Causal Association between Iritis or Uveitis and Glaucoma: A Two-Sample Mendelian Randomisation Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design Overview
2.2. Data Source
2.3. Selection of the Genetic Instrumental Variables
2.4. Mendelian Randomisation
3. Results
3.1. Genetic Instrumental Variables
3.2. Heterogeneity and Horizontal Pleiotropy of Instrumental Variables
3.3. Mendelian Randomisation for the Casual Association between Iritis and Glaucoma
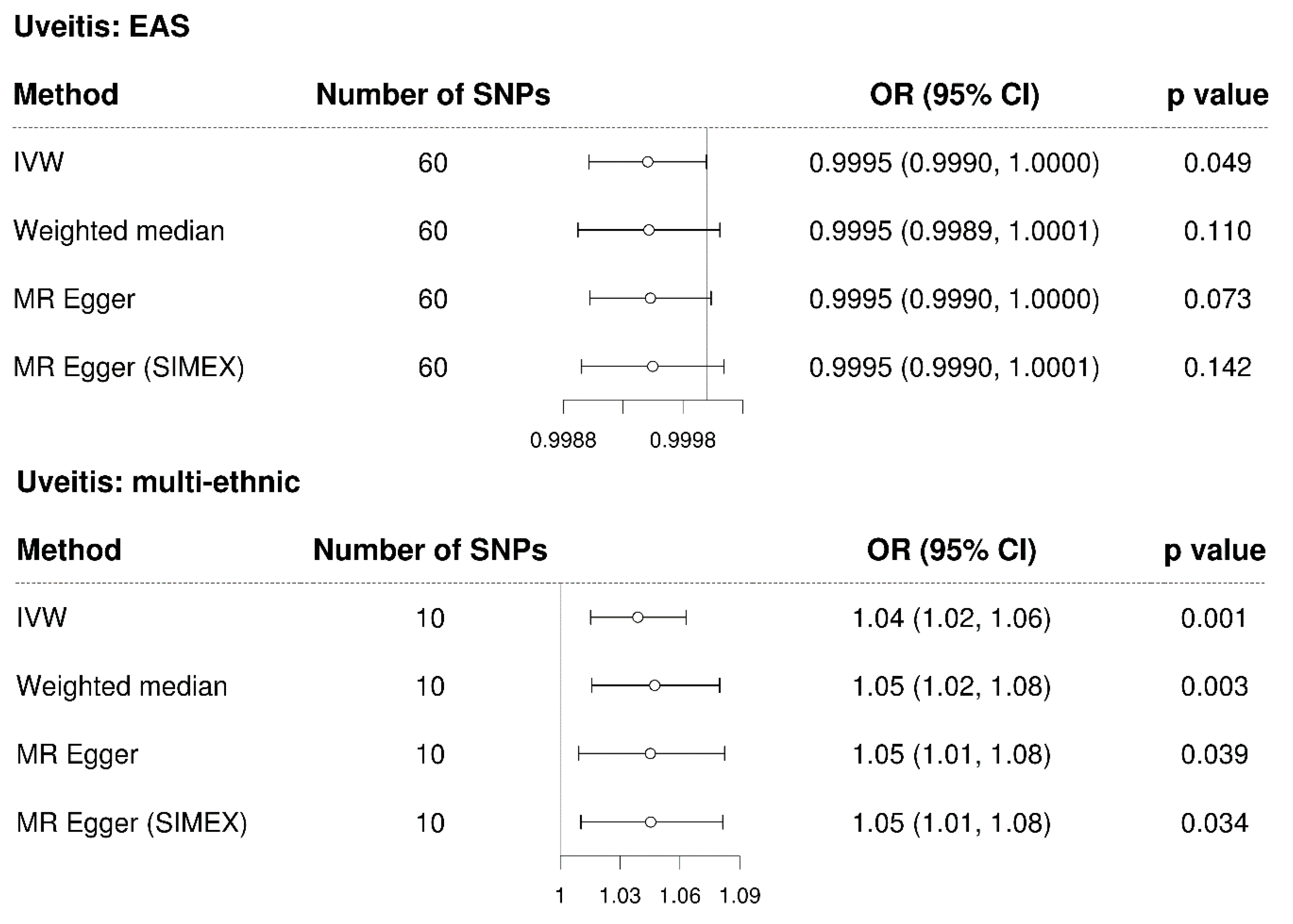
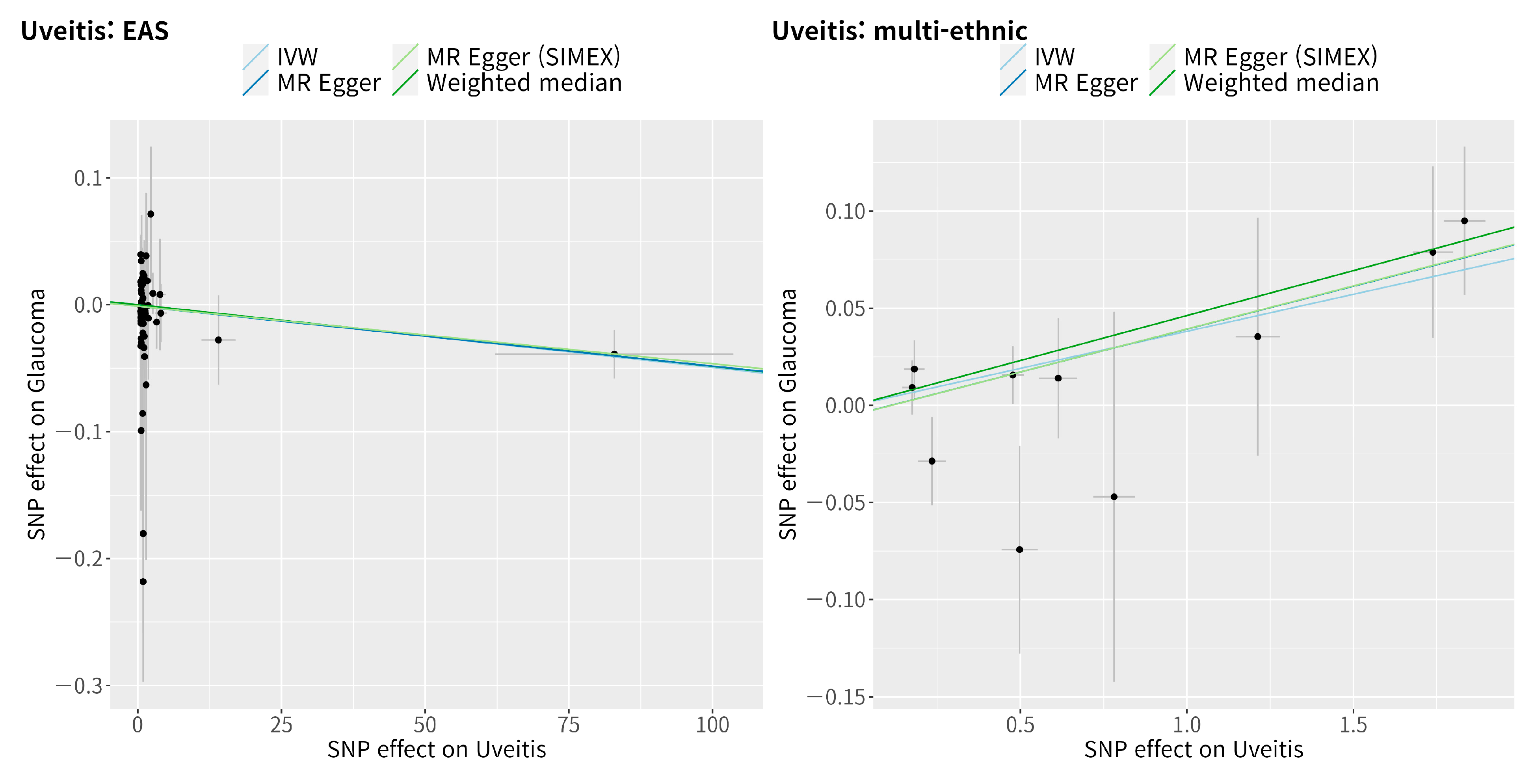
3.4. Mendelian Randomisation for the Casual Association between Uveitis and Glaucoma
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tsirouki, T.; Dastiridou, A.; Symeonidis, C.; Tounakaki, O.; Brazitikou, I.; Kalogeropoulos, C.; Androudi, S. A Focus on the Epidemiology of Uveitis. Ocul. Immunol. Inflamm. 2018, 26, 2–16. [Google Scholar] [CrossRef] [PubMed]
- Siddique, S.S.; Suelves, A.M.; Baheti, U.; Foster, C.S. Glaucoma and uveitis. Surv. Ophthalmol. 2013, 58, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Quigley, H.A. Glaucoma. Lancet 2011, 377, 1367–1377. [Google Scholar] [CrossRef] [PubMed]
- Tham, Y.C.; Li, X.; Wong, T.Y.; Quigley, H.A.; Aung, T.; Cheng, C.Y. Global prevalence of glaucoma and projections of glaucoma burden through 2040: A systematic review and meta-analysis. Ophthalmology 2014, 121, 2081–2090. [Google Scholar] [CrossRef]
- Hou, S.; Li, N.; Liao, X.; Kijlstra, A.; Yang, P. Uveitis genetics. Exp. Eye Res. 2020, 190, 107853. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, M.; Mizuki, N.; Ohno, S. Pathogenesis of Non-Infectious Uveitis Elucidated by Recent Genetic Findings. Front. Immunol. 2021, 12, 640473. [Google Scholar] [CrossRef]
- Su, G.; Zhong, Z.; Zhou, Q.; Du, L.; Ye, Z.; Li, F.; Zhuang, W.; Wang, C.; Liang, L.; Ji, Y.; et al. Identification of Novel Risk Loci for Behcet’s Disease-Related Uveitis in a Chinese Population in a Genome-Wide Association Study. Arthritis. Rheumatol. 2022, 74, 671–681. [Google Scholar] [CrossRef] [PubMed]
- Merayo-Lloves, J.; Power, W.J.; Rodriguez, A.; Pedroza-Seres, M.; Foster, C.S. Secondary glaucoma in patients with uveitis. Ophthalmologica 1999, 213, 300–304. [Google Scholar] [CrossRef]
- Takahashi, T.; Ohtani, S.; Miyata, K.; Miyata, N.; Shirato, S.; Mochizuki, M. A clinical evaluation of uveitis-associated secondary glaucoma. Jpn. J. Ophthalmol. 2002, 46, 556–562. [Google Scholar] [CrossRef]
