Identification of Two Homozygous Variants in MYBPC3 and SMYD1 Genes Associated with Severe Infantile Cardiomyopathy
Abstract
1. Introduction
2. Materials and Methods
2.1. Human Studies
2.2. Whole Exome Sequencing/Familial Genetic Testing
2.3. Histology and Ultrastructural Analysis
3. Results
3.1. Clinical Description of the Female Proband (P1)
3.2. Clinical Description of the Male Proband (P2)
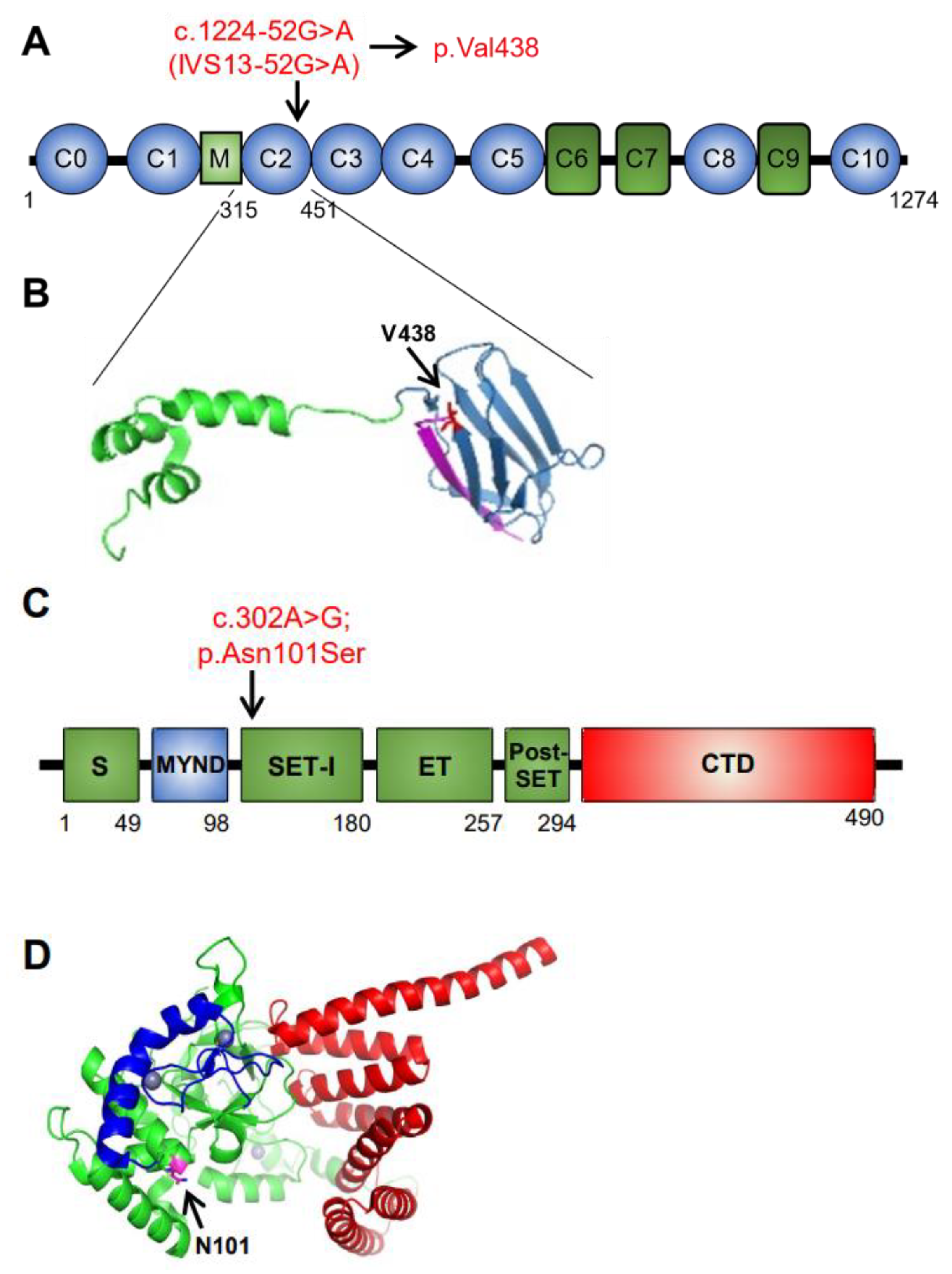
3.3. Identification of SMYD1 and MYBPC3 Variants in New Patients
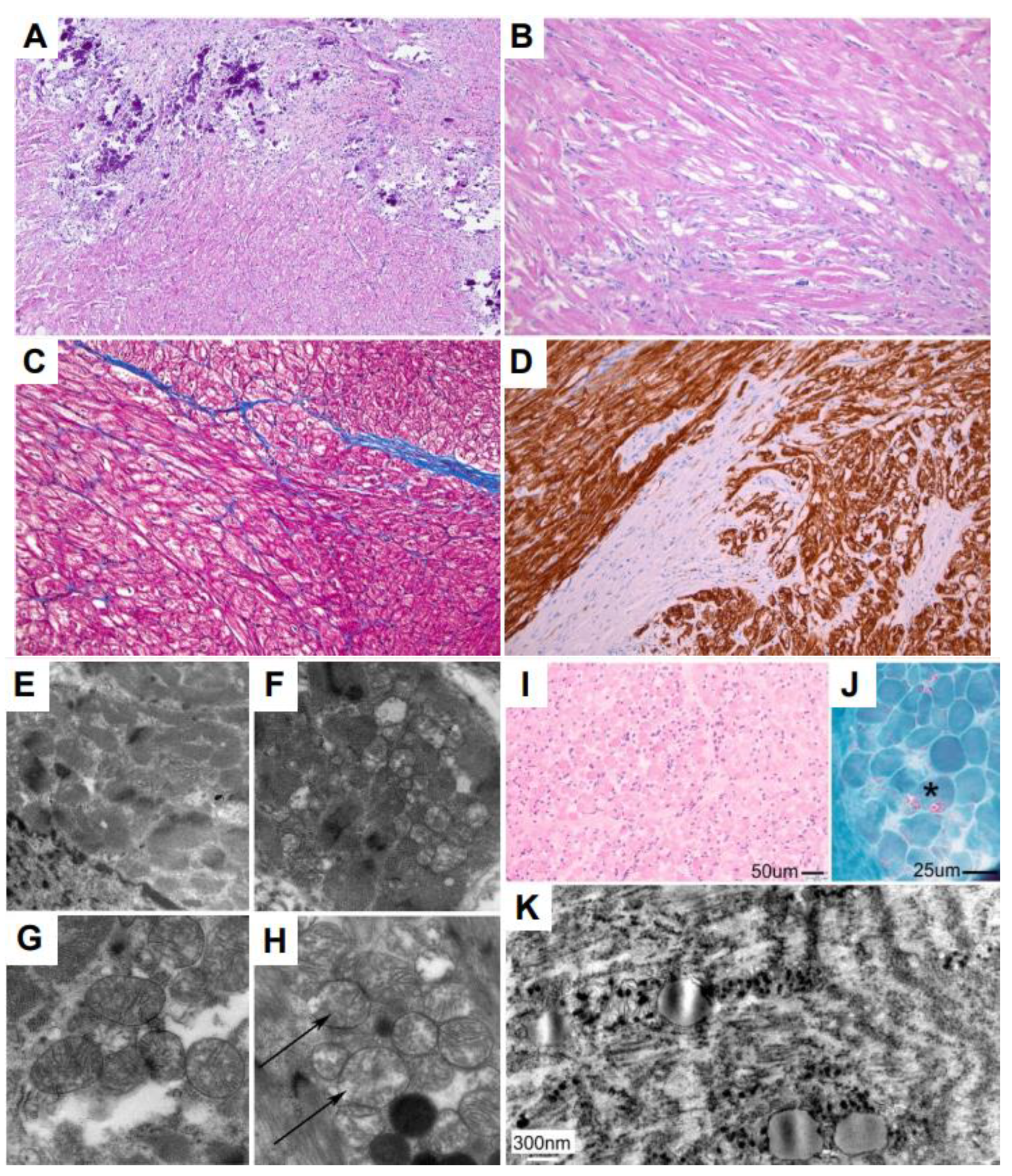
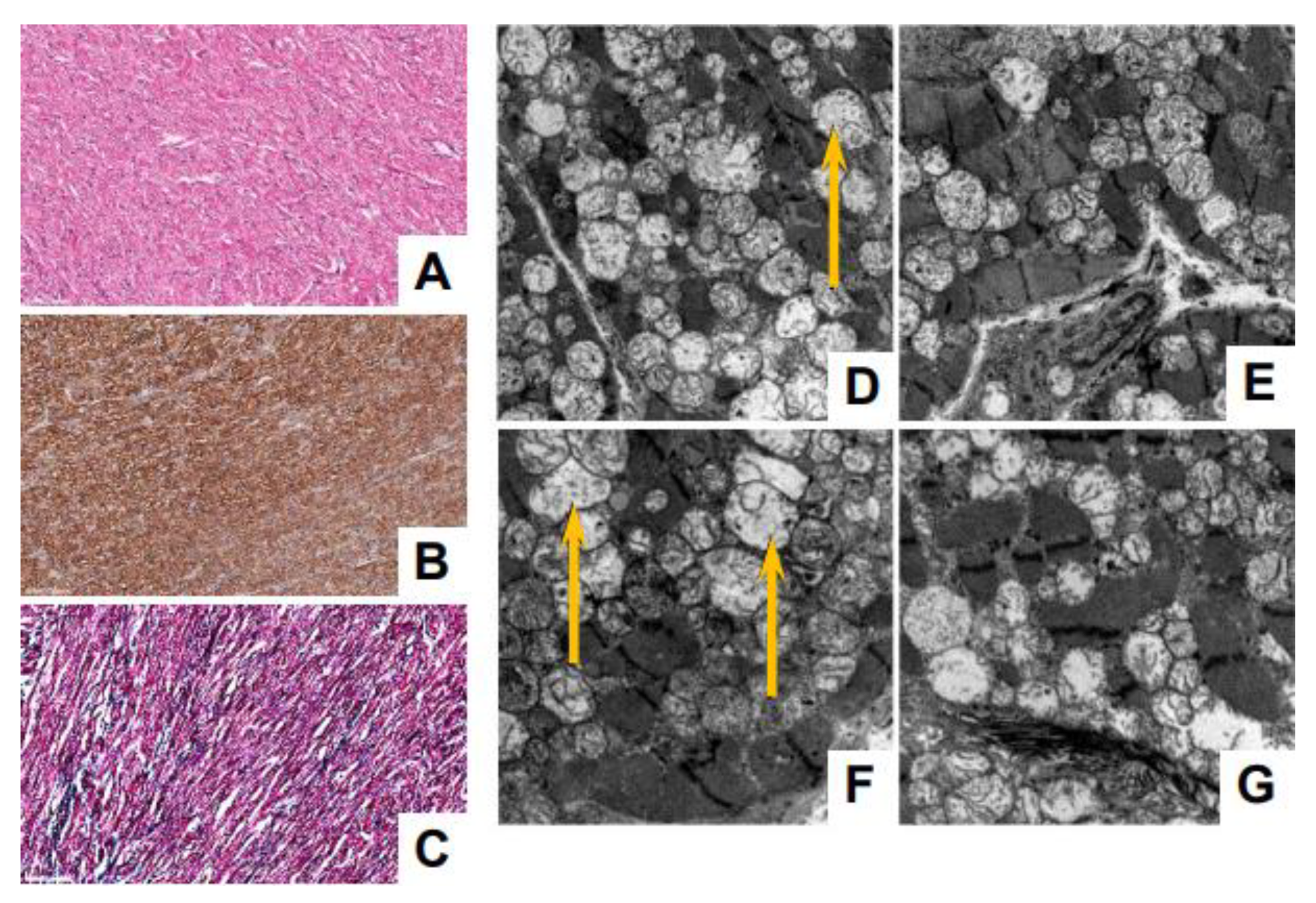
3.4. Myocardial and Skeletal Biopsy Pathology
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, T.M.; Hsu, D.T.; Kantor, P.; Towbin, J.A.; Ware, S.M.; Colan, S.D.; Chung, W.K.; Jefferies, J.L.; Rossano, J.W.; Castleberry, C.D.; et al. Pediatric Cardiomyopathies. Circ. Res. 2017, 121, 855–873. [Google Scholar] [CrossRef] [PubMed]
- Lipshultz, S.E.; Cochran, T.R.; Briston, D.A.; Brown, S.R.; Sambatakos, P.J.; Miller, T.L.; Carrillo, A.A.; Corcia, L.; Sanchez, J.E.; Diamond, M.B.; et al. Pediatric cardiomyopathies: Causes, epidemiology, clinical course, preventive strategies and therapies. Future Cardiol. 2013, 9, 817–848. [Google Scholar] [CrossRef] [PubMed]
- Konno, T.; Chang, S.; Seidman, J.G.; Seidman, C.E. Genetics of hypertrophic cardiomyopathy. Curr. Opin. Cardiol. 2010, 25, 205–209. [Google Scholar] [CrossRef]
- Fougerousse, F.; Delezoide, A.L.; Fiszman, M.Y.; Schwartz, K.; Beckmann, J.S.; Carrier, L. Cardiac myosin binding protein C gene is specifically expressed in heart during murine and human development. Circ. Res. 1998, 82, 130–133. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Govindan, S.; Lee, K.; Zhao, P.; Han, R.; Runte, K.E.; Craig, R.; Palmer, B.M.; Sadayappan, S. Cardiac myosin binding protein-C plays no regulatory role in skeletal muscle structure and function. PLoS ONE 2013, 8, e69671. [Google Scholar] [CrossRef]
- McClellan, G.; Kulikovskaya, I.; Winegrad, S. Changes in cardiac contractility related to calcium-mediated changes in phosphorylation of myosin-binding protein C. Biophys. J. 2001, 81, 1083–1092. [Google Scholar] [CrossRef]
