Genome-Wide Association Analysis of Grain Hardness in Common Wheat
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Grain Hardness (HI) Measurement and Grading
2.3. Statistical Analysis
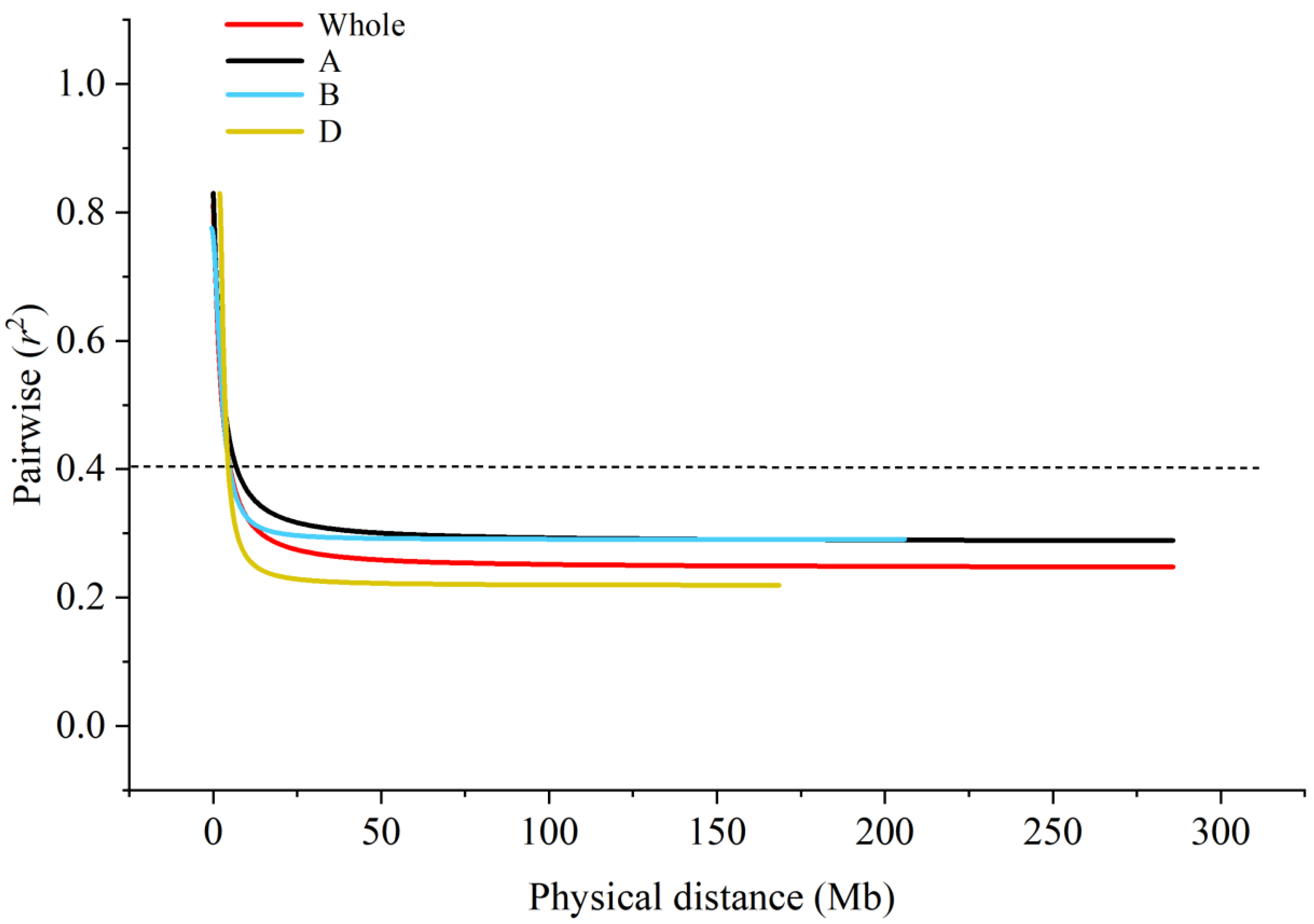
2.4. Chip Typing and Population Structure Analysis
2.5. Correlation Analysis
2.6. Candidate Gene Screening
3. Results
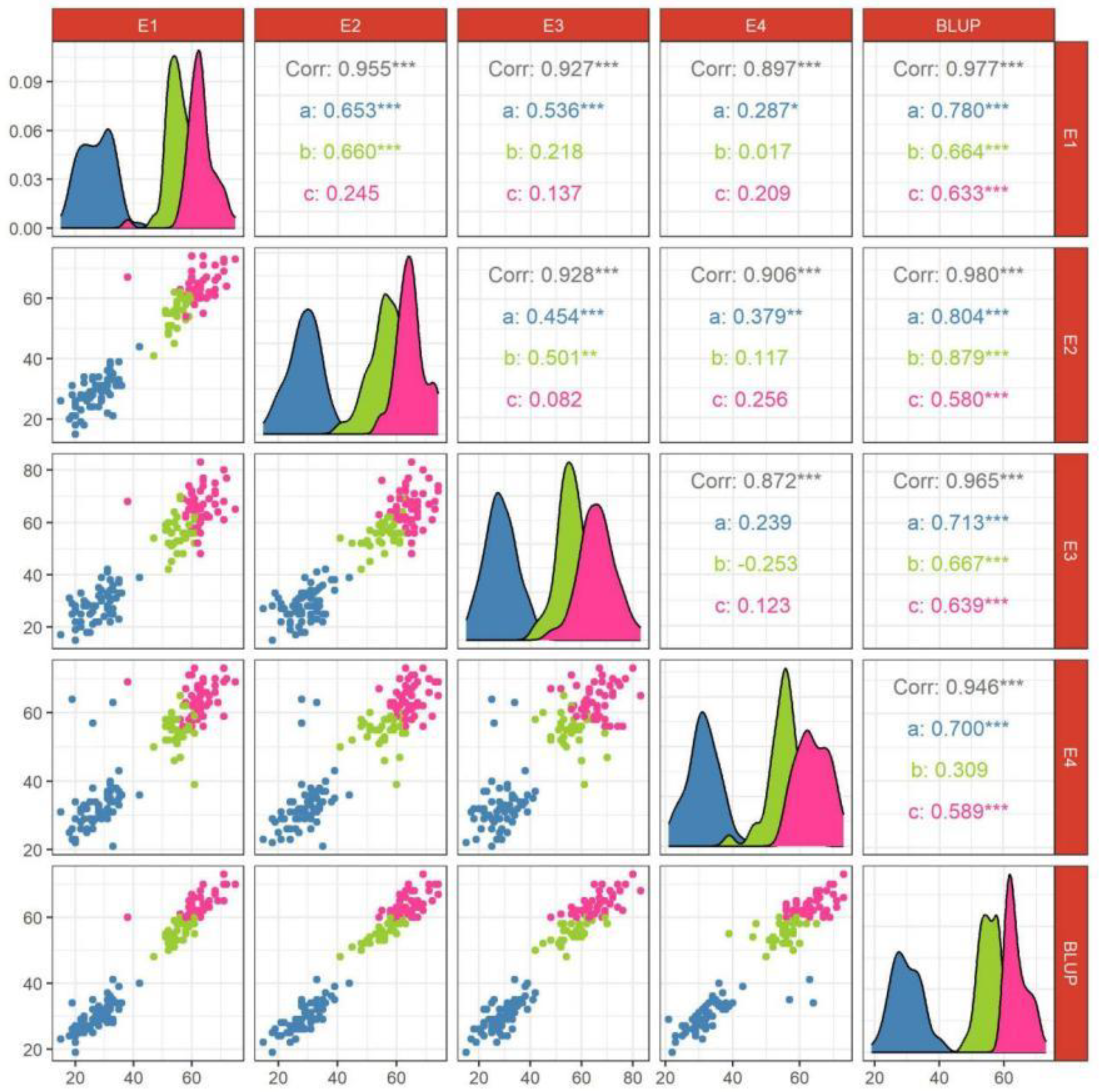
3.1. Phenotype Analysis
3.2. Variance and Correlation Analysis
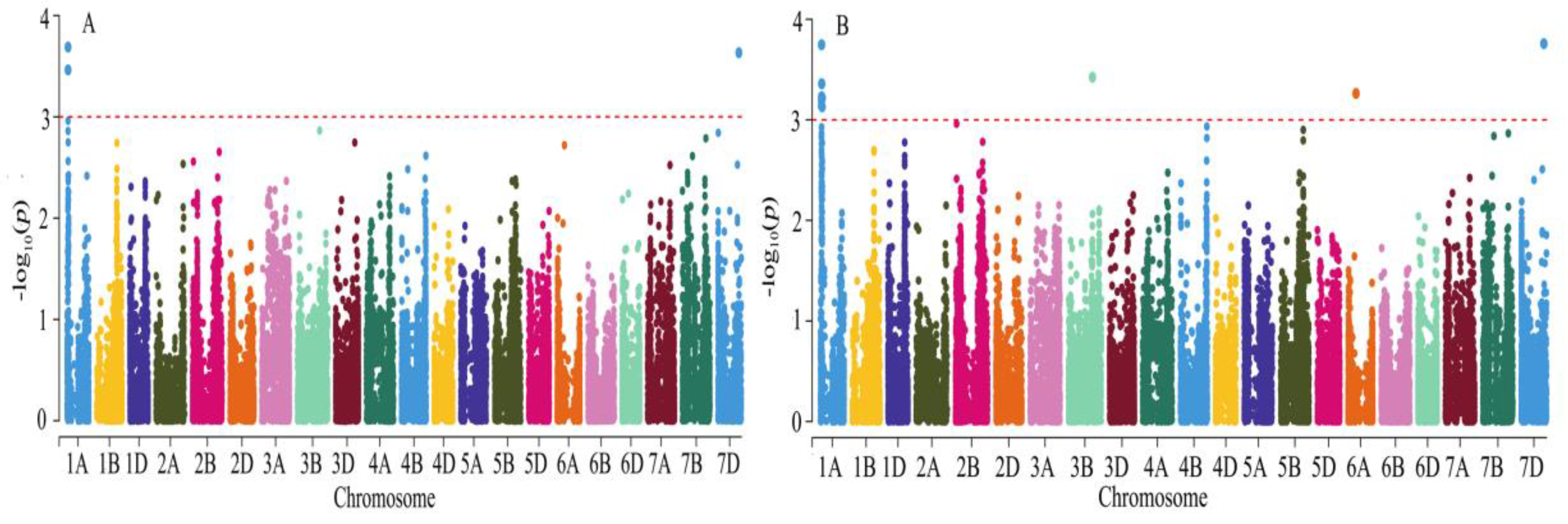
3.3. Grain Hardness Loci Identified by GWAS
3.4. Functional Prediction of Candidate Genes
4. Discussion
4.1. Phenotypic Variation of Wheat Grain Hardness
4.2. Genome-Wide Association Analysis of Wheat Grain Hardness
4.3. Candidate Gene Prediction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, H.; Jia, L.Z.; Ming, M.Y.; Zhao, C.; Ning, T.Y.; Jiao, N.Y. Current status and research advances on quality regionalization in the high quality wheat. J. Triticeae Crop. 2005, 3, 112–114. [Google Scholar]
- Zhou, Y.; Chen, Z.; Cheng, M.; Chen, J.; Zhu, T.T.; Wang, R.; Liu, Y.X.; Qi, P.F.; Chen, G.Y.; Jiang, Q.T.; et al. Uncovering the dispersion history, adaptive evolution and selection of wheat in China. Plant Biotech. 2018, 16, 280–291. [Google Scholar] [CrossRef]
- Hu, X.X.; Sun, L.J.; Zhou, G.Y.; Wu, L.N.; Lu, W.; Li, W.X.; Wang, S.; Yang, X.L.; Song, K.J.; Wang, B.J. Variations of wheat quality in china from 2006–2015. Sci. Agric. Sin. 2016, 49, 3063–3072. [Google Scholar]
- Wang, J.Y.; Yang, C.K.; Zhao, W.J.; Wang, Y.; Qiao, L.; Wu, B.B.; Zhao, J.J.; Zheng, X.W.; Wang, J.L.; Zheng, J. Genome-wide association study of grain hardness and novel Puroindoline alleles in common wheat. Mol. Breed. 2022, 40, 42. [Google Scholar] [CrossRef]
