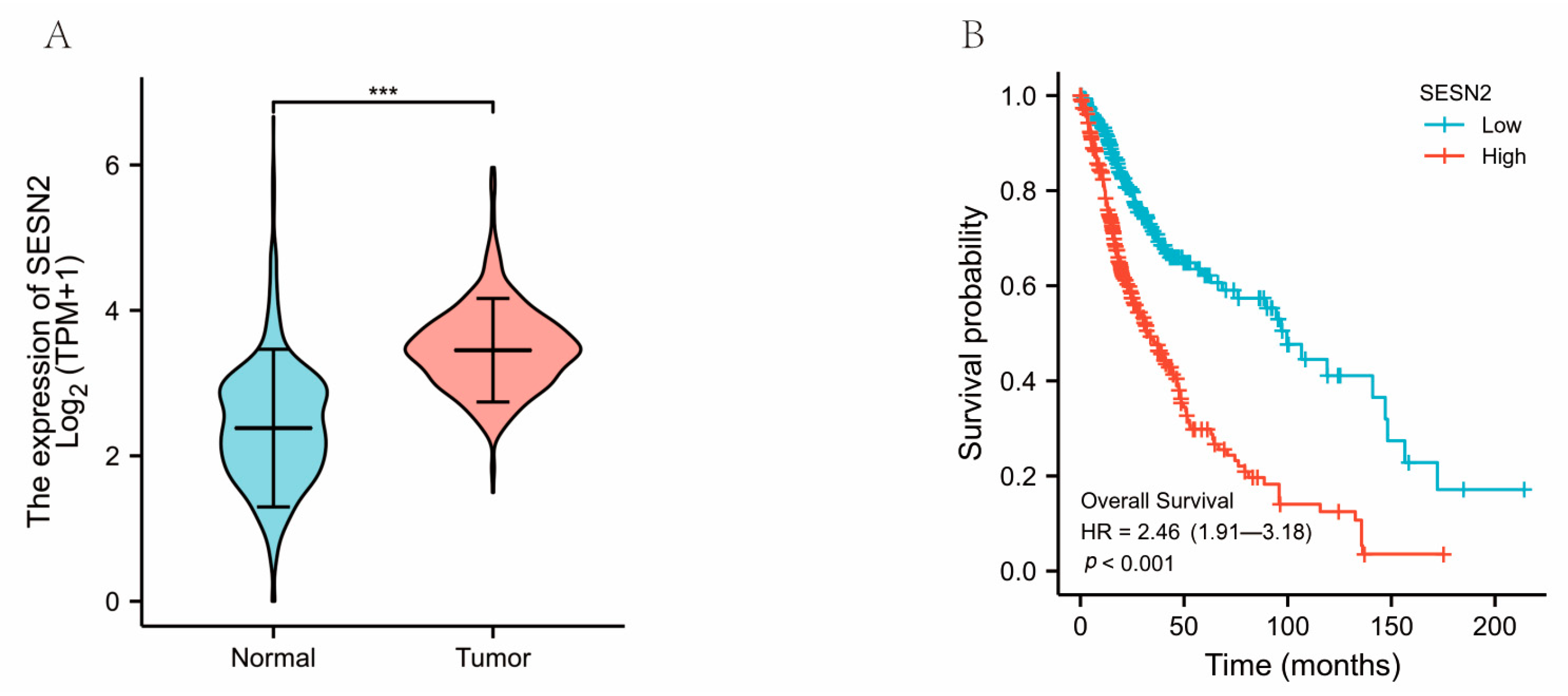
3.1. The Clinical Characteristics and SESN2 Expression of Glioma
3.2. Correlations of Clinical Characteristics with SESN2 Expression in Gliomas
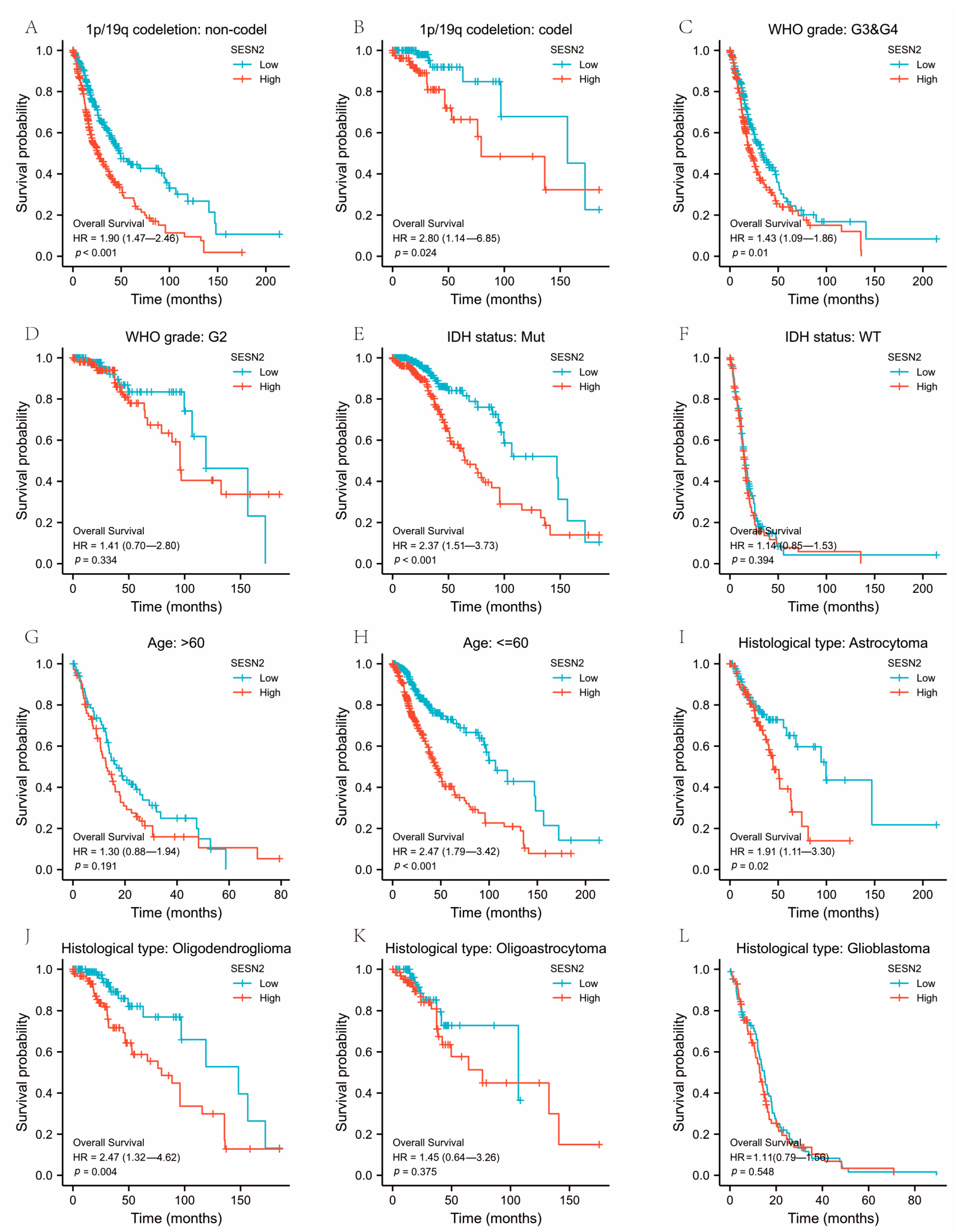
3.3. Associations between SESN2-Related Clinical Characteristics and Prognosis
3.4. Diagnostic Value of SESN2 in Glioma
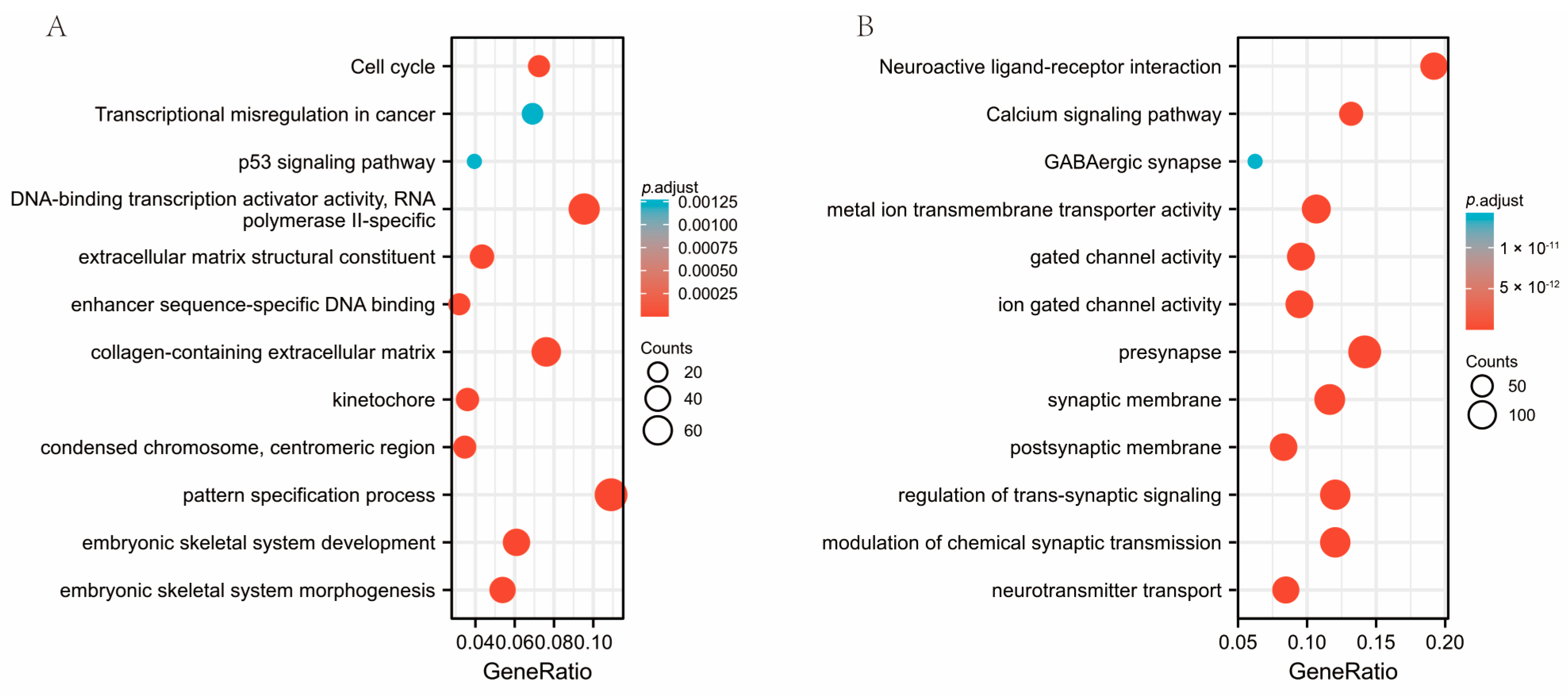
3.5. Co-Expression of Genes Responsible for SESN2 and Predicted Gene Functions
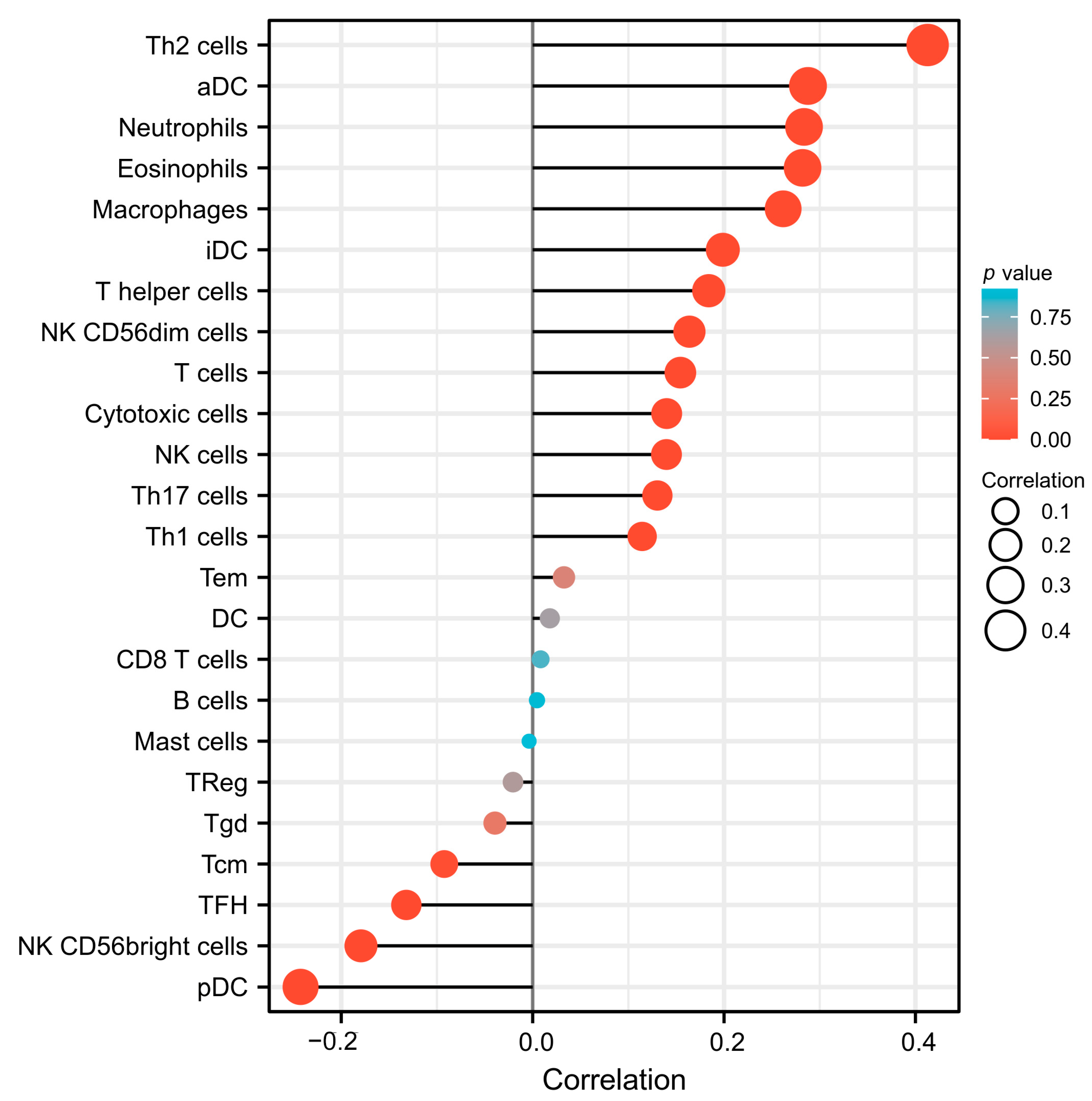
3.6. The Association between SESN2 and Immune Cell Infiltration in Glioma
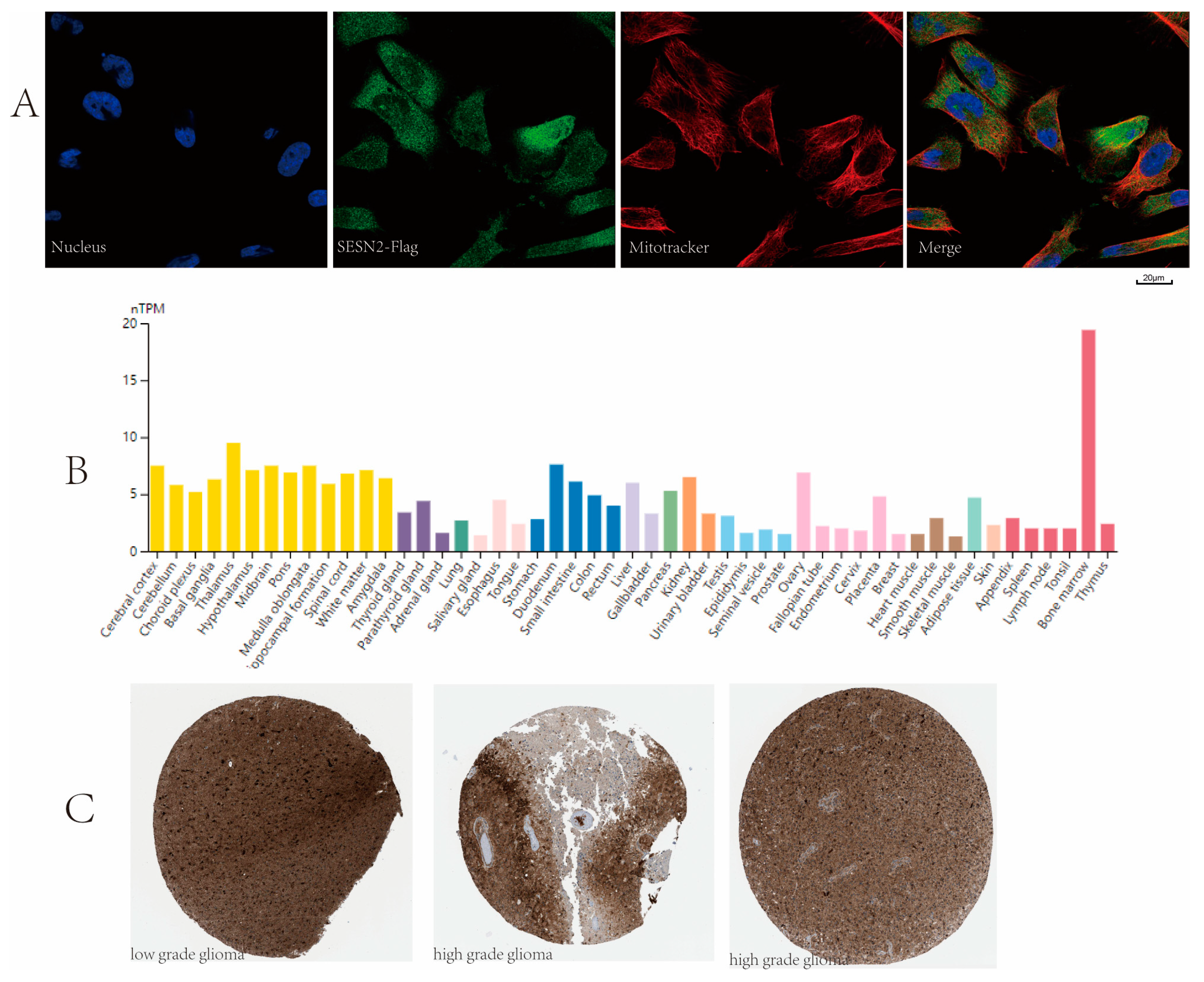
3.7. Subcellular Location and Protein Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Shergalis, A.; Bankhead, A., 3rd; Luesakul, U.; Muangsin, N.; Neamati, N. Current Challenges and Opportunities in Treating Glioblastoma. Pharmacol. Rev. 2018, 70, 412–445. [Google Scholar] [CrossRef] [PubMed]
- Wank, M.; Schilling, D.; Schmid, T.E.; Meyer, B.; Gempt, J.; Barz, M.; Schlegel, J.; Liesche, F.; Kessel, K.A.; Wiestler, B.; et al. Human Glioma Migration and Infiltration Properties as a Target for Personalized Radiation Medicine. Cancers 2018, 10, 456. [Google Scholar] [CrossRef] [PubMed]
- Venur, V.A.; Peereboom, D.M.; Ahluwalia, M.S. Current medical treatment of glioblastoma. Cancer Treat. Res. 2015, 163, 103–115. [Google Scholar] [CrossRef] [PubMed]
- DeWitt, J.C.; Mock, A.; Louis, D.N. The 2016 WHO classification of central nervous system tumors: What neurologists need to know. Curr. Opin. Neurol. 2017, 30, 643–649. [Google Scholar] [CrossRef]
- Bianconi, A.; Aruta, G.; Rizzo, F.; Salvati, L.F.; Zeppa, P.; Garbossa, D.; Cofano, F. Systematic Review on Tumor Microenvironment in Glial Neoplasm: From Understanding Pathogenesis to Future Therapeutic Perspectives. Int. J. Mol. Sci. 2022, 23, 4166. [Google Scholar] [CrossRef]
- Wei, J.; Chen, P.; Gupta, P.; Ott, M.; Zamler, D.; Kassab, C.; Bhat, K.P.; Curran, M.A.; de Groot, J.F.; Heimberger, A.B. Immune biology of glioma-associated macrophages and microglia: Functional and therapeutic implications. Neuro Oncol. 2020, 22, 180–194. [Google Scholar] [CrossRef]