- Sadayappan, S.; Osinska, H.; Klevitsky, R.; Lorenz, J.N.; Sargent, M.; Molkentin, J.D.; Seidman, C.E.; Seidman, J.G.; Robbins, J. Cardiac myosin binding protein C phosphorylation is cardioprotective. Proc. Natl. Acad. Sci. USA 2006, 103, 16918–16923. [Google Scholar] [CrossRef]
- Dhandapany, P.S.; Sadayappan, S.; Xue, Y.; Powell, G.T.; Rani, D.S.; Nallari, P.; Rai, T.S.; Khullar, M.; Soares, P.; Bahl, A.; et al. A common MYBPC3 (cardiac myosin binding protein C) variant associated with cardiomyopathies in South Asia. Nat. Genet. 2009, 41, 187–191. [Google Scholar] [CrossRef]
- Michie, K.A.; Kwan, A.H.; Tung, C.S.; Guss, J.M.; Trewhella, J. A Highly Conserved Yet Flexible Linker Is Part of a Polymorphic Protein-Binding Domain in Myosin-Binding Protein C. Structure 2016, 24, 2000–2007. [Google Scholar] [CrossRef]
- Carrier, L.; Mearini, G.; Stathopoulou, K.; Cuello, F. Cardiac myosin-binding protein C (MYBPC3) in cardiac pathophysiology. Gene 2015, 573, 188–197. [Google Scholar] [CrossRef]
- Behrens-Gawlik, V.; Mearini, G.; Gedicke-Hornung, C.; Richard, P.; Carrier, L. MYBPC3 in hypertrophic cardiomyopathy: From mutation identification to RNA-based correction. Pflug. Arch. 2014, 466, 215–223. [Google Scholar] [CrossRef]
- Niimura, H.; Patton, K.K.; McKenna, W.J.; Soults, J.; Maron, B.J.; Seidman, J.G.; Seidman, C.E. Sarcomere protein gene mutations in hypertrophic cardiomyopathy of the elderly. Circulation 2002, 105, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Van Driest, S.L.; Ommen, S.R.; Tajik, A.J.; Gersh, B.J.; Ackerman, M.J. Sarcomeric genotyping in hypertrophic cardiomyopathy. Mayo Clin. Proc. 2005, 80, 463–469. [Google Scholar] [CrossRef]
- Srivastava, A.; Garg, N.; Mittal, T.; Khanna, R.; Gupta, S.; Seth, P.K.; Mittal, B. Association of 25 bp deletion in MYBPC3 gene with left ventricle dysfunction in coronary artery disease patients. PLoS ONE 2011, 6, e24123. [Google Scholar] [CrossRef] [PubMed]
- Arif, M.; Nabavizadeh, P.; Song, T.; Desai, D.; Singh, R.; Bazrafshan, S.; Kumar, M.; Wang, Y.; Gilbert, R.J.; Dhandapany, P.S.; et al. Genetic, clinical, molecular, and pathogenic aspects of the South Asian-specific polymorphic MYBPC3(Delta25bp) variant. Biophys. Rev. 2020, 12, 1065–1084. [Google Scholar] [CrossRef]