- Herbert, H.M.; Viswanathan, A.; Jackson, H.; Lightman, S.L. Risk factors for elevated intraocular pressure in uveitis. J. Glaucoma 2004, 13, 96–99. [Google Scholar] [CrossRef]
- Weinreb, R.N.; Aung, T.; Medeiros, F.A. The pathophysiology and treatment of glaucoma: A review. JAMA 2014, 311, 1901–1911. [Google Scholar] [CrossRef] [PubMed]
- Alvarado, J.A.; Murphy, C.G. Outflow obstruction in pigmentary and primary open angle glaucoma. Arch. Ophthalmol. 1992, 110, 1769–1778. [Google Scholar] [CrossRef]
- Huang, X.F.; Brown, M.A. Progress in the genetics of uveitis. Genes. Immun. 2022, 23, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Wiggs, J.L.; Pasquale, L.R. Genetics of glaucoma. Hum. Mol. Genet. 2017, 26, R21–R27. [Google Scholar] [CrossRef] [PubMed]
- Gharahkhani, P.; Jorgenson, E.; Hysi, P.; Khawaja, A.P.; Pendergrass, S.; Han, X.; Ong, J.S.; Hewitt, A.W.; Segre, A.V.; Rouhana, J.M.; et al. Genome-wide meta-analysis identifies 127 open-angle glaucoma loci with consistent effect across ancestries. Nat. Commun. 2021, 12, 1258. [Google Scholar] [CrossRef] [PubMed]
- Youngblood, H.; Hauser, M.A.; Liu, Y. Update on the genetics of primary open-angle glaucoma. Exp. Eye Res. 2019, 188, 107795. [Google Scholar] [CrossRef]
- Sakurada, Y.; Mabuchi, F. Advances in glaucoma genetics. Prog. Brain Res. 2015, 220, 107–126. [Google Scholar] [CrossRef]
- Shin, J.H.; Lee, J.W.; Lim, S.H.; Yoon, B.W.; Lee, Y.; Seo, J.H. The microbiomes of the eyelid and buccal area of patients with uveitic glaucoma. BMC Ophthalmol. 2022, 22, 170. [Google Scholar] [CrossRef]
- Lee, J.W.; Lim, S.H.; Shin, J.H.; Lee, Y.; Seo, J.H. Differences in the eyelid and buccal microbiome between open-angle glaucoma and uveitic glaucoma. Acta Ophthalmol. 2022, 100, e770–e778. [Google Scholar] [CrossRef]
- Choquet, H.; Paylakhi, S.; Kneeland, S.C.; Thai, K.K.; Hoffmann, T.J.; Yin, J.; Kvale, M.N.; Banda, Y.; Tolman, N.G.; Williams, P.A.; et al. A multiethnic genome-wide association study of primary open-angle glaucoma identifies novel risk loci. Nat. Commun. 2018, 9, 2278. [Google Scholar] [CrossRef]
- Shin, H.T.; Yoon, B.W.; Seo, J.H. Analysis of risk allele frequencies of single nucleotide polymorphisms related to open-angle glaucoma in different ethnic groups. BMC Med. Genomics 2021, 14, 80. [Google Scholar] [CrossRef] [PubMed]
- Yoon, B.W.; Shin, H.T.; Seo, J.H. Risk Allele Frequency Analysis and Risk Prediction of Single-Nucleotide Polymorphisms for Prostate Cancer. Genes 2022, 13, 2039. [Google Scholar] [CrossRef]