- Ma, Q.; Long, W.; Xing, C.; Chu, J.; Luo, M.; Wang, H.Y.; Liu, Q.; Wang, R.F. Cancer Stem Cells and Immunosuppressive Microenvironment in Glioma. Front. Immunol. 2018, 9, 2924. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Tosolini, M.; Kirilovsky, A.; Waldner, M.; Obenauf, A.C.; Angell, H.; Fredriksen, T.; Lafontaine, L.; Berger, A.; et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013, 39, 782–795. [Google Scholar] [CrossRef]
- Lai, S.W.; Liu, Y.S.; Lu, D.Y.; Tsai, C.F. Melatonin Modulates the Microenvironment of Glioblastoma Multiforme by Targeting Sirtuin 1. Nutrients 2019, 11, 1343. [Google Scholar] [CrossRef]
- Dunn, G.P.; Bruce, A.T.; Ikeda, H.; Old, L.J.; Schreiber, R.D. Cancer immunoediting: From immunosurveillance to tumor escape. Nat. Immunol. 2002, 3, 991–998. [Google Scholar] [CrossRef]
- Chen, S.D.; Yang, J.L.; Lin, T.K.; Yang, D.I. Emerging Roles of Sestrins in Neurodegenerative Diseases: Counteracting Oxidative Stress and Beyond. J. Clin. Med. 2019, 8, 1001. [Google Scholar] [CrossRef]
- Chuang, Y.C.; Yang, J.L.; Yang, D.I.; Lin, T.K.; Liou, C.W.; Chen, S.D. Roles of Sestrin2 and Ribosomal Protein S6 in Transient Global Ischemia-Induced Hippocampal Neuronal Injury. Int. J. Mol. Sci. 2015, 16, 26406–26416. [Google Scholar] [CrossRef]
- Budanov, A.V.; Shoshani, T.; Faerman, A.; Zelin, E.; Kamer, I.; Kalinski, H.; Gorodin, S.; Fishman, A.; Chajut, A.; Einat, P.; et al. Identification of a novel stress-responsive gene Hi95 involved in regulation of cell viability. Oncogene 2002, 21, 6017–6031. [Google Scholar] [CrossRef]
- Bae, S.H.; Sung, S.H.; Oh, S.Y.; Lim, J.M.; Lee, S.K.; Park, Y.N.; Lee, H.E.; Kang, D.; Rhee, S.G. Sestrins activate Nrf2 by promoting p62-dependent autophagic degradation of Keap1 and prevent oxidative liver damage. Cell Metab. 2013, 17, 73–84. [Google Scholar] [CrossRef]