- Millat, G.; Lafont, E.; Nony, S.; Rouvet, I.; Bozon, D. Functional characterization of putative novel splicing mutations in the cardiomyopathy-causing genes. DNA Cell Biol. 2015, 34, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Bagnall, R.D.; Ingles, J.; Dinger, M.E.; Cowley, M.J.; Ross, S.B.; Minoche, A.E.; Lal, S.; Turner, C.; Colley, A.; Rajagopalan, S.; et al. Whole Genome Sequencing Improves Outcomes of Genetic Testing in Patients with Hypertrophic Cardiomyopathy. J. Am. Coll. Cardiol. 2018, 72, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Harper, A.R.; Bowman, M.; Hayesmoore, J.B.G.; Sage, H.; Salatino, S.; Blair, E.; Campbell, C.; Currie, B.; Goel, A.; McGuire, K.; et al. Reevaluation of the South Asian MYBPC3(Delta25bp) Intronic Deletion in Hypertrophic Cardiomyopathy. Circ. Genom. Precis. Med. 2020, 13, e002783. [Google Scholar] [CrossRef] [PubMed]
- Holliday, M.; Singer, E.S.; Ross, S.B.; Lim, S.; Lal, S.; Ingles, J.; Semsarian, C.; Bagnall, R.D. Transcriptome Sequencing of Patients with Hypertrophic Cardiomyopathy Reveals Novel Splice-Altering Variants in MYBPC3. Circ. Genom. Precis. Med. 2021, 14, e003202. [Google Scholar] [CrossRef]
- Lopes, L.R.; Barbosa, P.; Torrado, M.; Quinn, E.; Merino, A.; Ochoa, J.P.; Jager, J.; Futema, M.; Carmo-Fonseca, M.; Monserrat, L.; et al. Cryptic Splice-Altering Variants in MYBPC3 Are a Prevalent Cause of Hypertrophic Cardiomyopathy. Circ. Genom. Precis. Med. 2020, 13, e002905. [Google Scholar] [CrossRef]
- Torrado, M.; Maneiro, E.; Lamounier Junior, A.; Fernandez-Burriel, M.; Sanchez Giralt, S.; Martinez-Carapeto, A.; Cazon, L.; Santiago, E.; Ochoa, J.P.; McKenna, W.J.; et al. Identification of an elusive spliceogenic MYBPC3 variant in an otherwise genotype-negative hypertrophic cardiomyopathy pedigree. Sci. Rep. 2022, 12, 7284. [Google Scholar] [CrossRef] [PubMed]
- Sadayappan, S.; Puckelwartz, M.J.; McNally, E.M. South Asian-Specific MYBPC3(Delta25bp) Intronic Deletion and Its Role in Cardiomyopathies and Heart Failure. Circ. Genom. Precis. Med. 2020, 13, e002986. [Google Scholar] [CrossRef] [PubMed]
- Rea, S.; Eisenhaber, F.; O’Carroll, D.; Strahl, B.D.; Sun, Z.W.; Schmid, M.; Opravil, S.; Mechtler, K.; Ponting, C.P.; Allis, C.D.; et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 2000, 406, 593–599. [Google Scholar] [CrossRef]
- Sims, R.J., 3rd; Weihe, E.K.; Zhu, L.; O’Malley, S.; Harriss, J.V.; Gottlieb, P.D. m-Bop, a repressor protein essential for cardiogenesis, interacts with skNAC, a heart- and muscle-specific transcription factor. J. Biol. Chem. 2002, 277, 26524–26529. [Google Scholar] [CrossRef]
