Integrating Whole-Genome Resequencing and RNA Sequencing Data Reveals Selective Sweeps and Differentially Expressed Genes Related to Nervous System Changes in Luxi Gamecocks
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Sampling and Genome Resequencing Data
2.2. Mapping and SNP Calling
2.3. Detection of Selective Sweeps
2.4. RNA Extraction and Sequencing
2.5. Gene Quantification and Differential Gene Expression Analysis
2.6. Gene Ontology Enrichment and Pathway Analysis
3. Results
3.1. Overview of the Whole-genome Sequencing Data and the RNA-Sequencing Data
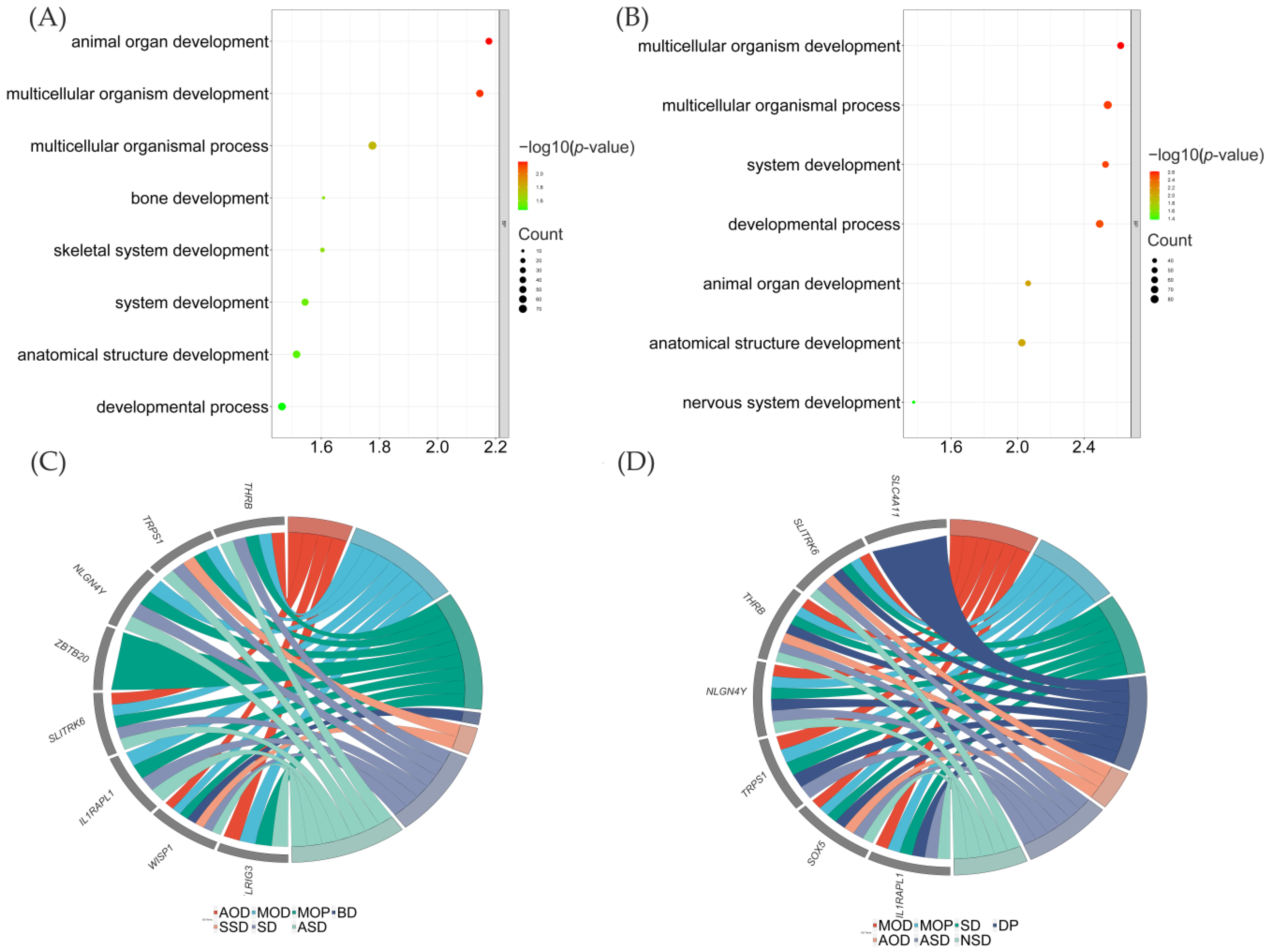
3.2. Selective Sweeps Revealed Genes Are Mostly Associated with Organism Development
3.3. Differentially Expressed Genes between LXFCs and TC/WL Chickens Were Significantly Associated with Nervous System in the Midbrain
3.4. Genes under Selective Sweeps Mostly Showed Differential Expression between LXFC and TC/WL Chickens in the Midbrain
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, W.; Li, H.; Guo, J. Analysis on the Genetic Diversity and the Origin of MtDNA in Chinese Game Chicken. Acta Vet. Zootech. Sin. 2009, 40, 1560–1563. [Google Scholar] [CrossRef]
- China national commission of Animal Genetic Resources. Animal Genetic Resource in China: Poultry; China Agriculture Press: Beijing, China, 2011; ISBN 978-7-109-15221-2. [Google Scholar]
- Liu, Y.-P.; Zhu, Q.; Yao, Y.-G. Genetic Relationship of Chinese and Japanese Gamecocks Revealed by MtDNA Sequence Variation. Biochem. Genet. 2006, 44, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Qu, L.J.; Li, X.Y.; Yang, N. Genetic Relationships among Different Breeds of Chinese Gamecocks Revealed by MtDNA Variation. Asian Australas. J. Anim. Sci. 2009, 22, 1085–1090. [Google Scholar] [CrossRef]
- Guo, X.; Fang, Q.; Ma, C.; Zhou, B.; Wan, Y.; Jiang, R. Whole-Genome Resequencing of Xishuangbanna Fighting Chicken to Identify Signatures of Selection. Genet. Sel. Evol. 2016, 48, 62. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Luo, C.; Wang, M.; Guo, L.; Chen, X.; Li, Z.; Zheng, M.; Folaniyi, B.S.; Luo, W.; Shu, D. Genome Diversity of Chinese Indigenous Chicken and the Selective Signatures in Chinese Gamecock Chicken. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- Komiyama, T.; Iwama, H.; Osada, N.; Nakamura, Y.; Kobayashi, H.; Tateno, Y.; Gojobori, T. Dopamine Receptor Genes and Evolutionary Differentiation in the Domestication of Fighting Cocks and Long-Crowing Chickens. PLoS ONE 2014, 9, e101778. [Google Scholar] [CrossRef]
- Komiyama, T.; Lin, M.; Ogura, A. ACGH Analysis to Estimate Genetic Variations among Domesticated Chickens. BioMed Res. Int. 2016, 2016, 1794329. [Google Scholar] [CrossRef] [PubMed]
- Komiyama, T.; Ikeo, K.; Gojobori, T. The Evolutionary Origin of Long-Crowing Chicken: Its Evolutionary Relationship with Fighting Cocks Disclosed by the MtDNA Sequence Analysis. Gene 2004, 333, 91–99. [Google Scholar] [CrossRef]
- Komiyama, T.; Yoshikawa, M.; Yokoyama, K.; Kobayashi, H. Analysis of the Source of Aggressiveness in Gamecocks. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef]
