Characterization of NDM-5-Producing Escherichia coli Strains Isolated from Pediatric Patients with Bloodstream Infections in a Chinese Hospital
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacteria Isolates and Clinical Data Collection
2.2. Strain Identification and β-Lactamase Genes Confirmation
2.3. Antimicrobial Susceptibility Testing
2.4. Conjugation Assay
2.5. DNA Extraction
2.6. Whole-Genome Sequencing and Bioinformatics Analysis
2.7. Ethical Statements
3. Results
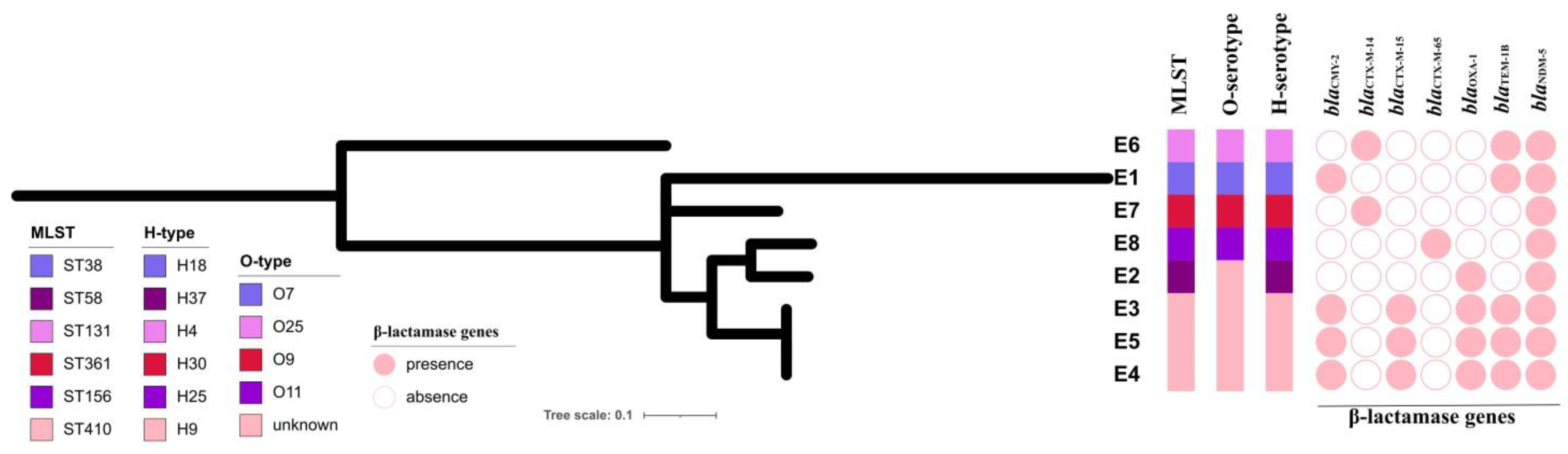
3.1. Clinical Information and Overview of the NDM-5-Producing E. coli Strains
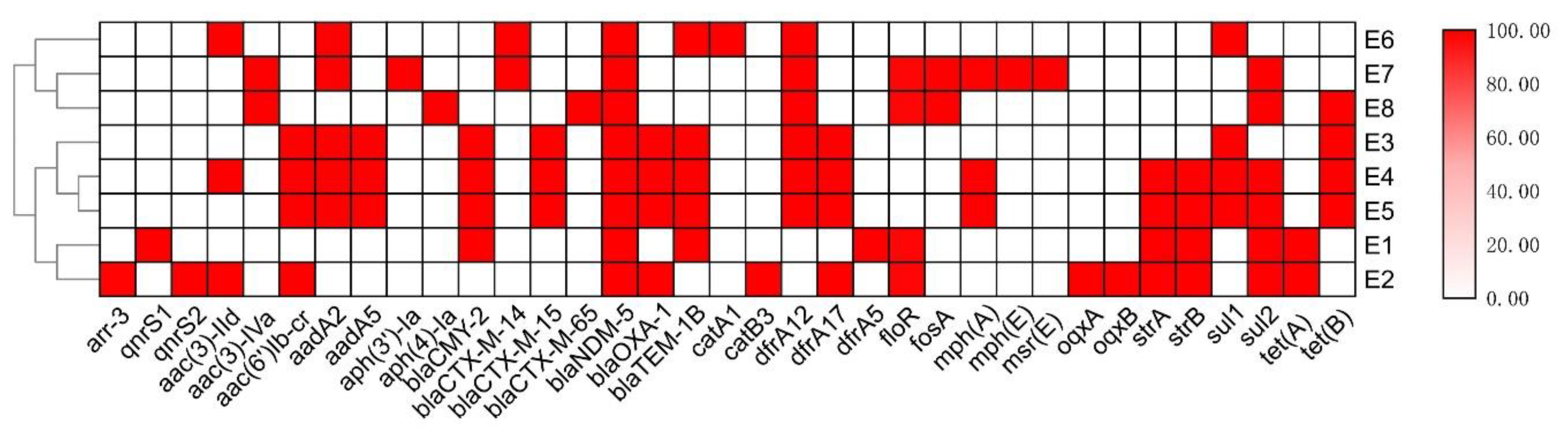
3.2. Antimicrobial Resistance Profiles of the NDM-5-Producing E. coli Strains
3.3. Genomic Characteristics of the NDM-5-Producing E. coli Strains
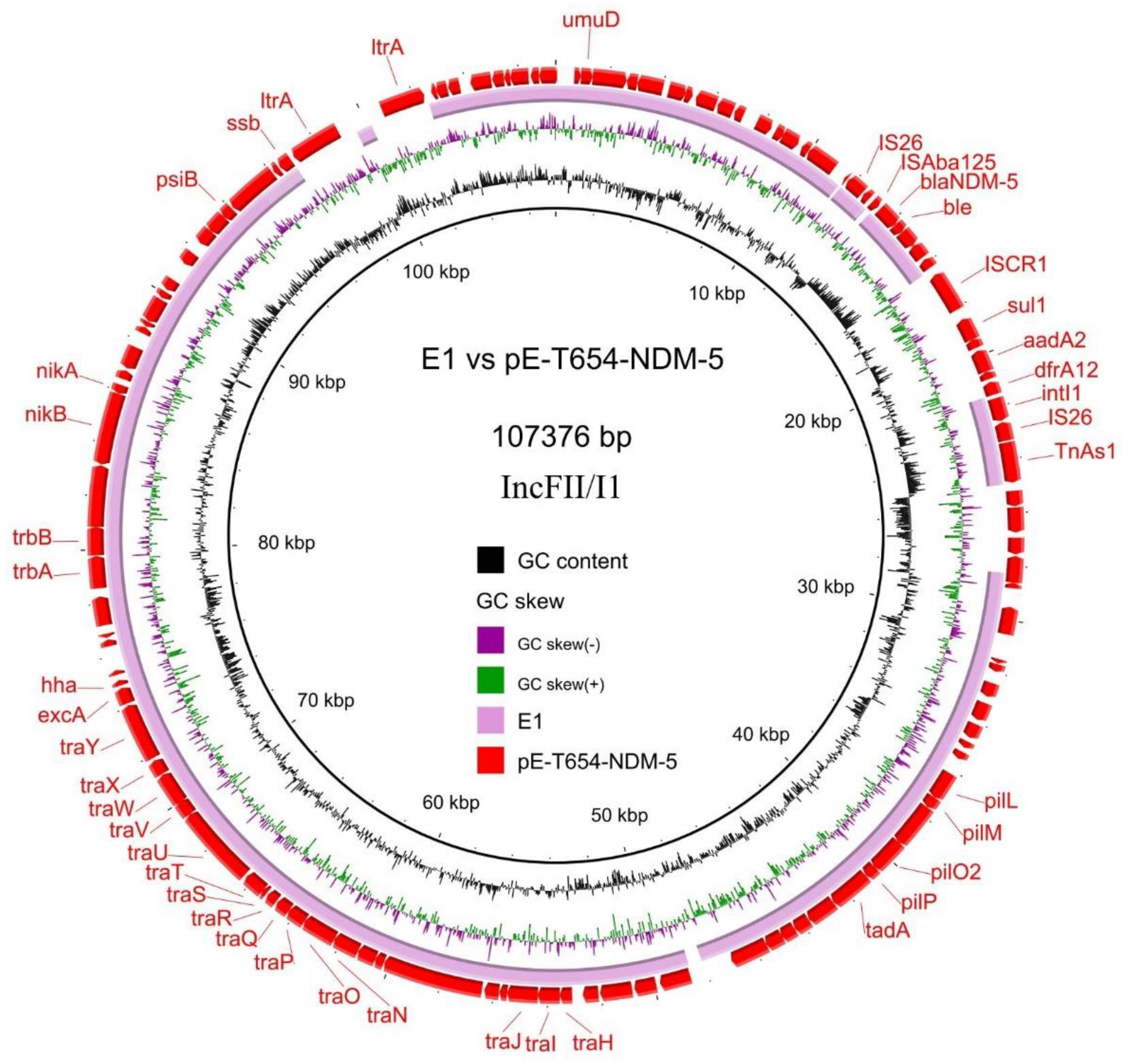
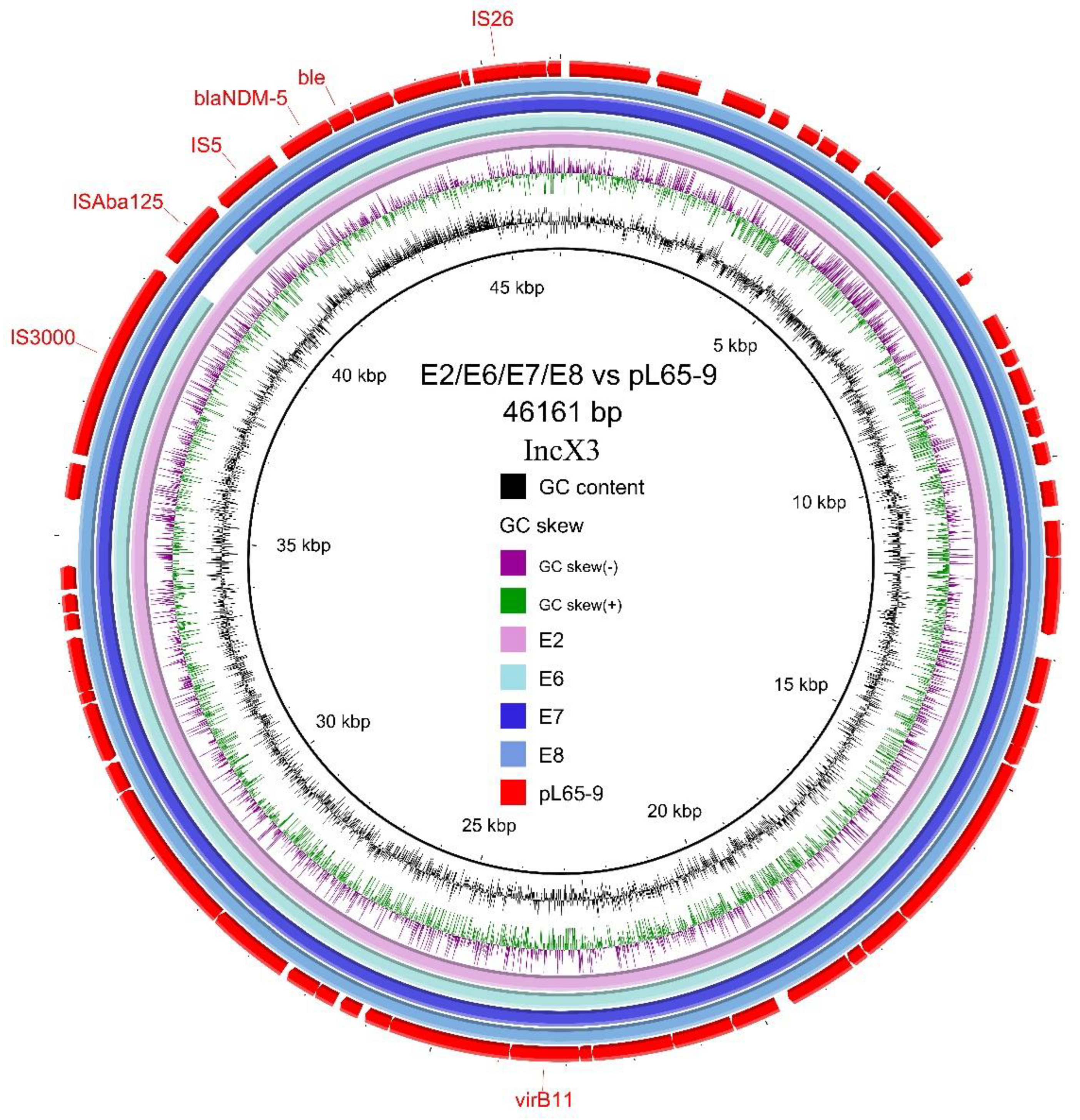
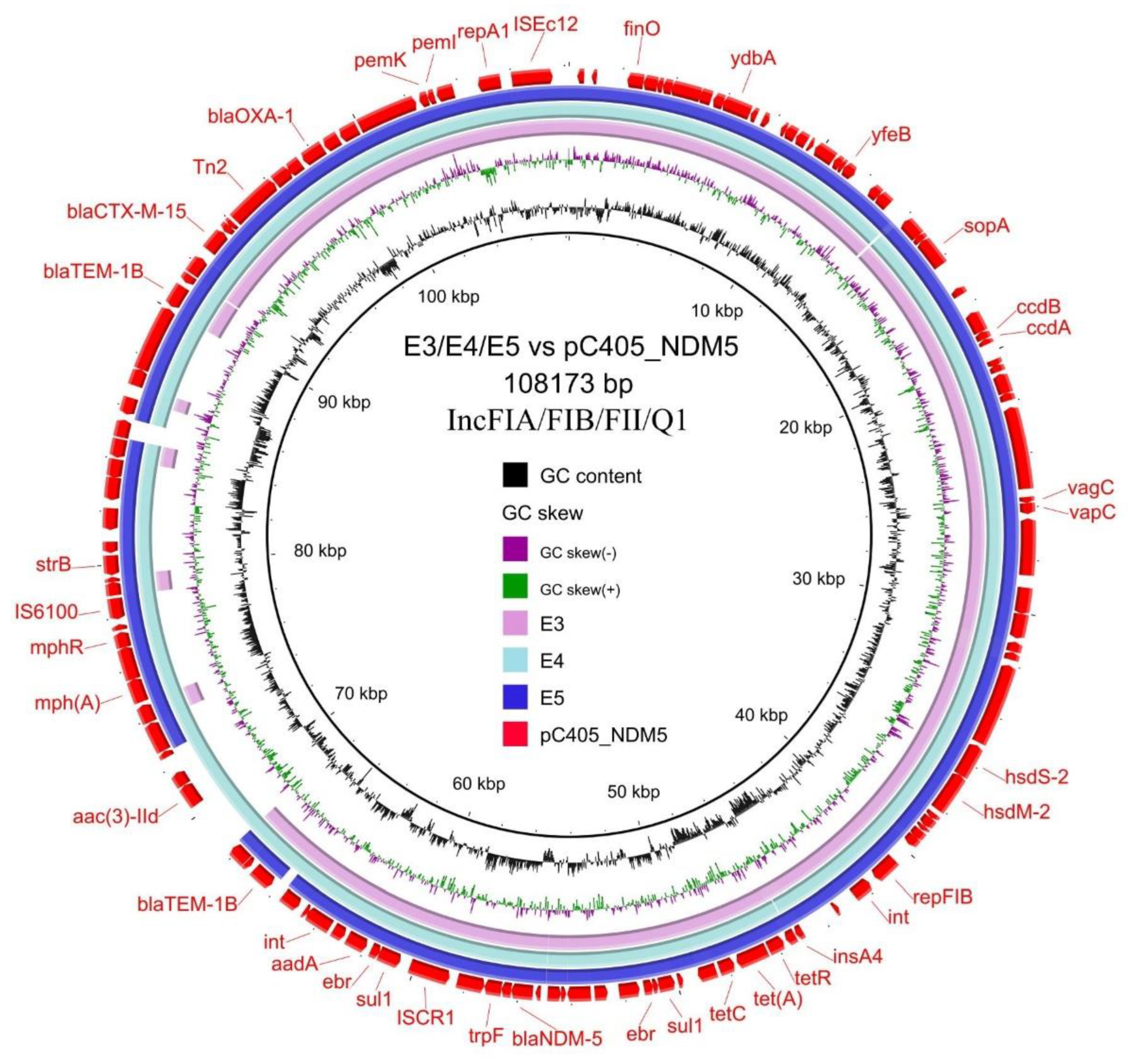
3.4. Transferability and Genetic Analysis of blaNDM-5-Carrying Plasmids
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Di Franco, S.; Alfieri, A.; Pace, M.C.; Sansone, P.; Pota, V.; Fittipaldi, C.; Fiore, M.; Passavanti, M.B. Blood Stream Infections from MDR Bacteria. Life 2021, 11, 575. [Google Scholar] [CrossRef] [PubMed]
- Rudd, K.E.; Johnson, S.C.; Agesa, K.M.; Shackelford, K.A.; Tsoi, D.; Kievlan, D.R.; Colombara, D.V.; Ikuta, K.S.; Kissoon, N.; Finfer, S.; et al. Global, regional, and national sepsis incidence and mortality, 1990–2017: Analysis for the Global Burden of Disease Study. Lancet 2020, 395, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Shakur, S.M.; Whitehall, J.; Mudgil, P. Pediatric bloodstream infections in metropolitan Australia. World J. Pediatr. WJP 2019, 15, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Malande, O.O.; Nuttall, J.; Pillay, V.; Bamford, C.; Eley, B. A ten-year review of ESBL and non-ESBL Escherichia coli bloodstream infections among children at a tertiary referral hospital in South Africa. PloS ONE 2019, 14, e0222675. [Google Scholar] [CrossRef]
- Zaidi, A.K.; Ganatra, H.A.; Syed, S.; Cousens, S.; Lee, A.C.; Black, R.; Bhutta, Z.A.; Lawn, J.E. Effect of case management on neonatal mortality due to sepsis and pneumonia. BMC Public Health 2011, 11, S13. [Google Scholar] [CrossRef] [PubMed]
- Fleischmann-Struzek, C.; Goldfarb, D.M.; Schlattmann, P.; Schlapbach, L.J.; Reinhart, K.; Kissoon, N. The global burden of paediatric and neonatal sepsis: A systematic review. Lancet Respir. Med. 2018, 6, 223–230. [Google Scholar] [CrossRef]
- Naylor, N.R.; Pouwels, K.B.; Hope, R.; Green, N.; Henderson, K.L.; Knight, G.M.; Atun, R.; Robotham, J.V.; Deeny, S.R. The health and cost burden of antibiotic resistant and susceptible Escherichia coli bacteraemia in the English hospital setting: A national retrospective cohort study. PLoS ONE 2019, 14, e0221944. [Google Scholar] [CrossRef]