- Du, S.J.; Tan, X.; Zhang, J. SMYD proteins: Key regulators in skeletal and cardiac muscle development and function. Anat. Rec. 2014, 297, 1650–1662. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Rotllant, J.; Li, H.; De Deyne, P.; Du, S.J. SmyD1, a histone methyltransferase, is required for myofibril organization and muscle contraction in zebrafish embryos. Proc. Natl. Acad. Sci. USA 2006, 103, 2713–2718. [Google Scholar] [CrossRef]
- Warren, J.S.; Tracy, C.M.; Miller, M.R.; Makaju, A.; Szulik, M.W.; Oka, S.I.; Yuzyuk, T.N.; Cox, J.E.; Kumar, A.; Lozier, B.K.; et al. Histone methyltransferase Smyd1 regulates mitochondrial energetics in the heart. Proc. Natl. Acad. Sci. USA 2018, 115, E7871–E7880. [Google Scholar] [CrossRef]
- Tracy, C.; Warren, J.S.; Szulik, M.; Wang, L.; Garcia, J.; Makaju, A.; Russell, K.; Miller, M.; Franklin, S. The Smyd Family of Methyltransferases: Role in Cardiac and Skeletal Muscle Physiology and Pathology. Curr. Opin. Physiol. 2018, 1, 140–152. [Google Scholar] [CrossRef]
- Gottlieb, P.D.; Pierce, S.A.; Sims, R.J.; Yamagishi, H.; Weihe, E.K.; Harriss, J.V.; Maika, S.D.; Kuziel, W.A.; King, H.L.; Olson, E.N.; et al. Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat. Genet. 2002, 31, 25–32. [Google Scholar] [CrossRef]
- Hwang, I.; Gottlieb, P.D. The Bop gene adjacent to the mouse CD8b gene encodes distinct zinc-finger proteins expressed in CTLs and in muscle. J. Immunol. 1997, 158, 1165–1174. [Google Scholar] [CrossRef]
- Rasmussen, T.L.; Ma, Y.; Park, C.Y.; Harriss, J.; Pierce, S.A.; Dekker, J.D.; Valenzuela, N.; Srivastava, D.; Schwartz, R.J.; Stewart, M.D.; et al. Smyd1 facilitates heart development by antagonizing oxidative and ER stress responses. PLoS ONE 2015, 10, e0121765. [Google Scholar] [CrossRef]
- Franklin, S.; Kimball, T.; Rasmussen, T.L.; Rosa-Garrido, M.; Chen, H.; Tran, T.; Miller, M.R.; Gray, R.; Jiang, S.; Ren, S.; et al. The chromatin-binding protein Smyd1 restricts adult mammalian heart growth. Am. J. Physiol. Heart Circ. Physiol. 2016, 311, H1234–H1247. [Google Scholar] [CrossRef]