- Chen, F.; Li, G.Y.; Geng, H.W.; Xia, L.Q.; Xia, X.C.; He, Z.H. Review and prospect of wheat kernel hardness and Its molecular genetics basis. Chin. Agric. Sci. 2005, 38, 1088–1094. [Google Scholar]
- General Administration of Quality Supervision, Inspection and Quarantine of the People’s Republic of China. National Standard of the People’s Republic of China: GB/T1351-2008[S]; Standards Press of China: Beijing, China, 2008. [Google Scholar]
- Wang, L.L.; Wang, S.J.; Shi, J. Specificities and research progress of hardness of wheat seeds. Cereals Oils 2006, 1, 19–22. [Google Scholar]
- Simmonds, D.H.; Barlow, K.K.; Wrigley, C.W. The biochemical basis of grain hardness in wheat. Cereal Chem. 1973, 50, 553–562. [Google Scholar]
- Morris, C.F.; Massa, A.N. Puroindoline genotype of the U.S. national institute of standards & technology reference material. Cereal Chem. 2003, 80, 674–678. [Google Scholar]
- Zhang, X.; Zhang, Y.; Gao, D.R.; Bie, T.D.; Zhang, B.Q. The development of weak-gluten wheat breeding and present situation of its production. J. Triticeae Crop. 2012, 32, 184–189. [Google Scholar]
- Li, S.B. Study on structural characters of grain endosperm of wheat. J. Northwest Sci.-Tech. Univ. 2022, 30, 7–10. [Google Scholar]
- Hu, W.J.; Zhang, X.; Liu, Q.; Fang, Z.W.; Gao, D.R. Genome-wide association study of grain hardness in common wheat. J. Triticeae Crop. 2021, 41, 157–163. [Google Scholar]
- Liu, J.; He, Z.H.; Zhao, Z.D.; Pena, R.J.; Rajaram, S. Wheat quality traits and quality parameters of cooked dry white Chinese noodles. Euphytica 2003, 131, 147–154. [Google Scholar] [CrossRef]
- Li, H.M.; Liang, H.; Tang, Z.X. QTL analysis for grain pentosans and hardness index in a chinese 1RS.1BL×NON-1RS.1BL wheat cross. Plant Mol. Biol. Rep. 2013, 31, 477–484. [Google Scholar] [CrossRef]