- Dellinger, R.P.; Levy, M.M.; Rhodes, A.; Annane, D.; Gerlach, H.; Opal, S.M.; Sevransky, J.E.; Sprung, C.L.; Douglas, I.S.; Jaeschke, R.; et al. Surviving Sepsis Campaign: International guidelines for management of severe sepsis and septic shock, 2012. Intensive Care Med. 2013, 39, 165–228. [Google Scholar] [CrossRef]
- Kissoon, N.; Carcillo, J.A.; Espinosa, V.; Argent, A.; Devictor, D.; Madden, M.; Singhi, S.; van der Voort, E.; Latour, J. World Federation of Pediatric Intensive Care and Critical Care Societies: Global Sepsis Initiative. Pediatr. Crit. Care Med. A J. Soc. Crit. Care Med. World Fed. Pediatr. Intensive Crit. Care Soc. 2011, 12, 494–503. [Google Scholar] [CrossRef]
- Holmes, C.L.; Anderson, M.T.; Mobley, H.L.T.; Bachman, M.A. Pathogenesis of Gram-Negative Bacteremia. Clin. Microbiol. Rev. 2021, 34, e00234-20. [Google Scholar] [CrossRef]
- Diekema, D.J.; Hsueh, P.R.; Mendes, R.E.; Pfaller, M.A.; Rolston, K.V.; Sader, H.S.; Jones, R.N. The Microbiology of Bloodstream Infection: 20-Year Trends from the SENTRY Antimicrobial Surveillance Program. Antimicrob. Agents Chemother. 2019, 63, e00355-19. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Yoon, E.J.; Hong, J.S.; Choi, M.H.; Kim, H.S.; Kim, Y.R.; Kim, Y.A.; Uh, Y.; Shin, K.S.; Shin, J.H.; et al. Major Bloodstream Infection-Causing Bacterial Pathogens and Their Antimicrobial Resistance in South Korea, 2017–2019: Phase I Report From Kor-GLASS. Front. Microbiol. 2021, 12, 799084. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Sun, Z.; Zhang, Z. Antimicrobial resistance of pathogens causing nosocomial bloodstream infection in Hubei Province, China, from 2014 to 2016: A multicenter retrospective study. BMC Public Health 2018, 18, 1121. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Zhang, Z.; Sun, Z. Antimicrobial resistance trends in bloodstream infections at a large teaching hospital in China: A 20-year surveillance study (1998–2017). Antimicrob. Resist. Infect. Control. 2019, 8, 86. [Google Scholar] [CrossRef]
- Randolph, A.G.; McCulloh, R.J. Pediatric sepsis: Important considerations for diagnosing and managing severe infections in infants, children, and adolescents. Virulence 2014, 5, 179–189. [Google Scholar] [CrossRef]
- Abernethy, J.; Guy, R.; Sheridan, E.A.; Hopkins, S.; Kiernan, M.; Wilcox, M.H.; Johnson, A.P.; Hope, R. Epidemiology of Escherichia coli bacteraemia in England: Results of an enhanced sentinel surveillance programme. J. Hosp. Infect. 2017, 95, 365–375. [Google Scholar] [CrossRef]
- Williamson, D.A.; Lim, A.; Wiles, S.; Roberts, S.A.; Freeman, J.T. Population-based incidence and comparative demographics of community-associated and healthcare-associated Escherichia coli bloodstream infection in Auckland, New Zealand, 2005–2011. BMC Infect. Dis. 2013, 13, 385. [Google Scholar] [CrossRef]
- Okomo, U.; Akpalu, E.N.K.; Le Doare, K.; Roca, A.; Cousens, S.; Jarde, A.; Sharland, M.; Kampmann, B.; Lawn, J.E. Aetiology of invasive bacterial infection and antimicrobial resistance in neonates in sub-Saharan Africa: A systematic review and meta-analysis in line with the STROBE-NI reporting guidelines. Lancet Infect. Dis. 2019, 19, 1219–1234. [Google Scholar] [CrossRef]