- Fan, L.L.; Ding, D.B.; Huang, H.; Chen, Y.Q.; Jin, J.Y.; Xia, K.; Xiang, R. A de novo mutation of SMYD1 (p.F272L) is responsible for hypertrophic cardiomyopathy in a Chinese patient. Clin. Chem. Lab. Med. 2019, 57, 532–539. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Xie, Y.; Zhang, H.; Feng, Z.; Huang, J.; Huang, J.; Hu, S.; Wei, Y. Multiple genetic variants in adolescent patients with left ventricular noncompaction cardiomyopathy. Int. J. Cardiol. 2020, 302, 117–123. [Google Scholar] [CrossRef]
- Gordon, D.M.; Cunningham, D.; Zender, G.; Lawrence, P.J.; Penaloza, J.S.; Lin, H.; Fitzgerald-Butt, S.M.; Myers, K.; Duong, T.; Corsmeier, D.J.; et al. Exome sequencing in multiplex families with left-sided cardiac defects has high yield for disease gene discovery. PLoS Genet. 2022, 18, e1010236. [Google Scholar] [CrossRef]
- Coyan, G.N.; Zinn, M.D.; West, S.C.; Sharma, M.S. Heart Transplantation from Biventricular Support in Infant with Novel SMYD1 Mutation. Pediatr. Cardiol. 2019, 40, 1745–1747. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Lipshultz, S.E.; Law, Y.M.; Asante-Korang, A.; Austin, E.D.; Dipchand, A.I.; Everitt, M.D.; Hsu, D.T.; Lin, K.Y.; Price, J.F.; Wilkinson, J.D.; et al. Cardiomyopathy in Children: Classification and Diagnosis: A Scientific Statement From the American Heart Association. Circulation 2019, 140, e9–e68. [Google Scholar] [CrossRef]
- Lipshultz, S.E.; Sleeper, L.A.; Towbin, J.A.; Lowe, A.M.; Orav, E.J.; Cox, G.F.; Lurie, P.R.; McCoy, K.L.; McDonald, M.A.; Messere, J.E.; et al. The incidence of pediatric cardiomyopathy in two regions of the United States. N. Engl. J. Med. 2003, 348, 1647–1655. [Google Scholar] [CrossRef] [PubMed]
- Hershberger, R.E.; Givertz, M.M.; Ho, C.Y.; Judge, D.P.; Kantor, P.F.; McBride, K.L.; Morales, A.; Taylor, M.R.G.; Vatta, M.; Ware, S.M.; et al. Genetic evaluation of cardiomyopathy: A clinical practice resource of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2018, 20, 899–909. [Google Scholar] [CrossRef]
- Hershberger, R.E.; Hedges, D.J.; Morales, A. Dilated cardiomyopathy: The complexity of a diverse genetic architecture. Nat. Rev. Cardiol. 2013, 10, 531–547. [Google Scholar] [CrossRef]