- Yoon, B.W.; Shin, H.T.; Seo, J. Risk Allele Frequency Analysis of Single-Nucleotide Polymorphisms for Vitamin D Concentrations in Different Ethnic Group. Genes 2021, 12, 1530. [Google Scholar] [CrossRef]
- Shin, H.T.; Yoon, B.W.; Seo, J.H. Comparison of risk allele frequencies of single nucleotide polymorphisms associated with age-related macular degeneration in different ethnic groups. BMC Ophthalmol. 2021, 21, 97. [Google Scholar] [CrossRef]
- Burgess, S.; Thompson, S.G. Multivariable Mendelian randomization: The use of pleiotropic genetic variants to estimate causal effects. Am. J. Epidemiol. 2015, 181, 251–260. [Google Scholar] [CrossRef]
- Burgess, S.; Thompson, S.G. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 2017, 32, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Hanyuda, A.; Goto, A.; Nakatochi, M.; Sutoh, Y.; Narita, A.; Nakano, S.; Katagiri, R.; Wakai, K.; Takashima, N.; Koyama, T.; et al. Association Between Glycemic Traits and Primary Open-Angle Glaucoma: A Mendelian Randomization Study in the Japanese Population. Am. J. Ophthalmol. 2022, 245, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Aschard, H.; Kang, J.H.; Lentjes, M.A.H.; Do, R.; Wiggs, J.L.; Khawaja, A.P.; Pasquale, L.R.; Modifiable Risk Factors for Glaucoma Collaboration. Intraocular Pressure, Glaucoma, and Dietary Caffeine Consumption: A Gene-Diet Interaction Study from the UK Biobank. Ophthalmology 2021, 128, 866–876. [Google Scholar] [CrossRef]
- Sakaue, S.; Kanai, M.; Tanigawa, Y.; Karjalainen, J.; Kurki, M.; Koshiba, S.; Narita, A.; Konuma, T.; Yamamoto, K.; Akiyama, M.; et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat. Genet. 2021, 53, 1415–1424. [Google Scholar] [CrossRef]
- Loh, M.; Zhang, W.; Ng, H.K.; Schmid, K.; Lamri, A.; Tong, L.; Ahmad, M.; Lee, J.J.; Ng, M.C.Y.; Petty, L.E.; et al. Identification of genetic effects underlying type 2 diabetes in South Asian and European populations. Commun. Biol. 2022, 5, 329. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Thompson, S.G.; Collaboration, C.C.G. Avoiding bias from weak instruments in Mendelian randomization studies. Int. J. Epidemiol. 2011, 40, 755–764. [Google Scholar] [CrossRef]
- Burgess, S.; Butterworth, A.; Thompson, S.G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 2013, 37, 658–665. [Google Scholar] [CrossRef] [PubMed]