- Wang, L.X.; Zhu, X.M.; Yao, Y.M. Sestrin2: Its Potential Role and Regulatory Mechanism in Host Immune Response in Diseases. Front. Immunol. 2019, 10, 2797. [Google Scholar] [CrossRef]
- Luo, C.; Zhao, S.; Zhang, M.; Gao, Y.; Wang, J.; Hanigan, M.D.; Zheng, N. SESN2 negatively regulates cell proliferation and casein synthesis by inhibition the amino acid-mediated mTORC1 pathway in cow mammary epithelial cells. Sci. Rep. 2018, 8, 3912. [Google Scholar] [CrossRef]
- Chen, S.; Yan, W.; Lang, W.; Yu, J.; Xu, L.; Xu, X.; Liu, Y.; Bao, H. SESN2 correlates with advantageous prognosis in hepatocellular carcinoma. Diagn. Pathol. 2017, 12, 13. [Google Scholar] [CrossRef]
- Buitrago-Molina, L.E.; Marhenke, S.; Longerich, T.; Sharma, A.D.; Boukouris, A.E.; Geffers, R.; Guigas, B.; Manns, M.P.; Vogel, A. The degree of liver injury determines the role of p21 in liver regeneration and hepatocarcinogenesis in mice. Hepatology 2013, 58, 1143–1152. [Google Scholar] [CrossRef]
- Seo, K.; Ki, S.H.; Park, E.Y.; Shin, S.M. 5-Fluorouracil inhibits cell migration by induction of Sestrin2 in colon cancer cells. Arch. Pharm. Res. 2017, 40, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.; Kim, D.Y.; Kang, S.H.; Yun, H.K.; Kim, J.L.; Kim, B.R.; Park, S.H.; Na, Y.J.; Jo, M.J.; Jeong, Y.A.; et al. Docosahexaenoic Acid Enhances Oxaliplatin-Induced Autophagic Cell Death via the ER Stress/Sesn2 Pathway in Colorectal Cancer. Cancers 2019, 11, 982. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Yin, K.; Falcon, D.M.; Xue, X. The interaction of Hemin and Sestrin2 modulates oxidative stress and colon tumor growth. Toxicol. Appl. Pharmacol. 2019, 374, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Shah, P.; Budanov, A.V.; Qiang, L.; Ming, M.; Aplin, A.; Sims, D.M.; He, Y.Y. Sestrin2 protein positively regulates AKT enzyme signaling and survival in human squamous cell carcinoma and melanoma cells. J. Biol. Chem. 2014, 289, 35806–35814. [Google Scholar] [CrossRef]