- Komiyama, T.; Ikeo, K.; Gojobori, T. Where Is the Origin of the Japanese Gamecocks? Gene 2003, 317, 195–202. [Google Scholar] [CrossRef]
- Komiyama, T.; Ikeo, K.; Tateno, Y.; Gojobori, T. Japanese Domesticated Chickens Have Been Derived from Shamo Traditional Fighting Cocks. Mol. Phylogenet. Evol. 2004, 33, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zheng, M.; Abdalla, B.A.; Zhang, Z.; Xu, Z.; Ye, Q.; Xu, H.; Luo, W.; Nie, Q.; Zhang, X. Genome-Wide Association Study of Aggressive Behaviour in Chicken. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Narvaes, R.; Martins de Almeida, R.M. Aggressive Behavior and Three Neurotransmitters: Dopamine, GABA, and Serotonin—A Review of the Last 10 Years. Psychol. Neurosci. 2014, 7, 601. [Google Scholar] [CrossRef]
- Seo, D.; Patrick, C.J.; Kennealy, P.J. Role of Serotonin and Dopamine System Interactions in the Neurobiology of Impulsive Aggression and Its Comorbidity with Other Clinical Disorders. Aggress. Violent Behav. 2008, 13, 383–395. [Google Scholar] [CrossRef]
- Cases, O.; Seif, I.; Grimsby, J.; Gaspar, P.; Chen, K.; Pournin, S.; Muller, U.; Aguet, M.; Babinet, C.; Shih, J.C.; et al. Aggressive Behavior and Altered Amounts of Brain Serotonin and Norepinephrine in Mice Lacking MAOA. Science 1995, 268, 1763–1766. [Google Scholar] [CrossRef]
- Olivier, B. Serotonin: A Never-Ending Story. Eur. J. Pharmacol. 2015, 753, 2–18. [Google Scholar] [CrossRef]
- Trifu, S.C.; Tudor, A.; Radulescu, I. Aggressive Behavior in Psychiatric Patients in Relation to Hormonal Imbalance. Exp. Ther. Med. 2020, 20, 3483–3487. [Google Scholar] [CrossRef]
- Haden, S.C.; Scarpa, A. The Noradrenergic System and Its Involvement in Aggressive Behaviors. Aggress. Violent Behav. 2007, 12, 1–15. [Google Scholar] [CrossRef]
- Katayama, K.; Yamada, K.; Ornthanalai, V.G.; Inoue, T.; Ota, M.; Murphy, N.P.; Aruga, J. Slitrk1-Deficient Mice Display Elevated Anxiety-like Behavior and Noradrenergic Abnormalities. Mol. Psychiatry 2010, 15, 177–184. [Google Scholar] [CrossRef]
- Dennis, R.; Cheng, H.W. The Dopaminergic System and Aggression in Laying Hens. Poult. Sci. 2011, 90, 2440–2448. [Google Scholar] [CrossRef]
- Smith, A.N.; Kabelik, D. The Effects of Dopamine Receptor 1 and 2 Agonists and Antagonists on Sexual and Aggressive Behaviors in Male Green Anoles. PLoS ONE 2017, 12, e0172041. [Google Scholar] [CrossRef]
- Wang, M.-S.; Thakur, M.; Peng, M.-S.; Jiang, Y.; Frantz, L.A.F.; Li, M.; Zhang, J.-J.; Wang, S.; Peters, J.; Otecko, N.O. 863 Genomes Reveal the Origin and Domestication of Chicken. Cell Res. 2020, 30, 693–701. [Google Scholar] [CrossRef]
- Zhang, Q.; Gou, W.; Wang, X.; Zhang, Y.; Ma, J.; Zhang, H.; Zhang, Y.; Zhang, H. Genome Resequencing Identifies Unique Adaptations of Tibetan Chickens to Hypoxia and High-Dose Ultraviolet Radiation in High-Altitude Environments. Genome Biol. Evol. 2016, 8, 765–776. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data [Online]. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 8 June 2022).