- Elseady, N.S.M.; Khamis, N.; AbdelGhani, S.; Rabea, H.M.; Elanany, M.G.; Nashat Alsheshtawi, K.; Abdelrahim, M.E.A. Antibiotic sensitivity/resistance pattern of hospital acquired blood stream infection in children cancer patients: A retrospective study. Int. J. Clin. Pract. 2021, 75, e14617. [Google Scholar] [CrossRef]
- Stoll, B.J.; Puopolo, K.M.; Hansen, N.I.; Sánchez, P.J.; Bell, E.F.; Carlo, W.A.; Cotten, C.M.; D’Angio, C.T.; Kazzi, S.N.J.; Poindexter, B.B.; et al. Early-Onset Neonatal Sepsis 2015 to 2017, the Rise of Escherichia coli, and the Need for Novel Prevention Strategies. JAMA Pediatr. 2020, 174, e200593. [Google Scholar] [CrossRef]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef] [PubMed]
- Saber, A.; Sebbar, N.K.; Sert, Y.; Alzaqri, N.; Hökelek, T.; El Ghayati, L.; Talbaoui, A.; Mague, J.T.; Baba, Y.F.; Urrutigoîty, M.; et al. Syntheses of N-substituted benzimidazolone derivatives: DFT calculations, Hirshfeld surface analysis, molecular docking studies and antibacterial activities. J. Mol. Struct. 2020, 1200, 127174. [Google Scholar] [CrossRef]
- Bouzian, Y.; Karrouchi, K.; Sert, Y.; Lai, C.-H.; Mahi, L.; Ahabchane, N.H.; Talbaoui, A.; Mague, J.T.; Essassi, E.M. Synthesis, spectroscopic characterization, crystal structure, DFT, molecular docking and in vitro antibacterial potential of novel quinoline derivatives. J. Mol. Struct. 2020, 1209, 127940. [Google Scholar] [CrossRef]
- Skjøt-Rasmussen, L.; Olsen, S.S.; Jensen, U.S.; Hammerum, A.M. Increasing consumption of antimicrobial agents in Denmark parallels increasing resistance in Escherichia coli bloodstream isolates. Int. J. Antimicrob. Agents 2012, 40, 86–88. [Google Scholar] [CrossRef]
- Tamma, P.D.; Goodman, K.E.; Harris, A.D.; Tekle, T.; Roberts, A.; Taiwo, A.; Simner, P.J. Comparing the Outcomes of Patients With Carbapenemase-Producing and Non-Carbapenemase-Producing Carbapenem-Resistant Enterobacteriaceae Bacteremia. Clin. Infect. Dis. 2017, 64, 257–264. [Google Scholar] [CrossRef]
- Pitout, J.D. Extraintestinal Pathogenic Escherichia coli: A Combination of Virulence with Antibiotic Resistance. Front. Microbiol. 2012, 3, 9. [Google Scholar] [CrossRef]
- Peirano, G.; Pitout, J.D.D. Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae: Update on Molecular Epidemiology and Treatment Options. Drugs 2019, 79, 1529–1541. [Google Scholar] [CrossRef]
- Naas, T.; Oueslati, S.; Bonnin, R.A.; Dabos, M.L.; Zavala, A.; Dortet, L.; Retailleau, P.; Iorga, B.I. Beta-lactamase database (BLDB)—Structure and function. J. Enzym. Inhib. Med. Chem. 2017, 32, 917–919. [Google Scholar] [CrossRef]
- Wang, S.; Zhao, S.Y.; Xiao, S.Z.; Gu, F.F.; Liu, Q.Z.; Tang, J.; Guo, X.K.; Ni, Y.X.; Han, L.Z. Antimicrobial Resistance and Molecular Epidemiology of Escherichia coli Causing Bloodstream Infections in Three Hospitals in Shanghai, China. PLoS ONE 2016, 11, e0147740. [Google Scholar] [CrossRef]