- Liu, S.Y.; Lee, Y.J.; Lee, T.C. Association of platelet-derived growth factor receptor β accumulation with increased oxidative stress and cellular injury in sestrin 2 silenced human glioblastoma cells. FEBS Lett. 2011, 585, 1853–1858. [Google Scholar] [CrossRef]
- Blum, A.; Wang, P.; Zenklusen, J.C. SnapShot: TCGA-Analyzed Tumors. Cell 2018, 173, 530. [Google Scholar] [CrossRef]
- Ceccarelli, M.; Barthel, F.P.; Malta, T.M.; Sabedot, T.S.; Salama, S.R.; Murray, B.A.; Morozova, O.; Newton, Y.; Radenbaugh, A.; Pagnotta, S.M.; et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell 2016, 164, 550–563. [Google Scholar] [CrossRef]
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018, 173, 400–416.e11. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef]
- Balachandran, V.P.; Gonen, M.; Smith, J.J.; DeMatteo, R.P. Nomograms in oncology: More than meets the eye. Lancet Oncol. 2015, 16, e173–e180. [Google Scholar] [CrossRef]
- Brentnall, A.R.; Cuzick, J. Use of the concordance index for predictors of censored survival data. Stat. Methods Med. Res. 2018, 27, 2359–2373. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Cioffi, G.; Gittleman, H.; Patil, N.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2012-2016. Neuro Oncol. 2019, 21 (Suppl. S5), v1–v100. [Google Scholar] [CrossRef]
- Smoll, N.R.; Gautschi, O.P.; Schatlo, B.; Schaller, K.; Weber, D.C. Relative survival of patients with supratentorial low-grade gliomas. Neuro Oncol. 2012, 14, 1062–1069. [Google Scholar] [CrossRef]
- Ohgaki, H.; Kleihues, P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J. Neuropathol. Exp. Neurol. 2005, 64, 479–489. [Google Scholar] [CrossRef]
- Bleeker, F.E.; Molenaar, R.J.; Leenstra, S. Recent advances in the molecular understanding of glioblastoma. J. Neurooncol. 2012, 108, 11–27. [Google Scholar] [CrossRef]
- Saaid, A.; Monticelli, M.; Ricci, A.A.; Orlando, G.; Botta, C.; Zeppa, P.; Bianconi, A.; Osella-Abate, S.; Bruno, F.; Pellerino, A.; et al. Prognostic Analysis of the IDH1 G105G (rs11554137) SNP in IDH-Wildtype Glioblastoma. Genes 2022, 13, 1439. [Google Scholar] [CrossRef]