- Li, H.; Durbin, R. Fast and Accurate Long-Read Alignment with Burrows–Wheeler Transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Poplin, R.; Ruano-Rubio, V.; DePristo, M.A.; Fennell, T.J.; Carneiro, M.O.; Van der Auwera, G.A.; Kling, D.E.; Gauthier, L.D.; Levy-Moonshine, A.; Roazen, D.; et al. Scaling Accurate Genetic Variant Discovery to Tens of Thousands of Samples. BioRxiv 2018, 201178. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Ayres, D.L.; Darling, A.; Zwickl, D.J.; Beerli, P.; Holder, M.T.; Lewis, P.O.; Huelsenbeck, J.P.; Ronquist, F.; Swofford, D.L.; Cummings, M.P.; et al. BEAGLE: An Application Programming Interface and High-Performance Computing Library for Statistical Phylogenetics. Syst. Biol. 2012, 61, 170–173. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A Program for Annotating and Predicting the Effects of Single Nucleotide Polymorphisms, SnpEff: SNPs in the Genome of Drosophila Melanogaster Strain W1118; Iso-2; Iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-Generation PLINK: Rising to the Challenge of Larger and Richer Datasets. Gigascience 2015, 4, s13742-015. [Google Scholar] [CrossRef]
- Lewontin, R.C.; Krakauer, J. Distribution of Gene Frequency as a Test of the Theory of the Selective Neutrality of Polymorphisms. Genetics 1973, 74, 175–195. [Google Scholar] [CrossRef]
- Fariello, M.I.; Boitard, S.; Naya, H.; SanCristobal, M.; Servin, B. Detecting Signatures of Selection through Haplotype Differentiation among Hierarchically Structured Populations. Genetics 2013, 193, 929–941. [Google Scholar] [CrossRef] [PubMed]
- Tajima, F. Statistical Method for Testing the Neutral Mutation Hypothesis by DNA Polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Voight, B.F.; Kudaravalli, S.; Wen, X.; Pritchard, J.K. A Map of Recent Positive Selection in the Human Genome. PLoS Biol. 2006, 4, e72. [Google Scholar] [CrossRef]
- Sabeti, P.C.; Varilly, P.; Fry, B.; Lohmueller, J.; Hostetter, E.; Cotsapas, C.; Xie, X.; Byrne, E.H.; McCarroll, S.A.; Gaudet, R.; et al. Genome-Wide Detection and Characterization of Positive Selection in Human Populations. Nature 2007, 449, 913–918. [Google Scholar] [CrossRef] [PubMed]