- Devanga Ragupathi, N.K.; Veeraraghavan, B.; Muthuirulandi Sethuvel, D.P.; Anandan, S.; Vasudevan, K.; Neeravi, A.R.; Daniel, J.L.K.; Sathyendra, S.; Iyadurai, R.; Mutreja, A. First Indian report on genome-wide comparison of multidrug-resistant Escherichia coli from blood stream infections. PLoS ONE 2020, 15, e0220428. [Google Scholar] [CrossRef]
- Harris, P.N.A.; Ben Zakour, N.L.; Roberts, L.W.; Wailan, A.M.; Zowawi, H.M.; Tambyah, P.A.; Lye, D.C.; Jureen, R.; Lee, T.H.; Yin, M.; et al. Whole genome analysis of cephalosporin-resistant Escherichia coli from bloodstream infections in Australia, New Zealand and Singapore: High prevalence of CMY-2 producers and ST131 carrying blaCTX-M-15 and blaCTX-M-27. J. Antimicrob. Chemother. 2018, 73, 634–642. [Google Scholar] [CrossRef] [PubMed]
- Nicolas-Chanoine, M.H.; Bertrand, X.; Madec, J.Y. Escherichia coli ST131, an intriguing clonal group. Clin. Microbiol. Rev. 2014, 27, 543–574. [Google Scholar] [CrossRef] [PubMed]
- Roer, L.; Overballe-Petersen, S.; Hansen, F.; Schønning, K.; Wang, M.; Røder, B.L.; Hansen, D.S.; Justesen, U.S.; Andersen, L.P.; Fulgsang-Damgaard, D.; et al. Escherichia coli Sequence Type 410 Is Causing New International High-Risk Clones. mSphere 2018, 3, e00337-18. [Google Scholar] [CrossRef] [PubMed]
- Dahbi, G.; Mora, A.; Mamani, R.; López, C.; Alonso, M.P.; Marzoa, J.; Blanco, M.; Herrera, A.; Viso, S.; García-Garrote, F.; et al. Molecular epidemiology and virulence of Escherichia coli O16:H5-ST131: Comparison with H30 and H30-Rx subclones of O25b:H4-ST131. Int. J. Med. Microbiol. IJMM 2014, 304, 1247–1257. [Google Scholar] [CrossRef] [PubMed]
- Falgenhauer, L.; Imirzalioglu, C.; Ghosh, H.; Gwozdzinski, K.; Schmiedel, J.; Gentil, K.; Bauerfeind, R.; Kämpfer, P.; Seifert, H.; Michael, G.B.; et al. Circulation of clonal populations of fluoroquinolone-resistant CTX-M-15-producing Escherichia coli ST410 in humans and animals in Germany. Int. J. Antimicrob. Agents 2016, 47, 457–465. [Google Scholar] [CrossRef]
- Rodríguez-Baño, J.; Gutiérrez-Gutiérrez, B.; Machuca, I.; Pascual, A. Treatment of Infections Caused by Extended-Spectrum-Beta-Lactamase-, AmpC-, and Carbapenemase-Producing Enterobacteriaceae. Clin. Microbiol. Rev. 2018, 31, e00079-17. [Google Scholar] [CrossRef] [PubMed]
- Bonomo, R.A.; Burd, E.M.; Conly, J.; Limbago, B.M.; Poirel, L.; Segre, J.A.; Westblade, L.F. Carbapenemase-Producing Organisms: A Global Scourge. Clin. Infect. Dis. 2018, 66, 1290–1297. [Google Scholar] [CrossRef]
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef]
- Dortet, L.; Poirel, L.; Nordmann, P. Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. BioMed Res. Int. 2014, 2014, 249856. [Google Scholar] [CrossRef]
- Cui, J.; Li, F.; Shi, Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef]