- Li, T.; Jiang, D.; Wu, K. p62 promotes bladder cancer cell growth by activating KEAP1/NRF2-dependent antioxidative response. Cancer Sci. 2020, 111, 1156–1164. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)1. Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef]
- Reuss, D.E.; Kratz, A.; Sahm, F.; Capper, D.; Schrimpf, D.; Koelsche, C.; Hovestadt, V.; Bewerunge-Hudler, M.; Jones, D.T.; Schittenhelm, J.; et al. Adult IDH wild type astrocytomas biologically and clinically resolve into other tumor entities. Acta Neuropathol. 2015, 130, 407–417. [Google Scholar] [CrossRef]
- Suzuki, H.; Aoki, K.; Chiba, K.; Sato, Y.; Shiozawa, Y.; Shiraishi, Y.; Shimamura, T.; Niida, A.; Motomura, K.; Ohka, F.; et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat. Genet. 2015, 47, 458–468. [Google Scholar] [CrossRef]
- Rhee, S.G.; Bae, S.H. The antioxidant function of sestrins is mediated by promotion of autophagic degradation of Keap1 and Nrf2 activation and by inhibition of mTORC1. Free Radic. Biol. Med. 2015, 88 Pt B, 205–211. [Google Scholar] [CrossRef]
- Wu, C.L.; Chen, S.D.; Yin, J.H.; Hwang, C.S.; Yang, D.I. Nuclear Factor-kappaB-Dependent Sestrin2 Induction Mediates the Antioxidant Effects of BDNF Against Mitochondrial Inhibition in Rat Cortical Neurons. Mol. Neurobiol. 2016, 53, 4126–4142. [Google Scholar] [CrossRef]
- Lin, M.Y.; Chang, Y.C.; Wang, S.Y.; Yang, M.H.; Chang, C.H.; Hsiao, M.; Kitsis, R.N.; Lee, Y.J. OncomiR miR-182-5p Enhances Radiosensitivity by Inhibiting the Radiation-Induced Antioxidant Effect through SESN2 in Head and Neck Cancer. Antioxidants 2021, 10, 1808. [Google Scholar] [CrossRef]
- Kumar, A.; Shaha, C. SESN2 facilitates mitophagy by helping Parkin translocation through ULK1 mediated Beclin1 phosphorylation. Sci. Rep. 2018, 8, 615. [Google Scholar] [CrossRef]