- Szmatoła, T.; Gurgul, A.; Jasielczuk, I.; Ząbek, T.; Ropka-Molik, K.; Litwińczuk, Z.; Bugno-Poniewierska, M. A Comprehensive Analysis of Runs of Homozygosity of Eleven Cattle Breeds Representing Different Production Types. Animals 2019, 9, 1024. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The Variant Call Format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Szpiech, Z.A.; Hernandez, R.D. Selscan: An Efficient Multithreaded Program to Perform EHH-Based Scans for Positive Selection. Mol. Biol. Evol. 2014, 31, 2824–2827. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-sequencing Data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. G: Profiler: A Web Server for Functional Enrichment Analysis and Conversions of Gene Lists (2019 Update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- Yagi, T.; Takeichi, M. Cadherin Superfamily Genes: Functions, Genomic Organization, and Neurologic Diversity. Genes Dev. 2000, 14, 1169–1180. [Google Scholar] [CrossRef] [PubMed]
- Junghof, J.; Kogure, Y.; Yu, T.; Verdugo-Sivianes, E.M.; Narita, M.; Lucena-Cacace, A.; Yoshida, Y. CDH18 Is a Fetal Epicardial Biomarker Regulating Differentiation towards Vascular Smooth Muscle Cells. npj Regen. Med. 2022, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Grados, M.; Walkup, J. A New Gene for Tourette’s Syndrome: A Window into Causal Mechanisms? Trends Genet. 2006, 22, 291–293. [Google Scholar] [CrossRef]
- Miranda, D.M.; Wigg, K.; Kabia, E.M.; Feng, Y.; Sandor, P.; Barr, C.L. Association of SLITRK1 to Gilles de La Tourette Syndrome. Am. J. Med. Genet. 2009, 150B, 483–486. [Google Scholar] [CrossRef]
- Tekin, M.; Chioza, B.A.; Matsumoto, Y.; Diaz-Horta, O.; Cross, H.E.; Duman, D.; Kokotas, H.; Moore-Barton, H.L.; Sakoori, K.; Ota, M.; et al. SLITRK6 Mutations Cause Myopia and Deafness in Humans and Mice. J. Clin. Investig. 2013, 123, 2094–2102. [Google Scholar] [CrossRef]