- Gazza, L.; Taddei, F.; Corbellini, M.; Cacciatori, P.; Pogna, N.E. Genetic and environmental factors affecting grain texture in common wheat. J. Cereal Sci. 2008, 47, 52–58. [Google Scholar] [CrossRef]
- Aoun, M.; Carter, A.H.; Ward, B.P.; Morris, C.F. Genome-wide association mapping of the ‘super-soft’ kernel texture in white winter wheat. Theor. Appl. Genet. 2021, 134, 2547–2559. [Google Scholar] [CrossRef]
- Sun, X.; Liu, T.; Ning, T.Y.; Liu, K.; Duan, X.X.; Wang, X.R.; Wang, Q.L.; An, Y.L.; Guan, X.; Tian, J.C. Genetic dissection of wheat kernel hardness using conditional QTL mapping of kernel size and protein-related traits. Plant Mol. Biol. Rep. 2018, 36, 1–12. [Google Scholar] [CrossRef]
- Wang, G.M.; Leonard, J.M.; Ross, A.S.; Peterson, C.J.; Zemetra, R.S.; Campbell, K.G.; Riera-Lizarazu, O. Identification of genetic factors controlling kernel hardness and related traits in a recombinant inbred population derived from a soft × ‘extra-soft’ wheat (Triticum aestivum, L.) cross. Theor. Appl. Genet. 2012, 124, 207–221. [Google Scholar] [CrossRef]
- Tu, M.; Li, Y. Toward the genetic basis and multiple QTLS of kernel hardness in wheat. Plants 2020, 9, 1631. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Marza, F.; Ma, H.; Carver, B.F.; Bai, G.H. Mapping quantitative trait loci for quality factors in an inter-class cross of US and Chinese wheat. Theor. Appl. Genet. 2010, 120, 1041. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cui, F.; Ding, A.M.; Zhao, C.H.; Wang, X.Q.; Wang, L.; Bao, Y.G.; Qi, X.L.; Li, X.F.; Gao, J.R.; et al. QTL detection of seven quality traits in wheat using two related recombinant inbred line populations. Euphytica 2012, 183, 207–226. [Google Scholar] [CrossRef]
- Maria, I.K.; Alecia, M.S.; Deven, R.S.; Daniel, Z.M.; Craig, F. Mapping kernel texture in a soft durum (Triticum turgidum subsp. durum) wheat population. J. Cereal Sci. 2019, 85, 20–26. [Google Scholar]