- Bowden, J.; Del Greco, M.F.; Minelli, C.; Davey Smith, G.; Sheehan, N.; Thompson, J. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat. Med. 2017, 36, 1783–1802. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, Y.A.; Seo, J.H. Causal Association of Obesity and Dyslipidemia with Type 2 Diabetes: A Two-Sample Mendelian Randomization Study. Genes 2022, 13, 2407. [Google Scholar] [CrossRef]
- Bowden, J.; Davey Smith, G.; Haycock, P.C.; Burgess, S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet. Epidemiol. 2016, 40, 304–314. [Google Scholar] [CrossRef] [PubMed]
- Bowden, J.; Davey Smith, G.; Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015, 44, 512–525. [Google Scholar] [CrossRef]
- Bowden, J.; Del Greco, M.F.; Minelli, C.; Davey Smith, G.; Sheehan, N.A.; Thompson, J.R. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: The role of the I2 statistic. Int. J. Epidemiol. 2016, 45, 1961–1974. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Davey Smith, G.; Davies, N.M.; Dudbridge, F.; Gill, D.; Glymour, M.M.; Hartwig, F.P.; Holmes, M.V.; Minelli, C.; Relton, C.L.; et al. Guidelines for performing Mendelian randomization investigations. Wellcome Open Res. 2019, 4, 186. [Google Scholar] [CrossRef]
- Greco, M.F.; Minelli, C.; Sheehan, N.A.; Thompson, J.R. Detecting pleiotropy in Mendelian randomisation studies with summary data and a continuous outcome. Stat. Med. 2015, 34, 2926–2940. [Google Scholar] [CrossRef]
- Verbanck, M.; Chen, C.Y.; Neale, B.; Do, R. Publisher Correction: Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018, 50, 1196. [Google Scholar] [CrossRef]
- Garcia-Aparicio, A.; Garcia de Yebenes, M.J.; Oton, T.; Munoz-Fernandez, S. Prevalence and Incidence of Uveitis: A Systematic Review and Meta-analysis. Ophthalmic. Epidemiol. 2021, 28, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Kanda, T.; Shibata, M.; Taguchi, M.; Ishikawa, S.; Harimoto, K.; Takeuchi, M. Prevalence and aetiology of ocular hypertension in acute and chronic uveitis. Br. J. Ophthalmol. 2014, 98, 932–936. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Aparicio, A.; Alonso Martin, L.; Quiros Zamorano, R.; Lopez Lancho, R.; Del Olmo Perez, L.; Sanchez Fernandez, S.; Garcia de Yebenes, M.J.; Jimenez Escribano, R.; Gonzalez Del Valle, F.; Munoz-Fernandez, S. Complications of uveitis in a Spanish population, UveCAM study. Arch. Soc. Esp. Oftalmol. (Engl. Ed.) 2022, 97, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, C.; Huang, Y.; Guan, P.; Huang, D.; Yu, H.; Yang, X.; Liu, L. Mendelian randomization analyses in ocular disease: A powerful approach to causal inference with human genetic data. J. Transl. Med. 2022, 20, 621. [Google Scholar] [CrossRef]