- Ware, S.M.; Wilkinson, J.D.; Tariq, M.; Schubert, J.A.; Sridhar, A.; Colan, S.D.; Shi, L.; Canter, C.E.; Hsu, D.T.; Webber, S.A.; et al. Genetic Causes of Cardiomyopathy in Children: First Results From the Pediatric Cardiomyopathy Genes Study. J. Am. Heart Assoc. 2021, 10, e017731. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, M.; Elliott, P. The genetics of hypertrophic cardiomyopathy. Glob. Cardiol. Sci. Pract. 2018, 2018, 36. [Google Scholar] [CrossRef]
- Helms, A.S.; Thompson, A.D.; Glazier, A.A.; Hafeez, N.; Kabani, S.; Rodriguez, J.; Yob, J.M.; Woolcock, H.; Mazzarotto, F.; Lakdawala, N.K.; et al. Spatial and Functional Distribution of MYBPC3 Pathogenic Variants and Clinical Outcomes in Patients With Hypertrophic Cardiomyopathy. Circ. Genom. Precis. Med. 2020, 13, 396–405. [Google Scholar] [CrossRef] [PubMed]
- Wessels, M.W.; Herkert, J.C.; Frohn-Mulder, I.M.; Dalinghaus, M.; van den Wijngaard, A.; de Krijger, R.R.; Michels, M.; de Coo, I.F.; Hoedemaekers, Y.M.; Dooijes, D. Compound heterozygous or homozygous truncating MYBPC3 mutations cause lethal cardiomyopathy with features of noncompaction and septal defects. Eur. J. Hum. Genet. 2015, 23, 922–928. [Google Scholar] [CrossRef]
- Sadayappan, S.; de Tombe, P.P. Cardiac myosin binding protein-C: Redefining its structure and function. Biophys. Rev. 2012, 4, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Charron, P.; Dubourg, O.; Desnos, M.; Bennaceur, M.; Carrier, L.; Camproux, A.C.; Isnard, R.; Hagege, A.; Langlard, J.M.; Bonne, G.; et al. Clinical features and prognostic implications of familial hypertrophic cardiomyopathy related to the cardiac myosin-binding protein C gene. Circulation 1998, 97, 2230–2236. [Google Scholar] [CrossRef]
- Redin, C.; Pavlidou, D.C.; Bhuiyan, Z.; Porretta, A.P.; Monney, P.; Bedoni, N.; Maurer, F.; Sekarski, N.; Atallah, I.; Emeline, D.; et al. The <<Amish>> NM_000256.3:c.3330+2T>G splice variant in MYBPC3 associated with hypertrophic cardiomyopathy is an ancient Swiss mutation. Eur. J. Med. Genet. 2022, 65, 104627. [Google Scholar] [CrossRef]
- Zahka, K.; Kalidas, K.; Simpson, M.A.; Cross, H.; Keller, B.B.; Galambos, C.; Gurtz, K.; Patton, M.A.; Crosby, A.H. Homozygous mutation of MYBPC3 associated with severe infantile hypertrophic cardiomyopathy at high frequency among the Amish. Heart 2008, 94, 1326–1330. [Google Scholar] [CrossRef]