- Morlet, T.; Rabinowitz, M.R.; Looney, L.R.; Riegner, T.; Greenwood, L.A.; Sherman, E.A.; Achilly, N.; Zhu, A.; Yoo, E.; O’Reilly, R.C.; et al. A Homozygous SLITRK6 Nonsense Mutation Is Associated with Progressive Auditory Neuropathy in Humans: SLITRK6 Is Associated With Human Auditory Neuropathy. Laryngoscope 2014, 124, E95–E103. [Google Scholar] [CrossRef]
- Yasumura, M.; Yoshida, T.; Yamazaki, M.; Abe, M.; Natsume, R.; Kanno, K.; Uemura, T.; Takao, K.; Sakimura, K.; Kikusui, T.; et al. IL1RAPL1 Knockout Mice Show Spine Density Decrease, Learning Deficiency, Hyperactivity and Reduced Anxiety-like Behaviours. Sci. Rep. 2015, 4, 6613. [Google Scholar] [CrossRef]
- Lencz, T.; Guha, S.; Liu, C.; Rosenfeld, J.; Mukherjee, S.; DeRosse, P.; John, M.; Cheng, L.; Zhang, C.; Badner, J.A.; et al. Genome-Wide Association Study Implicates NDST3 in Schizophrenia and Bipolar Disorder. Nat. Commun. 2013, 4, 2739. [Google Scholar] [CrossRef]
- Zhang, C.; Lu, W.; Wang, Z.; Ni, J.; Zhang, J.; Tang, W.; Fang, Y. A Comprehensive Analysis of NDST3 for Schizophrenia and Bipolar Disorder in Han Chinese. Transl. Psychiatry 2016, 6, e701. [Google Scholar] [CrossRef]
- Maiese, K. WISP1: Clinical Insights for a Proliferative and Restorative Member of the CCN Family. Curr. Neurovascular Res. 2014, 11, 378–389. [Google Scholar] [CrossRef]
- Park, H.; Lee, S.; Kim, H.-J.; Ju, Y.S.; Shin, J.-Y.; Hong, D.; von Grotthuss, M.; Lee, D.-S.; Park, C.; Kim, J.H.; et al. Comprehensive Genomic Analyses Associate UGT8 Variants with Musical Ability in a Mongolian Population. J. Med. Genet. 2012, 49, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Zaccariotto, E.; Cachón-González, M.B.; Wang, B.; Lim, S.; Hirth, B.; Park, H.; Fezoui, M.; Sardi, S.P.; Mason, P.; Barker, R.H.; et al. A Novel Brain-Penetrant Oral UGT8 Inhibitor Decreases in Vivo Galactosphingolipid Biosynthesis in Murine Krabbe Disease. Biomed. Pharmacother. 2022, 149, 112808. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Tanegashima, K.; Ro, H.; Dawid, I.B. Lrig3 Regulates Neural Crest Formation in Xenopus by Modulating Fgf and Wnt Signaling Pathways. Development 2008, 135, 1283–1293. [Google Scholar] [CrossRef] [PubMed]