- Gao, L.; Yang, J.; Song, S.J.; Xu, K.; Zhao, Y. Genome–wide association study of grain morphology in wheat. Euphytica 2021, 170, 217. [Google Scholar] [CrossRef]
- Yan, Y.L.; Shi, X.L.; Zhang, J.B.; Geng, H.W.; Xiao, J.; Lu, Z.F.; Ni, Z.F.; Cong, H. Genome-wide association study of grain quality related characteristics of spring wheat. Chin. Agric. Sci. 2021, 54, 4033–4047. [Google Scholar]
- Mangini, G.; Blanco, A.; Nigro, D.; Signorile, M.A.; Simeone, R. Candidate genes and quantitative trait loci for grain yield and seed size in durum wheat. Plants 2021, 10, 312. [Google Scholar] [CrossRef]
- Muhammad, A.; Li, J.; Hu, W.; Yu, J.S.; Wang, L.Q. Uncovering genomic regions controlling plant architectural traits in hexaploid wheat using different GWAS models. Sci. Rep. 2021, 11, 6767. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, S.; Wei, W.; Xie, H.Y.; Liu, K.; Zhang, C.; Wu, Z.Y.; Jiang, H.; Cao, J.J.; Zhao, L.X.; et al. Genome-wide association study of pre-harvest sprouting tolerance using a 90K SNP array in common wheat (Triticum aestivum L.). Theor. Appl. Genet. 2019, 132, 2947–2963. [Google Scholar] [CrossRef]
- Chemists, A. Approved Methods of the American Association of Cereal Chemists; AACC: Arnold, MD, USA, 1995. [Google Scholar]
- Bates, D.; Mchler, M.; Bolker, B.M.; Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Schloerke, B.; Crowley, J.; Di, C.; Wickham, H.; Briatte, A.F.; Marbach, A.M. GGally: Extension to ggplot2. Log. Et Anal. 2014, 10, 141–156. [Google Scholar]
- Wang, S.X.; Zhu, Y.L.; Zhang, H.P.; Chang, C.; Ma, C.X. Analysis of genetic diversity and relationship among wheat breeding parents by SSR markers. J. Triticeae Crop. 2014, 34, 621–627. [Google Scholar]