- Just, S.; Meder, B.; Berger, I.M.; Etard, C.; Trano, N.; Patzel, E.; Hassel, D.; Marquart, S.; Dahme, T.; Vogel, B.; et al. The myosin-interacting protein SMYD1 is essential for sarcomere organization. J. Cell Sci. 2011, 124, 3127–3136. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xu, J.; Bian, Y.H.; Rotllant, P.; Shen, T.; Chu, W.; Zhang, J.; Schneider, M.; Du, S.J. Smyd1b_tv1, a key regulator of sarcomere assembly, is localized on the M-line of skeletal muscle fibers. PLoS ONE 2011, 6, e28524. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhong, Y.; Wang, Z.; Gao, J.; Xu, J.; Chu, W.; Zhang, J.; Fang, S.; Du, S.J. Smyd1b is required for skeletal and cardiac muscle function in zebrafish. Mol. Biol. Cell 2013, 24, 3511–3521. [Google Scholar] [CrossRef] [PubMed]
- Ranjbarvaziri, S.; Kooiker, K.B.; Ellenberger, M.; Fajardo, G.; Zhao, M.; Vander Roest, A.S.; Woldeyes, R.A.; Koyano, T.T.; Fong, R.; Ma, N.; et al. Altered Cardiac Energetics and Mitochondrial Dysfunction in Hypertrophic Cardiomyopathy. Circulation 2021, 144, 1714–1731. [Google Scholar] [CrossRef] [PubMed]
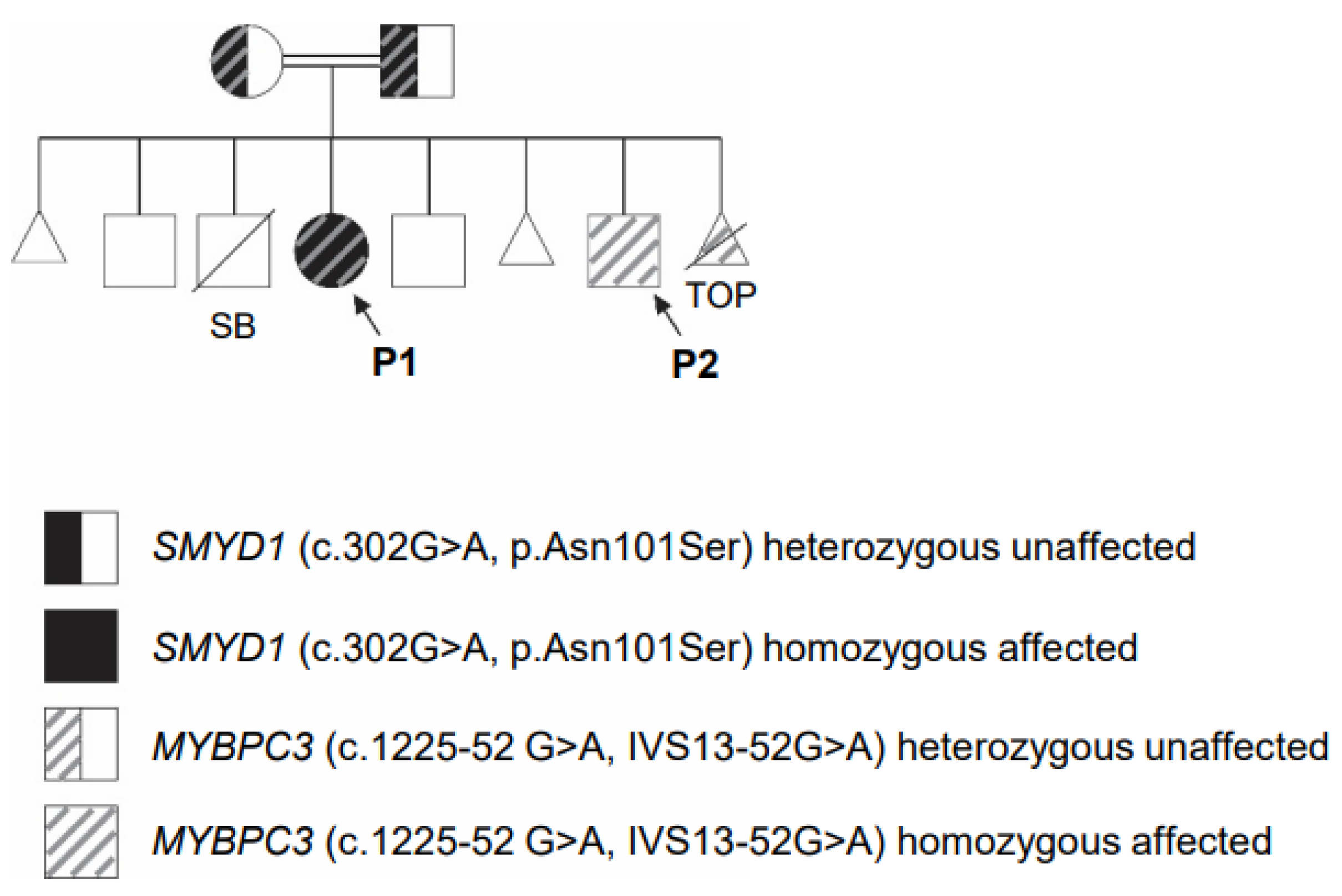
| Relationship | Affected (Infantile Cardiomyopathy) | MYBPC3 (c.1224-52G>A: IVS13-52G>A) | SMYD1 (c.302A>G; p.Asn101Ser) | Current Age (Years) |
|---|---|---|---|---|
| Father | No | Heterozygous | Heterozygous | 40 |
| Mother | No | Heterozygous | Heterozygous | 29 |
| Male | No | No | No | 7 |
| Female (P1) | Yes | Homozygous | Homozygous | 5 |
| Male | No | No | No | 4 |
| Male (P2) | Yes | Homozygous | No | 1 |
| TOP | Unknown | Homozygous | Not done | Pregnancy terminated |
| Gene | Mode of Inheritance | Variant | Reference Sequence | Classification | Result |
|---|---|---|---|---|---|
| SMYD1 | Unknown | c.302A>G, p.Asn101Ser | NM_198274.3 | Variant of uncertain significance | Homozygous |
| MYBPC3 | Autosomal Dominant/ Recessive | c.1224-52G>A, IVS13-52G>A | NM_000256.3 | Pathogenic Variant | Homozygous |
| Reference | Phenotype | Zygosity | Number of Patients | Patient’s Age (Years) |
|---|---|---|---|---|
| Bagnall et al. [17] | Hypertrophic cardiomyopathy | unknown | 3 | 23, 29, one unknown |
| Harper et al. [18] | Hypertrophic cardiomyopathy | heterozygous | OMGL * cohort: 32 of 2757 HCMR * cohort: 23 of 2636 | OMGL cohort: 54.5 +/− 16.3 HCMR cohort: 49.5 +/− 11.3 |
| Lopes et al. [20] | Hypertrophic cardiomyopathy | heterozygous | 18 | 10–72 |