- Abraira, V.E.; del Rio, T.; Tucker, A.F.; Slonimsky, J.; Keirnes, H.L.; Goodrich, L.V. Cross-Repressive Interactions between Lrig3 and Netrin 1 Shape the Architecture of the Inner Ear. Development 2008, 135, 4091–4099. [Google Scholar] [CrossRef]
- Ayka, A.; Şehirli, A.Ö. The Role of the SLC Transporters Protein in the Neurodegenerative Disorders. Clin. Psychopharmacol. Neurosci. 2020, 18, 174–187. [Google Scholar] [CrossRef] [PubMed]
- Wilcox, J.A.; Quadri, S. Replication of NTNG1 Association in Schizophrenia. Psychiatr. Genet. 2014, 24, 266–268. [Google Scholar] [CrossRef]
- Zhu, Y.; Yang, H.; Bi, Y.; Zhang, Y.; Zhen, C.; Xie, S.; Qin, H.; He, J.; Liu, L.; Liu, Y. Positive Association between NTNG1 and Schizophrenia in Chinese Han Population. J. Genet. 2011, 90, 499–502. [Google Scholar] [CrossRef]
- Mei, L.; Nave, K.-A. Neuregulin-ERBB Signaling in the Nervous System and Neuropsychiatric Diseases. Neuron 2014, 83, 27–49. [Google Scholar] [CrossRef] [PubMed]
- Ledonne, A.; Mercuri, N.B. On the Modulatory Roles of Neuregulins/ErbB Signaling on Synaptic Plasticity. IJMS 2019, 21, 275. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.K.; Choi, E.-J. Pathological Roles of MAPK Signaling Pathways in Human Diseases. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2010, 1802, 396–405. [Google Scholar] [CrossRef]
- Child, N.D.; Benarroch, E.E. MTOR: Its Role in the Nervous System and Involvement in Neurologic Disease. Neurology 2014, 83, 1562–1572. [Google Scholar] [CrossRef]
- Maiese, K.; Chong, Z.Z.; Shang, Y.C.; Wang, S. MTOR: On Target for Novel Therapeutic Strategies in the Nervous System. Trends Mol. Med. 2013, 19, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Lipton, J.O.; Sahin, M. The Neurology of MTOR. Neuron 2014, 84, 275–291. [Google Scholar] [CrossRef]
- Eastwood, S.L.; Harrison, P.J. Decreased MRNA Expression of Netrin-G1 and Netrin-G2 in the Temporal Lobe in Schizophrenia and Bipolar Disorder. Neuropsychopharmacology 2008, 33, 933–945. [Google Scholar] [CrossRef]