- Xu, M.; Li, S.; Zhu, J.; Luo, D.; Song, W.; Zhou, M. Plasma lipid levels and risk of primary open angle glaucoma: A genetic study using Mendelian randomization. BMC Ophthalmol. 2020, 20, 390. [Google Scholar] [CrossRef]
- Choquet, H.; Melles, R.B.; Yin, J.; Hoffmann, T.J.; Thai, K.K.; Kvale, M.N.; Banda, Y.; Hardcastle, A.J.; Tuft, S.J.; Glymour, M.M.; et al. A multiethnic genome-wide analysis of 44,039 individuals identifies 41 new loci associated with central corneal thickness. Commun. Biol. 2020, 3, 301. [Google Scholar] [CrossRef]
- Chong, R.S.; Li, H.; Cheong, A.J.; Fan, Q.; Koh, V.; Raghavan, L.; Nongpiur, M.E.; Cheng, C.Y. Mendelian Randomization Implicates Bidirectional Association Between Myopia and Primary Open Angle Glaucoma or Intraocular Pressure. Ophthalmology 2022. ahead of print. [Google Scholar] [CrossRef]
- Tekeli, O.; Elgin, U.; Takmaz, T.; Eksioglu, U.; Bas, Z.; Yarangumeli, A.; Karakurt, A.; Evren Kemer, O.; Mumcuoglu, T.; Aktas, Z.; et al. Characteristics of uveitic glaucoma in Turkish patients. Eur. J. Ophthalmol. 2021, 31, 1836–1843. [Google Scholar] [CrossRef]
- Chen, B.; Li, J.; He, C.; Li, D.; Tong, W.; Zou, Y.; Xu, W. Role of HLA-B27 in the pathogenesis of ankylosing spondylitis (Review). Mol. Med. Rep. 2017, 15, 1943–1951. [Google Scholar] [CrossRef]
- Mayr, W.R.; Grabner, G. HLA antigens-risk factors for primary open-angle glaucoma? Albrecht Von Graefes Arch Klin Exp Ophthalmol. 1978, 207, 291–297. [Google Scholar] [CrossRef]
- Wang, R.F.; Chu, R.Y.; Guo, B.K. HLA-A, B antigens and the relation between high myopia and high myopia with glaucoma. Chin. Med. J. 1986, 99, 775–778. [Google Scholar] [PubMed]
- Monnet, D.; Breban, M.; Hudry, C.; Dougados, M.; Brezin, A.P. Ophthalmic findings and frequency of extraocular manifestations in patients with HLA-B27 uveitis: A study of 175 cases. Ophthalmology 2004, 111, 802–809. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Huang, X.F.; Yang, M.M.; Cai, W.J.; Zheng, M.Q.; Mao, G.; Pang, C.P.; Jin, Z.B. CFI-rs7356506 is a genetic protective factor for acute anterior uveitis in Chinese patients. Br. J. Ophthalmol. 2014, 98, 1592–1596. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.R.; Chiariglione, M.; Arch, A.J. Whole-exome sequencing study identifies rare variants and genes associated with intraocular pressure and glaucoma. Nat. Commun. 2022, 13, 7376. [Google Scholar] [CrossRef]