- Sekar, R.; Mythreyee, M.; Srivani, S.; Sivakumaran, D.; Lallitha, S.; Saranya, S. Carbapenem-resistant Enterobacteriaceae in Pediatric Bloodstream Infections in Rural Southern India. Indian Pediatr. 2017, 54, 1021–1024. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Sun, L.; Zhu, Y.; Shen, M.; Tu, Y. Draft genome sequence of Escherichia coli ST977: A clinical multidrug-resistant strain harbouring bla(NDM-3) isolated from a bloodstream infection. J. Glob. Antimicrob. Resist. 2018, 13, 121–122. [Google Scholar] [CrossRef]
- Löfmark, S.; Sjöström, K.; Mäkitalo, B.; Edquist, P.; Tegmark Wisell, K.; Giske, C.G. Carbapenemase-producing Enterobacteriaceae in Sweden 2007-2013: Experiences from seven years of systematic surveillance and mandatory reporting. Drug Resist. Updates Rev. Comment. Antimicrob. Anticancer. Chemother. 2015, 20, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; González Molina, M.K.; Carmona Cartaya, Y.; Hart Casares, M.; Aung, M.S.; Kobayashi, N.; Quiñones Pérez, D. Multicenter Study of Carbapenemase-Producing Enterobacterales in Havana, Cuba, 2016-2021. Antibiotics 2022, 11, 514. [Google Scholar] [CrossRef]
- Villegas, M.V.; Pallares, C.J.; Escandón-Vargas, K.; Hernández-Gómez, C.; Correa, A.; Álvarez, C.; Rosso, F.; Matta, L.; Luna, C.; Zurita, J.; et al. Characterization and Clinical Impact of Bloodstream Infection Caused by Carbapenemase-Producing Enterobacteriaceae in Seven Latin American Countries. PLoS ONE 2016, 11, e0154092. [Google Scholar] [CrossRef]
- Lutgring, J.D.; Balbuena, R.; Reese, N.; Gilbert, S.E.; Ansari, U.; Bhatnagar, A.; Boyd, S.; Campbell, D.; Cochran, J.; Haynie, J.; et al. Antibiotic Susceptibility of NDM-Producing Enterobacterales Collected in the United States in 2017 and 2018. Antimicrob. Agents Chemother. 2020, 64, e00499-20. [Google Scholar] [CrossRef] [PubMed]
- Hornsey, M.; Phee, L.; Wareham, D.W. A novel variant, NDM-5, of the New Delhi metallo-β-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob. Agents Chemother. 2011, 55, 5952–5954. [Google Scholar] [CrossRef]
- Ahmad, N.; Ali, S.M.; Khan, A.U. Detection of New Delhi Metallo-β-Lactamase Variants NDM-4, NDM-5, and NDM-7 in Enterobacter aerogenes Isolated from a Neonatal Intensive Care Unit of a North India Hospital: A First Report. Microb. Drug Resist. 2018, 24, 161–165. [Google Scholar] [CrossRef]
- Cho, S.Y.; Huh, H.J.; Baek, J.Y.; Chung, N.Y.; Ryu, J.G.; Ki, C.S.; Chung, D.R.; Lee, N.Y.; Song, J.H. Klebsiella pneumoniae co-producing NDM-5 and OXA-181 carbapenemases, South Korea. Emerg. Infect. Dis. 2015, 21, 1088–1089. [Google Scholar] [CrossRef]
- Wailan, A.M.; Paterson, D.L.; Caffery, M.; Sowden, D.; Sidjabat, H.E. Draft Genome Sequence of NDM-5-Producing Escherichia coli Sequence Type 648 and Genetic Context of blaNDM-5 in Australia. Genome Announc. 2015, 3. [Google Scholar] [CrossRef]