- Chichinadze, K.; Chichinadze, N.; Lazarashvili, A. Hormonal and Neurochemical Mechanisms of Aggression and a New Classification of Aggressive Behavior. Aggress. Violent Behav. 2011, 16, 461–471. [Google Scholar] [CrossRef]
- Umukoro, S.; Aladeokin, A.C.; Eduviere, A.T. Aggressive Behavior: A Comprehensive Review of Its Neurochemical Mechanisms and Management. Aggress. Violent Behav. 2013, 18, 195–203. [Google Scholar] [CrossRef]
- Bertelsen, B.; Melchior, L.; Jensen, L.R.; Groth, C.; Glenthøj, B.; Rizzo, R.; Debes, N.M.; Skov, L.; Brøndum-Nielsen, K.; Paschou, P.; et al. Intragenic Deletions Affecting Two Alternative Transcripts of the IMMP2L Gene in Patients with Tourette Syndrome. Eur. J. Hum. Genet. 2014, 22, 1283–1289. [Google Scholar] [CrossRef] [PubMed]
- IMGSAC; Maestrini, E.; Pagnamenta, A.T.; Lamb, J.A.; Bacchelli, E.; Sykes, N.H.; Sousa, I.; Toma, C.; Barnby, G.; Butler, H.; et al. High-Density SNP Association Study and Copy Number Variation Analysis of the AUTS1 and AUTS5 Loci Implicate the IMMP2L–DOCK4 Gene Region in Autism Susceptibility. Mol. Psychiatry 2010, 15, 954–968. [Google Scholar] [CrossRef] [PubMed]
- Adell, A.; Celada, P.; Abellán, M.T.; Artigas, F. Origin and Functional Role of the Extracellular Serotonin in the Midbrain Raphe Nuclei. Brain Res. Rev. 2002, 39, 154–180. [Google Scholar] [CrossRef] [PubMed]
- Våge, J.; Wade, C.; Biagi, T.; Fatjó, J.; Amat, M.; Lindblad-Toh, K.; Lingaas, F. Association of Dopamine-and Serotonin-Related Genes with Canine Aggression. Genes Brain Behav. 2010, 9, 372–378. [Google Scholar] [CrossRef] [PubMed]
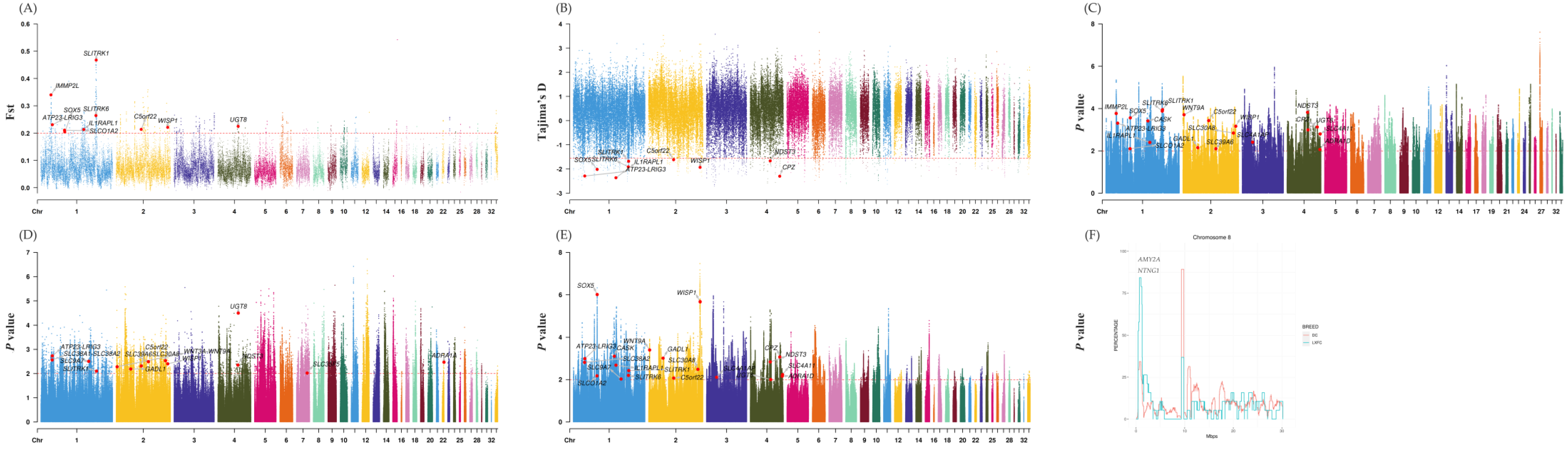
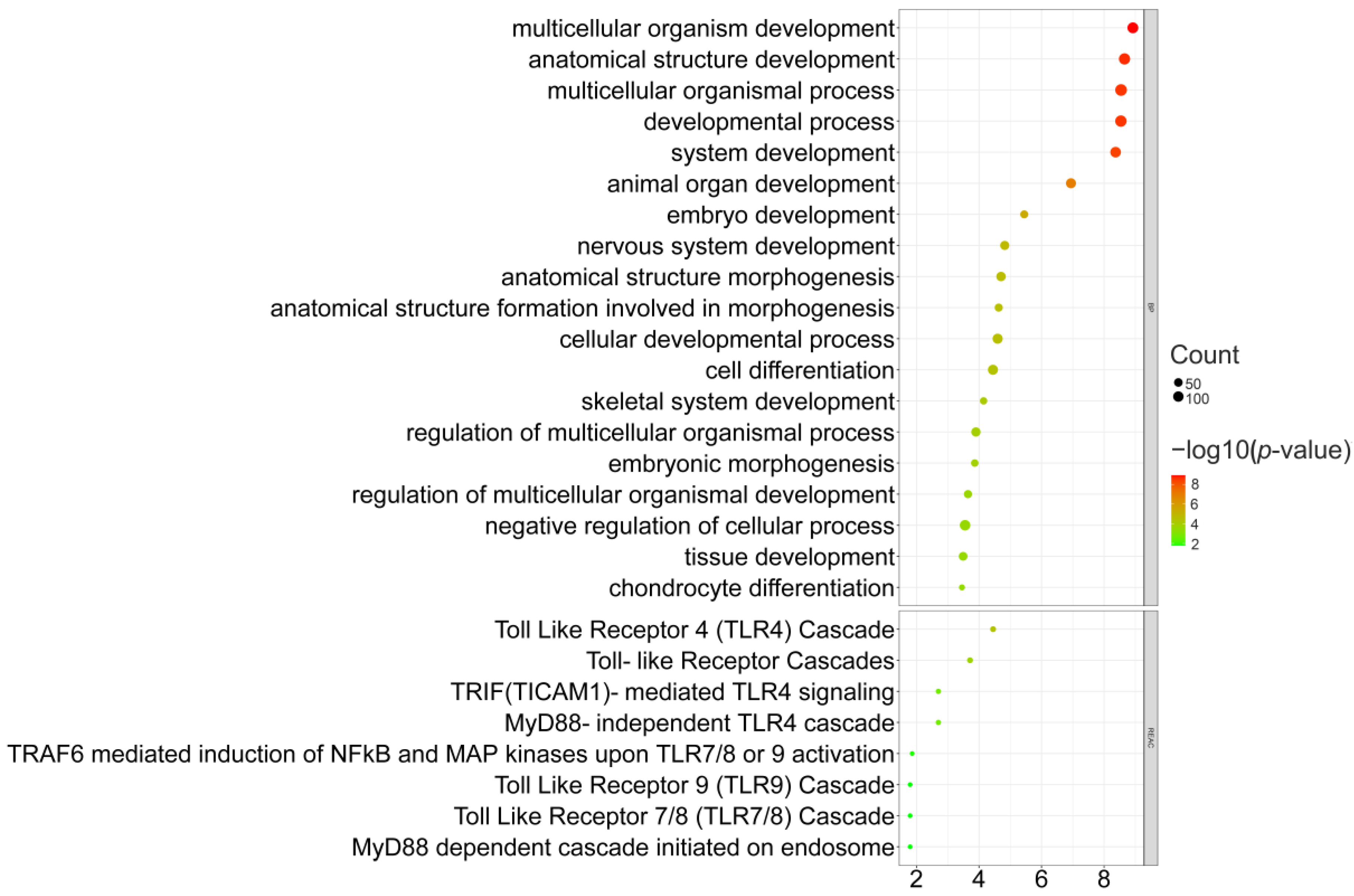
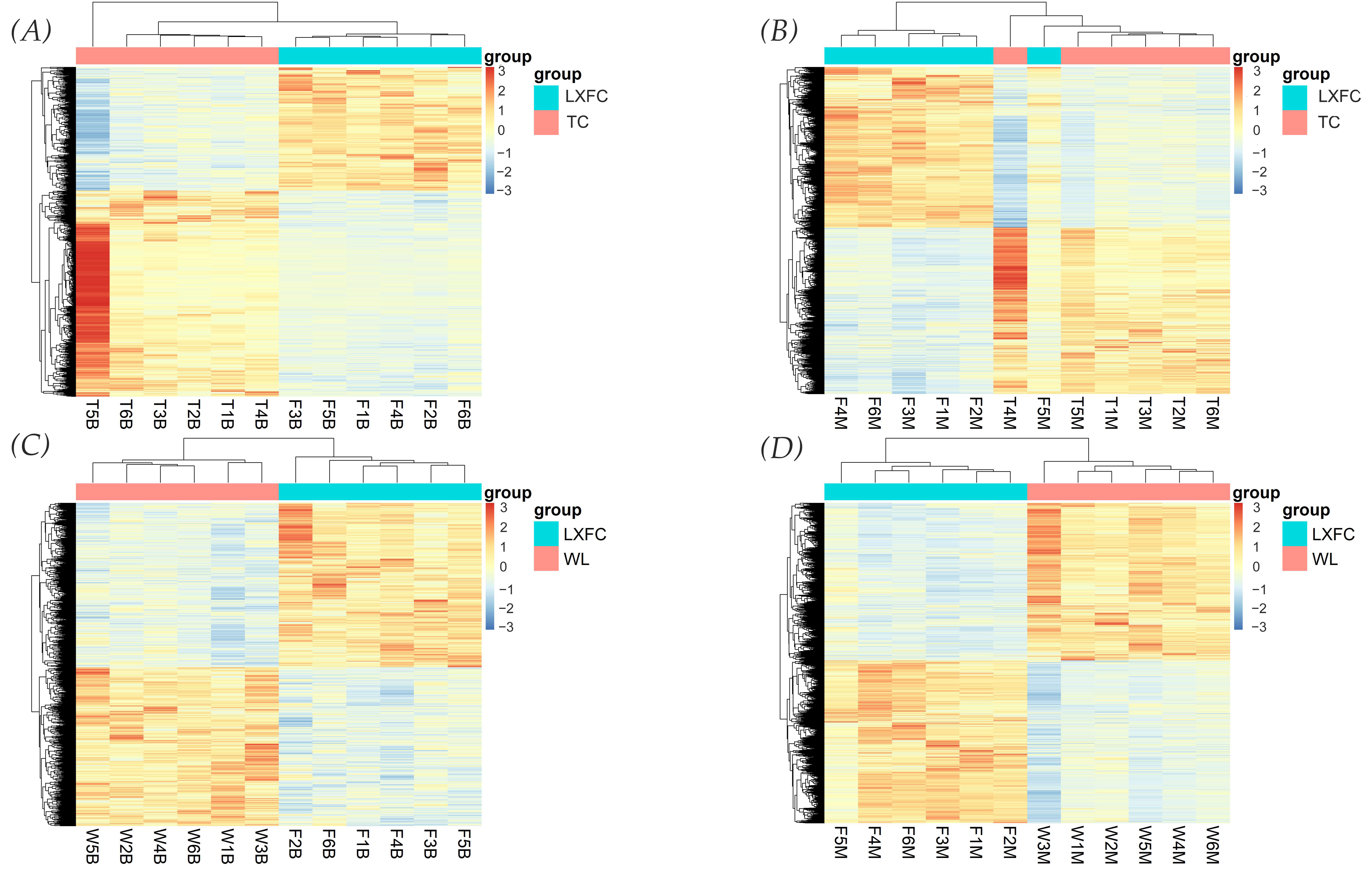

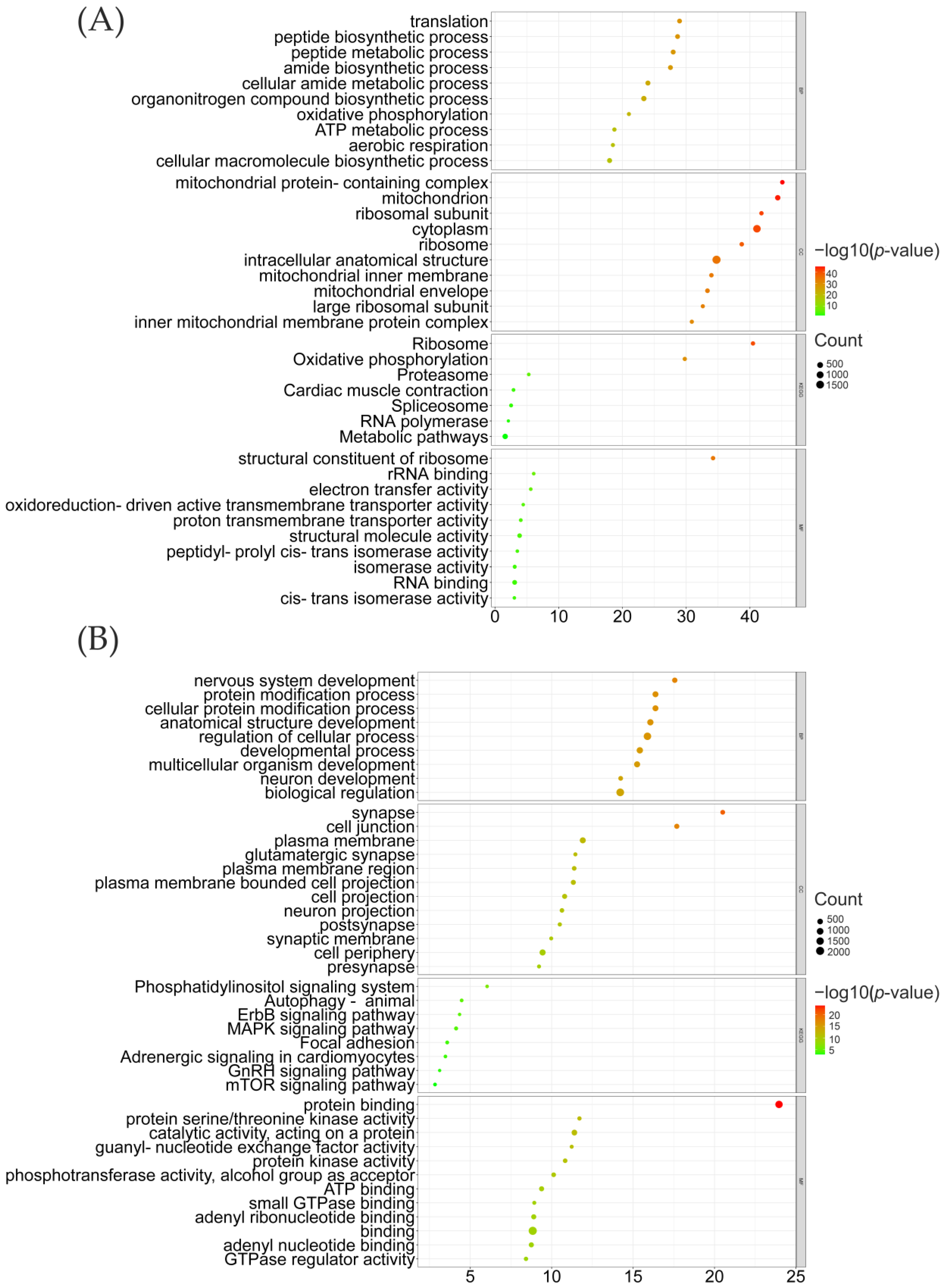
| Population | Abbreviation | Numbers |
|---|---|---|
| Luxi gamecock | LXFC | 19 |
| Shandong native chicken | SDNC | 30 |
| Xishuangbanna gamecock | YNFC | 16 |
| Yunnan native chicken | YNNC | 22 |
| Tibetan chicken | TC | 18 |
| Rhode Island red | RDH | 20 |
| Gene | Full Name | Fst | Tajima’s D | hapFLK | iHS | XP-EHH |
|---|---|---|---|---|---|---|