- Malik, B.; Devine, H.; Patani, R.; La Spada, A.R.; Hanna, M.G.; Greensmith, L. Gene expression analysis reveals early dysregulation of disease pathways and links Chmp7 to pathogenesis of spinal and bulbar muscular atrophy. Sci. Rep. 2019, 9, 3539. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wei, J.; Li, Z.; Tian, Y.; Du, C. Bioinformatical analysis of gene expression signatures of different glioma subtypes. Oncol. Lett. 2018, 15, 2807–2814. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; He, F.; Zhang, W.; Chen, W.; Yu, B. Bioinformatics analysis of microarray data to reveal the pathogenesis of diffuse intrinsic pontine glioma. Biol. Res. 2018, 51, 26. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Xiong, P.; Luo, Y.; Bu, X.; Qian, S.; Zhong, W. Bioinformatics analysis of the molecular mechanism of diffuse intrinsic pontine glioma. Oncol. Lett. 2016, 12, 2524–2530. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, D.; Wang, L.; Yu, J.; Du, D.; Chen, Y.; Gao, P.; Wang, D.M.; Yu, J.; Zhang, F.; et al. Distinct genomic aberrations between low-grade and high-grade gliomas of Chinese patients. PLoS ONE 2013, 8, e57168. [Google Scholar] [CrossRef]
- Venkataramani, V.; Tanev, D.I.; Strahle, C.; Studier-Fischer, A.; Fankhauser, L.; Kessler, T.; Körber, C.; Kardorff, M.; Ratliff, M.; Xie, R.; et al. Glutamatergic synaptic input to glioma cells drives brain tumour progression. Nature 2019, 573, 532–538. [Google Scholar] [CrossRef]
- Inácio, R.F.; Zanon, R.G.; Castro, M.V.; Souza, H.M.; Bajgelman, M.C.; Verinaud, L.; Oliveira, A.L. Astroglioma conditioned medium increases synaptic elimination and correlates with major histocompatibility complex of class I (MHC I) upregulation in PC12Cells. Neurosci. Lett. 2016, 634, 160–167. [Google Scholar] [CrossRef]
- de Groot, M.; Aronica, E.; Heimans, J.J.; Reijneveld, J.C. Synaptic vesicle protein 2A predicts response to levetiracetam in patients with glioma. Neurology 2011, 77, 532–539. [Google Scholar] [CrossRef]
- Qi, Y.; Wang, Z.; Wu, F.; Yin, B.; Jiang, T.; Qiang, B.; Yuan, J.; Han, W.; Peng, X. Long noncoding RNA HOXD-AS2 regulates cell cycle to promote glioma progression. J. Cell Biochem. 2019, 120, 8343–8351. [Google Scholar] [CrossRef]