- Rojas, L.J.; Hujer, A.M.; Rudin, S.D.; Wright, M.S.; Domitrovic, T.N.; Marshall, S.H.; Hujer, K.M.; Richter, S.S.; Cober, E.; Perez, F.; et al. NDM-5 and OXA-181 Beta-Lactamases, a Significant Threat Continues To Spread in the Americas. Antimicrob. Agents Chemother. 2017, 61, e00454-17. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, N.M.; Zakaria, A.S.; Edward, E.A. Genomic Characterization of International High-Risk Clone ST410 Escherichia coli Co-Harboring ESBL-Encoding Genes and bla(NDM-5) on IncFIA/IncFIB/IncFII/IncQ1 Multireplicon Plasmid and Carrying a Chromosome-Borne bla(CMY-2) from Egypt. Antibiotics 2022, 11, 1031. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Xia, W.; Liu, G.; Huang, X.; Tang, C.; Liu, C.; Xu, Y.; Ni, F.; Mei, Y.; Pan, S. Characterization Of bla (NDM-5)-Positive Escherichia coli Prevalent In A University Hospital In Eastern China. Infect. Drug Resist. 2019, 12, 3029–3038. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, X.; Xie, Y.; Bi, D.; Sun, J.; Li, J.; Tai, C.; Deng, Z.; Ou, H.Y. ICEberg 2.0: An updated database of bacterial integrative and conjugative elements. Nucleic Acids Res. 2019, 47, D660–D665. [Google Scholar] [CrossRef]
- Zou, D.; Huang, Y.; Zhao, X.; Liu, W.; Dong, D.; Li, H.; Wang, X.; Huang, S.; Wei, X.; Yan, X.; et al. A novel New Delhi metallo-β-lactamase variant, NDM-14, isolated in a Chinese Hospital possesses increased enzymatic activity against carbapenems. Antimicrob. Agents Chemother. 2015, 59, 2450–2453. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing. Approved Standard. CLSI Document M100. 2022. Available online: https://clsi.org/standards/products/microbiology/documents/m100/ (accessed on 1 November 2022).
- Taylor, D.E.; De Grandis, S.A.; Karmali, M.A.; Fleming, P.C. Transmissible plasmids from Campylobacter jejuni. Antimicrob. Agents Chemother. 1981, 19, 831–835. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- Kleinheinz, K.A.; Joensen, K.G.; Larsen, M.V. Applying the ResFinder and VirulenceFinder web-services for easy identification of acquired antibiotic resistance and E. coli virulence genes in bacteriophage and prophage nucleotide sequences. Bacteriophage 2014, 4, e27943. [Google Scholar] [CrossRef]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A language and environment for statistical computing. 2014, 1, 201. [Google Scholar]
- Bessonov, K.; Laing, C.; Robertson, J.; Yong, I.; Ziebell, K.; Gannon, V.P.J.; Nichani, A.; Arya, G.; Nash, J.H.E.; Christianson, S. ECTyper: In silico Escherichia coli serotype and species prediction from raw and assembled whole-genome sequence data. Microb. Genom. 2021, 7, 000728. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Snippy: Rapid Haploid Variant Calling and Core Genome Alignment. Available online: https://github.com/tseemann/snippy (accessed on 1 January 2022).
- Seemann, T.; Page, A.; Klotzl, F. Snp-Dists: Pairwise SNP Distance Matrix from A FASTA Sequence Alignment. 2018. Available online: https://github.com/tseemann/snp-dists (accessed on 1 January 2022).