| CDH18 | cadherin 18 | √ | √ | √ | √ | √ |
| SLITRK1 | SLIT and NTRK-like family member 1 | √ | - | √ | √ | √ |
| SLITRK6 | SLIT and NTRK-like family member 6 | √ | √ | √ | √ | - |
| NDST3 | N-deacetylase and N-sulfotransferase 3 | - | √ | √ | √ | √ |
| ATP23 | ATP23 metallopeptidase and ATP synthase assembly factor homolog | √ | - | √ | √ | √ |
| LRIG3 | leucine-rich repeats and immunoglobulin-like domains 3 | √ | - | √ | √ | √ |
| IL1RAPL1 | glutamate decarboxylase-like 1 | √ | √ | √ | - | √ |
| GADL1 | interleukin 1 receptor accessory protein-like 1 | - | √ | √ | √ | √ |
| C5orf22 | chromosome 2 open reading frame, human C5orf22 | √ | - | √ | √ | √ |
| UGT8 | UDP glycosyltransferase 8 | √ | - | √ | √ | √ |
| WISP1 | WNT1-inducible signaling pathway protein 1 | √ | - | √ | √ | √ |
| WNT9A | Wnt family member 9A | - | √ | √ | √ | √ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, J.; Chang, Y.; Li, J.; Bao, H.; Wu, C. Integrating Whole-Genome Resequencing and RNA Sequencing Data Reveals Selective Sweeps and Differentially Expressed Genes Related to Nervous System Changes in Luxi Gamecocks. Genes 2023, 14, 584. https://doi.org/10.3390/genes14030584
Zhou J, Chang Y, Li J, Bao H, Wu C. Integrating Whole-Genome Resequencing and RNA Sequencing Data Reveals Selective Sweeps and Differentially Expressed Genes Related to Nervous System Changes in Luxi Gamecocks. Genes. 2023; 14(3):584. https://doi.org/10.3390/genes14030584
Chicago/Turabian StyleZhou, Jieke, Ying Chang, Junying Li, Haigang Bao, and Changxin Wu. 2023. "Integrating Whole-Genome Resequencing and RNA Sequencing Data Reveals Selective Sweeps and Differentially Expressed Genes Related to Nervous System Changes in Luxi Gamecocks" Genes 14, no. 3: 584. https://doi.org/10.3390/genes14030584
APA StyleZhou, J., Chang, Y., Li, J., Bao, H., & Wu, C. (2023). Integrating Whole-Genome Resequencing and RNA Sequencing Data Reveals Selective Sweeps and Differentially Expressed Genes Related to Nervous System Changes in Luxi Gamecocks. Genes, 14(3), 584. https://doi.org/10.3390/genes14030584