- Wang, J.; Zhang, Z. GAPIT Version 3: Boosting Power and Accuracy for Genomic Association and Prediction. Genom. Proteom. Bioinf. 2021, 19, 629–640. [Google Scholar] [CrossRef]
- Xiao, Y.N.; Li, G.K.; Li, K.; Yu, Y.T.; Li, G.Y.; Li, W.; Gao, Y.S.; Hu, J.G. Genome-wide association study of kernel volume and weight in sweet corn. J. China Agric. Univ. 2022, 27, 12–25. [Google Scholar]
- Hao, S.; Lou, H.; Wang, H.; Shi, J.; Liu, D.; Baogerile; Tao, J.; Miao, S.; Pei, Q.; Yu, L.; et al. Genome-Wide Association Study Reveals the Genetic Basis of Five Quality Traits in Chinese Wheat. Front. Plant. 2022, 13, 835306. [Google Scholar] [CrossRef]
- Casagrande, C.R.; Mezzomo, H.C.; Borem, A.; Nardino, M. Choosing parent tropical wheat genotypes through genetic dissimilarity based on REML/BLUP. Crop. Breed. Appl. Biot. 2020, 20, 1–10. [Google Scholar] [CrossRef]
- Yan, Y.L.; Zhang, H.; Zhang, J.B.; Shi, X.L.; Geng, H.W.; Xiao, J.; Lu, Z.F.; Ni, Z.F.; Cong, H. Genome-wide association study of grain traits of spring wheat. J. Triticeae Crop. 2022, 42, 1182–1191. [Google Scholar]
- Mergoum, M.; Harilal, A.; Simsek, A.S.; Alamri, B.; Bassi, A. Agronomic and Quality Qtl Mapping in Spring Wheat. Indian J. Genet. Pl. Br. 2013, 1, 19–33. [Google Scholar]
- Liu, P.X.; Ma, X.F.; Wan, H.S.; Zheng, J.M.; Luo, J.T.; Pu, Z.G. Comparative proteomic analysis of two wheat genotypes with contrasting grain softness index. Acta Agron. Sin. 2020, 46, 1275–1282. [Google Scholar]
- Xu, T.; Xia, D.J.; Wan, J.; Jiang, S.H.; Song, J.H. Research progress of F-box protein involved in plant stress. Biotechnol. Bull. 2021, 37, 205–211. [Google Scholar]
- Ezeilo, U.R.; Zakaria, I.I.; Huyop, F.; Wahab, R.A. Enzymatic breakdown of lignocellulosic biomass: The role of glycosyl hydrolases and lytic polysaccharide monooxygenases. Biotechnol. Biotechnol. Equip. 2017, 31, 647–662. [Google Scholar] [CrossRef]