- Treangen, T.J.; Ondov, B.D.; Koren, S.; Phillippy, A.M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014, 15, 524. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef]
- Kelly, A.M.; Mathema, B.; Larson, E.L. Carbapenem-resistant Enterobacteriaceae in the community: A scoping review. Int. J. Antimicrob. Agents 2017, 50, 127–134. [Google Scholar] [CrossRef]
- Lipworth, S.; Vihta, K.D.; Davies, T.; Wright, S.; Tabirao, M.; Chau, K.; Vaughan, A.; Kavanagh, J.; Barker, L.; George, S.; et al. Molecular epidemiology and antimicrobial resistance phenotype of paediatric bloodstream infections caused by Gram-negative bacteria. Commun. Med. 2022, 2, 101. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhao, F.; Chen, J. Analysis of drug resistance genes of integrons in clinical isolates of Escherichia coli from elderly bloodstream infections. Cell. Mol. Biol. (Noisy-Le-Grand Fr.) 2022, 68, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Foglia, F.; Della Rocca, M.T.; Melardo, C.; Nastri, B.M.; Manfredini, M.; Montella, F.; De Filippis, A.; Finamore, E.; Galdiero, M. Bloodstream infections and antibiotic resistance patterns: A six-year surveillance study from southern Italy. Pathog. Glob. Health 2022, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cuzon, G.; Bonnin, R.A.; Nordmann, P. First identification of novel NDM carbapenemase, NDM-7, in Escherichia coli in France. PLoS ONE 2013, 8, e61322. [Google Scholar] [CrossRef]
- Yu, H.; Ma, D.; Liu, B.; Yang, S.; Lin, Q.; Yu, R.; Jia, X.; Niu, S.; Zhang, Q.; Huang, S. Differences in the Distribution of Species, Carbapenemases, Sequence Types, Antimicrobial Heteroresistance and Mortality Rates Between Pediatric and Adult Carbapenemase-Producing Enterobacterales in Bloodstream Infections. Front. Med. 2022, 9, 827474. [Google Scholar] [CrossRef]
- Dortet, L.; Cuzon, G.; Nordmann, P. Dissemination of carbapenemase-producing Enterobacteriaceae in France, 2012. J. Antimicrob. Chemother. 2014, 69, 623–627. [Google Scholar] [CrossRef]
- Zheng, W.; Yue, M.; Zhang, J.; Ruan, Z. Coexistence of two bla(CTX-M-14) genes in a bla(NDM-5)-carrying multidrug-resistant Escherichia coli strain recovered from a bloodstream infection in China. J. Glob. Antimicrob. Resist. 2021, 26, 11–14. [Google Scholar] [CrossRef]
- Huang, J.; Ma, S.; Yu, Q.; Fu, M.; Shao, L.; Shan, X.; Li, X. Whole genome sequence of an Escherichia coli ST410 isolate co-harbouring bla(NDM-5), bla(OXA-1), bla(CTX-M-15), bla(CMY-2), aac(3)-IIa and aac(6′)-Ib-cr genes isolated from a patient with bloodstream infection in China. J. Glob. Antimicrob. Resist. 2019, 19, 354–355. [Google Scholar] [CrossRef]
- Bi, R.; Kong, Z.; Qian, H.; Jiang, F.; Kang, H.; Gu, B.; Ma, P. High Prevalence of bla (NDM) Variants Among Carbapenem-Resistant Escherichia coli in Northern Jiangsu Province, China. Front Microbiol. 2018, 9, 2704. [Google Scholar] [CrossRef]
- Park, Y.; Choi, Q.; Kwon, G.C.; Koo, S.H. Emergence and transmission of New Delhi metallo-beta-lactamase-5-producing Escherichia coli Sequence Type 361 in a Tertiary Hospital in South Korea. J. Clin. Lab. Anal. 2020, 34, e23041. [Google Scholar] [CrossRef]
- Tang, B.; Chang, J.; Cao, L.; Luo, Q.; Xu, H.; Lyu, W.; Qian, M.; Ji, X.; Zhang, Q.; Xia, X.; et al. Characterization of an NDM-5 carbapenemase-producing Escherichia coli ST156 isolate from a poultry farm in Zhejiang, China. BMC Microbiol. 2019, 19, 82. [Google Scholar] [CrossRef] [PubMed]
- Bonnin, R.A.; Poirel, L.; Carattoli, A.; Nordmann, P. Characterization of an IncFII plasmid encoding NDM-1 from Escherichia coli ST131. PLoS ONE 2012, 7, e34752. [Google Scholar] [CrossRef] [PubMed]
- Paskova, V.; Medvecky, M.; Skalova, A.; Chudejova, K.; Bitar, I.; Jakubu, V.; Bergerova, T.; Zemlickova, H.; Papagiannitsis, C.C.; Hrabak, J. Characterization of NDM-Encoding Plasmids From Enterobacteriaceae Recovered From Czech Hospitals. Front. Microbiol. 2018, 9, 1549. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, H.; Li, Y.; Hou, Y.; Hao, C. Drug susceptibility and molecular epidemiology of Escherichia coli in bloodstream infections in Shanxi, China. PeerJ 2021, 9, e12371. [Google Scholar] [CrossRef] [PubMed]
- Quan, J.; Zhao, D.; Liu, L.; Chen, Y.; Zhou, J.; Jiang, Y.; Du, X.; Zhou, Z.; Akova, M.; Yu, Y. High prevalence of ESBL-producing Escherichia coli and Klebsiella pneumoniae in community-onset bloodstream infections in China. J. Antimicrob. Chemother. 2017, 72, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Nicolas-Chanoine, M.H.; Blanco, J.; Leflon-Guibout, V.; Demarty, R.; Alonso, M.P.; Caniça, M.M.; Park, Y.J.; Lavigne, J.P.; Pitout, J.; Johnson, J.R. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 2008, 61, 273–281. [Google Scholar] [CrossRef]
- Falgenhauer, L.; Waezsada, S.E.; Gwozdzinski, K.; Ghosh, H.; Doijad, S.; Bunk, B.; Spröer, C.; Imirzalioglu, C.; Seifert, H.; Irrgang, A.; et al. Chromosomal Locations of mcr-1 and bla CTX-M-15 in Fluoroquinolone-Resistant Escherichia coli ST410. Emerg. Infect. Dis. 2016, 22, 1689–1691. [Google Scholar] [CrossRef]
- Gu, J.N.; Chen, L.; Weng, X.B.; Yang, X.Y.; Pan, D.M. Clinical and Microbiological Characteristics of a Community-Acquired Carbapenem-Resistant Escherichia coli ST410 Isolate Harbouring blaNDM-5-Encoding IncX3-Type Plasmid From Blood. Front. Med. 2021, 8, 658058. [Google Scholar] [CrossRef]
- Ma, Z.; Zeng, Z.; Liu, J.; Liu, C.; Pan, Y.; Zhang, Y.; Li, Y. Emergence of IncHI2 Plasmid-Harboring blaNDM-5 from Porcine Escherichia coli Isolates in Guangdong, China. Pathogens 2021, 10, 954. [Google Scholar] [CrossRef]