- Zhou, W.; Ouyang, J.; Li, J.; Liu, F.; An, T.; Cheng, L.; Kuo, Z.C.; Zhang, C.; He, Y. MRPS17 promotes invasion and metastasis through PI3K/AKT signal pathway and could be potential prognostic marker for gastric cancer. J. Cancer 2021, 12, 4849–4861. [Google Scholar] [CrossRef]
- Sadler, T.; Scarpa, M.; Rieder, F.; West, G.; Stylianou, E. Cytokine-induced chromatin modifications of the type I collagen alpha 2 gene during intestinal endothelial-to-mesenchymal transition. Inflamm. Bowel Dis. 2013, 19, 1354–1364. [Google Scholar] [CrossRef]
- Cheng, Y.; Liu, C.; Liu, Y.; Su, Y.; Wang, S.; Jin, L.; Wan, Q.; Liu, Y.; Li, C.; Sang, X.; et al. Immune Microenvironment Related Competitive Endogenous RNA Network as Powerful Predictors for Melanoma Prognosis Based on WGCNA Analysis. Front. Oncol. 2020, 10, 577072. [Google Scholar] [CrossRef]
- Platten, M.; Weller, M.; Wick, W. Shaping the glioma immune microenvironment through tryptophan metabolism. CNS Oncol. 2012, 1, 99–106. [Google Scholar] [CrossRef]
- Constant, S.L.; Bottomly, K. Induction of Th1 and Th2 CD4+ T cell responses: The alternative approaches. Annu. Rev. Immunol. 1997, 15, 297–322. [Google Scholar] [CrossRef]
- Li, G.; Hu, Y.S.; Li, X.G.; Zhang, Q.L.; Wang, D.H.; Gong, S.F. Expression and switching of TH1/TH2 type cytokines gene in human gliomas. Chin. Med. Sci. J. 2005, 20, 268–272. [Google Scholar]
- Moriyama, M.; Tanaka, A.; Maehara, T.; Furukawa, S.; Nakashima, H.; Nakamura, S. T helper subsets in Sjögren’s syndrome and IgG4-related dacryoadenitis and sialoadenitis: A critical review. J. Autoimmun. 2014, 51, 81–88. [Google Scholar] [CrossRef]
- Pei, J.P.; Zhang, C.D.; Yusupu, M.; Zhang, C.; Dai, D.Q. Screening and Validation of the Hypoxia-Related Signature of Evaluating Tumor Immune Microenvironment and Predicting Prognosis in Gastric Cancer. Front. Immunol. 2021, 12, 705511. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Yuan, Y.; Chen, C.; Lin, J.; Ma, Q.; Liu, G.; Gao, Y.; Huang, Y.; Chen, L.; Chen, L.Z.; et al. Durable complete response to neoantigen-loaded dendritic-cell vaccine following anti-PD-1 therapy in metastatic gastric cancer. NPJ Precis. Oncol. 2022, 6, 34. [Google Scholar] [CrossRef] [PubMed]