| Type | Environment | Min | Max | Mean | SD | CV% |
|---|---|---|---|---|---|---|
| Hard wheat | E1 | 38.00 | 75.00 | 62.86 | 5.42 | 8.62 |
| E2 | 54.00 | 74.00 | 64.57 | 4.53 | 7.01 | |
| E3 | 48.00 | 83.00 | 66.08 | 6.97 | 10.55 | |
| E4 | 56.00 | 73.00 | 63.88 | 4.75 | 7.44 | |
| BLUP | 60.19 | 72.64 | 63.95 | 3.26 | 5.10 | |
| Mix wheat | E1 | 47.00 | 61.00 | 55.14 | 3.38 | 6.12 |
| E2 | 41.00 | 63.00 | 55.49 | 5.11 | 9.21 | |
| E3 | 42.00 | 70.00 | 55.78 | 5.74 | 10.28 | |
| E4 | 39.00 | 65.00 | 55.05 | 4.74 | 8.60 | |
| BLUP | 47.99 | 59.95 | 55.18 | 2.95 | 5.34 | |
| Soft wheat | E1 | 15.00 | 42.00 | 27.19 | 5.66 | 20.81 |
| E2 | 15.00 | 44.00 | 28.90 | 5.71 | 19.77 | |
| E3 | 15.00 | 42.00 | 28.44 | 6.12 | 21.53 | |
| E4 | 21.00 | 64.00 | 32.69 | 7.99 | 24.44 | |
| BLUP | 19.44 | 40.91 | 29.74 | 4.64 | 15.59 |
| Type | Genotype | Environment | Genotype × Environment | H2 |
|---|---|---|---|---|
| Hardness wheat | 1.68 * | 3.62 | 0.98 | 0.63 |
| Mix wheat | 1.77 * | 0.02 | 0.70 | 0.71 |
| Soft wheat | 3.02 *** | 13.11 *** | 0.853 | 0.81 |
| Classification Standard | SNP | Chr. | Position (Mb) | p Value | R2 (%) | Environment |
|---|---|---|---|---|---|---|
| Hardness Index | AX-111028882 | 1A | 27.39–31.94 | 1.10 × 10−4–9.74 × 10−4 | 7.63–10.66 | E1, E2, E3, E4, BLUP |
| AX-110005500 | 7D | 568.20 | 1.17 × 10−4–8.84 × 10−4 | 7.52–10.57 | E1, E2, E3, E4, BLUP | |
| Assignment | AX-108789085 | 1A | 24.86–32.43 | 9.75 × 10−5–9.44 × 10−4 | 7.77–10.86 | E1, E2, E3, BLUP |
| AX-108899745 | 1A | 42.55–49.23 | 9.35 × 10−5–9.31 × 10−4 | 7.72–11.01 | E1, E2, E3, BLUP | |
| AX-109815102 | 3B | 606.68 | 1.24 × 10−4–3.77 × 10−4 | 9.00–10.61 | E1, E2, BLUP | |