- Xu, L.; Wang, P.; Cheng, J.; Qin, S.; Xie, W. Characterization of a novel bla (NDM-5)-harboring IncFII plasmid and an mcr-1-bearing IncI2 plasmid in a single Escherichia coli ST167 clinical isolate. Infect. Drug Resist. 2019, 12, 511–519. [Google Scholar] [CrossRef]
- Villa, L.; García-Fernández, A.; Fortini, D.; Carattoli, A. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 2010, 65, 2518–2529. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Xiao, X.; Li, Y.; Liu, Y.; Li, R.; Wang, Z. Emergence of IncX3 Plasmid-Harboring bla (NDM-) (5) Dominated by Escherichia coli ST48 in a Goose Farm in Jiangsu, China. Front. Microbiol. 2019, 10, 2002. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Xie, L.; Wang, X.; Han, L.; Guo, X.; Ni, Y.; Qu, H.; Sun, J. Further Spread of bla NDM-5 in Enterobacteriaceae via IncX3 Plasmids in Shanghai, China. Front. Microbiol. 2016, 7, 424. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Fu, Y.; Shen, M.; Huang, D.; Du, X.; Hu, Q.; Zhou, Y.; Wang, D.; Yu, Y. Dissemination of bla(NDM-5) gene via an IncX3-type plasmid among non-clonal Escherichia coli in China. Antimicrob. Resist. Infect. Control. 2018, 7, 59. [Google Scholar] [CrossRef] [PubMed]
| Strain | Age * | Gender | Underlying Diseases | Clinical Department | Antibiotic Treatment | Outcome | Year of Isolation |
|---|---|---|---|---|---|---|---|
| E1 | 3 Y | male | acute myeloid leukemia | hematology | meropenem + amikacin | recovery | 2019 |
| E2 | 13 Y | male | acute lymphoblastic leukemia, post allo-HSCT | hematology | meropenem + amikacin, tigecycline | automatically discharged | 2017 |
| E3 | 1 M | female | neonatal sepsis, premature infant | neonatology | meropenem | recovery | 2020 |
| E4 | 9 D | female | necrotizing enterocolitis, neonatal sepsis | neonatology | meropenem | recovery | 2020 |
| E5 | 4 M | male | congenital atresia of biliary tract | gastroenterology | imipenem | recovery | 2016 |
| E6 | 11 Y | male | T lymphoblastic leukemia/lymphoma | hematology | tigecycline + amikacin | recovery | 2020 |
| E7 | 3 Y | female | acute lymphoblastic leukemia | hematology | meropenem + amikacin | recovery | 2017 |
| E8 | 12 D | male | neonatal sepsis, premature infant | neonatology | meropenem | recovery | 2017 |
| Strains | Description | MLST | Serotype a | Minimum Inhibitory Concentration (μg/mL) b | Conjugation Efficiency c | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | SAM | ATM | SXT | CIP | TZP | GEN | FEP | CRO | CAZ | CTT | TOB | IPM | LVX | |||||
| E1 | donor | ST38 | O7:H8 | ≥32 | ≥32/16 | 2 | ≥16/304 | 1 | 64/4 | ≤1 | ≥64 | ≥64 | ≥64 | ≥64 | ≤1 | ≥16 | 1 | NA |
| E2 | donor | ST58 | O?:H37 | ≥32 | ≥32/16 | ≤1 | ≥16/304 | 2 | 64/4 | ≥16 | 16 | ≥64 | ≥64 | ≥64 | ≥16 | ≥16 | 1 | NA |
| E3 | donor | ST410 | O?:H9 | ≥32 | ≥32/16 | ≥64 | ≥16/304 | ≥4 | ≥128/4 | ≤1 | ≥64 | ≥64 | ≥64 | ≥64 | ≥16 | 4 | ≥8 | NA |
| E4 | donor | ST410 | O?:H9 | ≥32 | ≥32/16 | ≥64 | ≥16/304 | ≥4 | ≥128/4 | ≤1 | ≥64 | ≥64 | ≥64 | ≥64 | ≥16 | 8 | ≥8 | NA |
| E5 | donor | ST410 | O?:H9 | ≥32 | ≥32/16 | 4 | ≥16/304 | 0.5 | ≥128/4 | ≥16 | ≥64 | ≥64 | ≥64 | ≥64 | 8 | ≥16 | 1 | NA |
| E6 | donor | ST131 | O25:H4 | ≥32 | ≥32/16 | ≥64 | ≥16/304 | ≥4 | ≥128/4 | ≥16 | ≥64 | ≥64 | ≥64 | ≥64 | ≥16 | ≥16 | ≥8 | NA |
| E7 | donor | ST361 | O9:H30 | ≥32 | ≥32/16 | 16 | ≥16/304 | ≥4 | ≥128/4 | ≥16 | ≥64 | ≥64 | ≥64 | ≥64 | ≥16 | ≥16 | ≥8 | NA |
| E8 | donor | ST156 | O11:H25 | ≥32 | ≥32/16 | 16 | ≥16/304 | 2 | ≥128/4 | ≤1 | ≥64 | ≥64 | ≥64 | ≥64 | ≤1 | ≥16 | 2 | NA |
| E1-TC | transconjugant | ST80 | O75:H7 | ≥32 | ≥32/16 | ≤1 | ≤1/19 | ≤0.25 | 64/4 | ≥16 | 16 | ≥64 | ≥64 | ≥64 | ≤1 | ≥16 | 0.5 | 2.5 × 10−3 |
| E6-TC | transconjugant | ST80 | O75:H7 | ≥32 | ≥32/16 | 16 | ≥16/304 | 1 | ≥128/4 | ≥16 | ≥64 | ≥64 | ≥64 | ≥64 | ≥16 | 16 | 1 | 2.9 × 10−6 |
| E7-TC | transconjugant | ST80 | O75:H7 | ≥32 | ≥32/16 | 16 | ≥16/304 | ≤0.25 | ≥128/4 | ≥16 | ≥64 | ≥64 | ≥64 | ≥64 | ≥16 | ≥16 | 0.5 | 3.5 × 10−6 |
| E8-TC | transconjugant | ST80 | O75:H7 | ≥32 | ≥32/16 | ≤1 | ≤1/19 | ≤0.25 | 64/4 | ≤1 | 16 | ≥64 | ≥64 | ≥64 | ≤1 | ≥16 | 0.5 | 1.6 × 10−6 |
| EC600 | recipient | ST80 | O75:H7 | 16 | 8/4 | ≤1 | ≤1/19 | ≤0.25 | ≤4/4 | ≤1 | ≤1 | ≤1 | ≤1 | ≤4 | ≤1 | ≤1 | 0.5 | NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, L.; Hu, H.; Xu, C.; Zhou, M.; Li, Y.; Li, Y.; Wu, S.; Dong, N. Characterization of NDM-5-Producing Escherichia coli Strains Isolated from Pediatric Patients with Bloodstream Infections in a Chinese Hospital. Genes 2023, 14, 520. https://doi.org/10.3390/genes14020520
Huang L, Hu H, Xu C, Zhou M, Li Y, Li Y, Wu S, Dong N. Characterization of NDM-5-Producing Escherichia coli Strains Isolated from Pediatric Patients with Bloodstream Infections in a Chinese Hospital. Genes. 2023; 14(2):520. https://doi.org/10.3390/genes14020520
Chicago/Turabian StyleHuang, Lili, Hongye Hu, Chen Xu, Mi Zhou, Yuanyuan Li, Yunbing Li, Shuyan Wu, and Ning Dong. 2023. "Characterization of NDM-5-Producing Escherichia coli Strains Isolated from Pediatric Patients with Bloodstream Infections in a Chinese Hospital" Genes 14, no. 2: 520. https://doi.org/10.3390/genes14020520
APA StyleHuang, L., Hu, H., Xu, C., Zhou, M., Li, Y., Li, Y., Wu, S., & Dong, N. (2023). Characterization of NDM-5-Producing Escherichia coli Strains Isolated from Pediatric Patients with Bloodstream Infections in a Chinese Hospital. Genes, 14(2), 520. https://doi.org/10.3390/genes14020520