- Cooper, M.A.; Fehniger, T.A.; Caligiuri, M.A. The biology of human natural killer-cell subsets. Trends Immunol. 2001, 22, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Munitz, A.; Levi-Schaffer, F. Eosinophils: ‘New’ roles for ‘old’ cells. Allergy 2004, 59, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.P.; Rothenberg, M.E. Eosinophils and cancer. Cancer Immunol. Res. 2014, 2, 1–8. [Google Scholar] [CrossRef]
- Curran, C.S.; Bertics, P.J. Eosinophils in glioblastoma biology. J. Neuroinflamm. 2012, 9, 11. [Google Scholar] [CrossRef]


| Characteristic | Levels | Overall |
|---|---|---|
| WHO grade, n (%) | G2 | 224 (35.3%) |
| G3 | 243 (38.3%) | |
| G4 | 168 (26.5%) | |
| IDH status, n (%) | WT | 246 (35.9%) |
| Mut | 440 (64.1%) | |
| Histological type, n (%) | Astrocytoma | 195 (28%) |
| Glioblastoma | 168 (24.1%) | |
| Oligoastrocytoma | 134 (19.3%) | |
| Oligodendroglioma | 199 (28.6%) | |
| Age, n (%) | ≤60 | 553 (79.5%) |
| >60 | 143 (20.5%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, L.; Liu, Z.; Wang, H.; Lu, J.; Xu, J.; Meng, Y.; Huang, K.; Liu, B. SESN2 Could Be a Potential Marker for Diagnosis and Prognosis in Glioma. Genes 2023, 14, 701. https://doi.org/10.3390/genes14030701
Xu L, Liu Z, Wang H, Lu J, Xu J, Meng Y, Huang K, Liu B. SESN2 Could Be a Potential Marker for Diagnosis and Prognosis in Glioma. Genes. 2023; 14(3):701. https://doi.org/10.3390/genes14030701
Chicago/Turabian StyleXu, Lingdan, Zelin Liu, Huihui Wang, Jiyuan Lu, Jia Xu, Yucheng Meng, Ke Huang, and Bin Liu. 2023. "SESN2 Could Be a Potential Marker for Diagnosis and Prognosis in Glioma" Genes 14, no. 3: 701. https://doi.org/10.3390/genes14030701
APA StyleXu, L., Liu, Z., Wang, H., Lu, J., Xu, J., Meng, Y., Huang, K., & Liu, B. (2023). SESN2 Could Be a Potential Marker for Diagnosis and Prognosis in Glioma. Genes, 14(3), 701. https://doi.org/10.3390/genes14030701