| AX-111673206 | 4B | 656.90–661.26 | 3.60 × 10−4–9.83 × 10−4 | 7.63–9.00 | E1, E2, E4 | |
| AX-110005500 | 7D | 568.20 | 8.12 × 10−5–8.55 × 10−4 | 7.91–11.12 | E1, E2, E3, BLUP |
| SNP | Chr. | Position (Mb) | Gene | Gene Annotation or Coding Protein | Previously Reported |
|---|---|---|---|---|---|
| AX-108789085 | 1A | 24.86–32.43 | TraesCS1A01G045700 | Glutathione S-transferase | 2013_Mergoum_3 [13] |
| AX-108899745 | 1A | 42.55–49.23 | TraesCS1A01G062500 | Protease inhibitor/seed storage/lipid transfer protein family protein | qHA1A.1 [34] |
| AX-109815102 | 3B | 606.68 | TraesCS3B01G386000 | O-Glycosyl hydrolases family 17 protein | - |
| AX-111673206 | 4B | 656.90–661.26 | TraesCS4B01G372800 | F-box family protein | qHA4B.3 [27] |
| AX-110005500 | 7D | 568.20 | TraesCS7D01G448100 | Protease inhibitor/seed storage/lipid transfer family protein | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, X.; Lu, M.; Cao, J.; Pan, X.; Lu, J.; Zhao, L.; Zhang, H.; Chang, C.; Wang, J.; Ma, C. Genome-Wide Association Analysis of Grain Hardness in Common Wheat. Genes 2023, 14, 672. https://doi.org/10.3390/genes14030672
He X, Lu M, Cao J, Pan X, Lu J, Zhao L, Zhang H, Chang C, Wang J, Ma C. Genome-Wide Association Analysis of Grain Hardness in Common Wheat. Genes. 2023; 14(3):672. https://doi.org/10.3390/genes14030672
Chicago/Turabian StyleHe, Xianfang, Maoang Lu, Jiajia Cao, Xu Pan, Jie Lu, Li Zhao, Haiping Zhang, Cheng Chang, Jianlai Wang, and Chuanxi Ma. 2023. "Genome-Wide Association Analysis of Grain Hardness in Common Wheat" Genes 14, no. 3: 672. https://doi.org/10.3390/genes14030672
APA StyleHe, X., Lu, M., Cao, J., Pan, X., Lu, J., Zhao, L., Zhang, H., Chang, C., Wang, J., & Ma, C. (2023). Genome-Wide Association Analysis of Grain Hardness in Common Wheat. Genes, 14(3), 672. https://doi.org/10.3390/genes14030672