| Traits | Data Source | No. of Participants | Population | No. of Variants | Reference |
|---|---|---|---|---|---|
| Iritis | BBJ Project | 175,653 | East Asian | 13,429,677 | [29] |
| BBJ Project + UKB | 656,395 | East Asian + European | 25,844,196 | [29] | |
| Uveitis | BBJ Project | 174,725 | East Asian | 13,429,518 | [29] |
| BBJ Project + UKB | 655,467 | East Asian + European | 25,844,095 | [29] | |
| Glaucoma(Open-angle glaucoma) | GERA cohort + UKB | 240,302 (12,315 cases + 227,987 control) | Multiethnic: 214,102 European 5103 African unspecified 3571 Other mixed ancestry 1847 African American or Afro-Caribbean 5189 Hispanic or Latin American 5370 East Asian 5120 South Asian | 7,760,820 | [20] |
| Exposure | Heterogeneity | Horizontal Pleiotropy | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cochran’s Q Test from IVW | Rucker’s Q’ Test from MR-Egger | MR-PRESSO Global Test | MR–Egger | MR–Egger (SIMEX) | ||||||
| N | F | I2 (%) | p-Value | p-Value | p-Value | Intercept, β (SE) | p-Value | Intercept, β (SE) | p-Value | |
| Iritis | 2 | 41588 | 96.33 | 0.359 | ||||||
| Iritis * | 11 | 7704 | 99.11 | 0.587 | 0.558 | 0.659 | −0.009 (0.011) | 0.435 | −0.009 (0.010) | 0.389 |
| Uveitis | 60 | 6433 | 85.64 | 0.148 | 0.131 | 0.521 | −0.001 (0.003) | 0.688 | −0.001 (0.003) | 0.682 |
| Uveitis * | 10 | 8396 | 99.05 | 0.558 | 0.480 | 0.644 | −0.005 (0.01) | 0.644 | −0.005 (0.010) | 0.629 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seo, J.H.; Lee, Y. Causal Association between Iritis or Uveitis and Glaucoma: A Two-Sample Mendelian Randomisation Study. Genes 2023, 14, 642. https://doi.org/10.3390/genes14030642
Seo JH, Lee Y. Causal Association between Iritis or Uveitis and Glaucoma: A Two-Sample Mendelian Randomisation Study. Genes. 2023; 14(3):642. https://doi.org/10.3390/genes14030642
Chicago/Turabian StyleSeo, Je Hyun, and Young Lee. 2023. "Causal Association between Iritis or Uveitis and Glaucoma: A Two-Sample Mendelian Randomisation Study" Genes 14, no. 3: 642. https://doi.org/10.3390/genes14030642
APA StyleSeo, J. H., & Lee, Y. (2023). Causal Association between Iritis or Uveitis and Glaucoma: A Two-Sample Mendelian Randomisation Study. Genes, 14(3), 642. https://doi.org/10.3390/genes14030642