| Holliday et al. [19] | Nonobstructive hypertrophic cardiomyopathy | heterozygous | 1 | 48 |
| Torrado et al. [21] | Hypertrophic cardiomyopathy | unknown | HIC * cohort: 12 of 9611 | 15–69, three unknown |
| Nucleotide/Amino Acid Change | SMYD1 Protein Domain | Zygosity | Phenotype | gnomAD MAF * | PolyPhen-2 | SIFT | Reference |
|---|---|---|---|---|---|---|---|
| c.814T>C p.Phe272Leu | Post-SET | heterozygous | Atrial enlargement, hypertrophic cardiomyopathy | 0.000003979 | Possibly damaging | Deleterious | [33] |
| c.302A>G p.Asn101Ser | SET-I | homozygous | Biventricular heart failure | 0.00003589 | Benign | Tolerated | [36], this report |
| c.675delA p.Lys225Asnfs*8 | SET | heterozygous | Left ventricular noncompaction cardiomyopathy, arrhythmia | 0.00009899 | No PolyPhen-2 prediction available | No SIFT prediction available | [34] |
| c. 1321C>T p.Arg441Trp | CTD | heterozygous | Aortic valve stenosis and bicuspid aortic valve | 0.006564 | Possibly damaging | Deleterious | [35] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szulik, M.W.; Reyes-Múgica, M.; Marker, D.F.; Gomez, A.M.; Zinn, M.D.; Walsh, L.K.; Ochoa, J.P.; Franklin, S.; Ghaloul-Gonzalez, L. Identification of Two Homozygous Variants in MYBPC3 and SMYD1 Genes Associated with Severe Infantile Cardiomyopathy. Genes 2023, 14, 659. https://doi.org/10.3390/genes14030659
Szulik MW, Reyes-Múgica M, Marker DF, Gomez AM, Zinn MD, Walsh LK, Ochoa JP, Franklin S, Ghaloul-Gonzalez L. Identification of Two Homozygous Variants in MYBPC3 and SMYD1 Genes Associated with Severe Infantile Cardiomyopathy. Genes. 2023; 14(3):659. https://doi.org/10.3390/genes14030659
Chicago/Turabian StyleSzulik, Marta W., Miguel Reyes-Múgica, Daniel F. Marker, Ana M. Gomez, Matthew D. Zinn, Leslie K. Walsh, Juan Pablo Ochoa, Sarah Franklin, and Lina Ghaloul-Gonzalez. 2023. "Identification of Two Homozygous Variants in MYBPC3 and SMYD1 Genes Associated with Severe Infantile Cardiomyopathy" Genes 14, no. 3: 659. https://doi.org/10.3390/genes14030659
APA StyleSzulik, M. W., Reyes-Múgica, M., Marker, D. F., Gomez, A. M., Zinn, M. D., Walsh, L. K., Ochoa, J. P., Franklin, S., & Ghaloul-Gonzalez, L. (2023). Identification of Two Homozygous Variants in MYBPC3 and SMYD1 Genes Associated with Severe Infantile Cardiomyopathy. Genes, 14(3), 659. https://doi.org/10.3390/genes14030659






