Independent Associated SNPs at SORCS3 and Its Protein Interactors for Multiple Brain-Related Disorders and Traits
Abstract
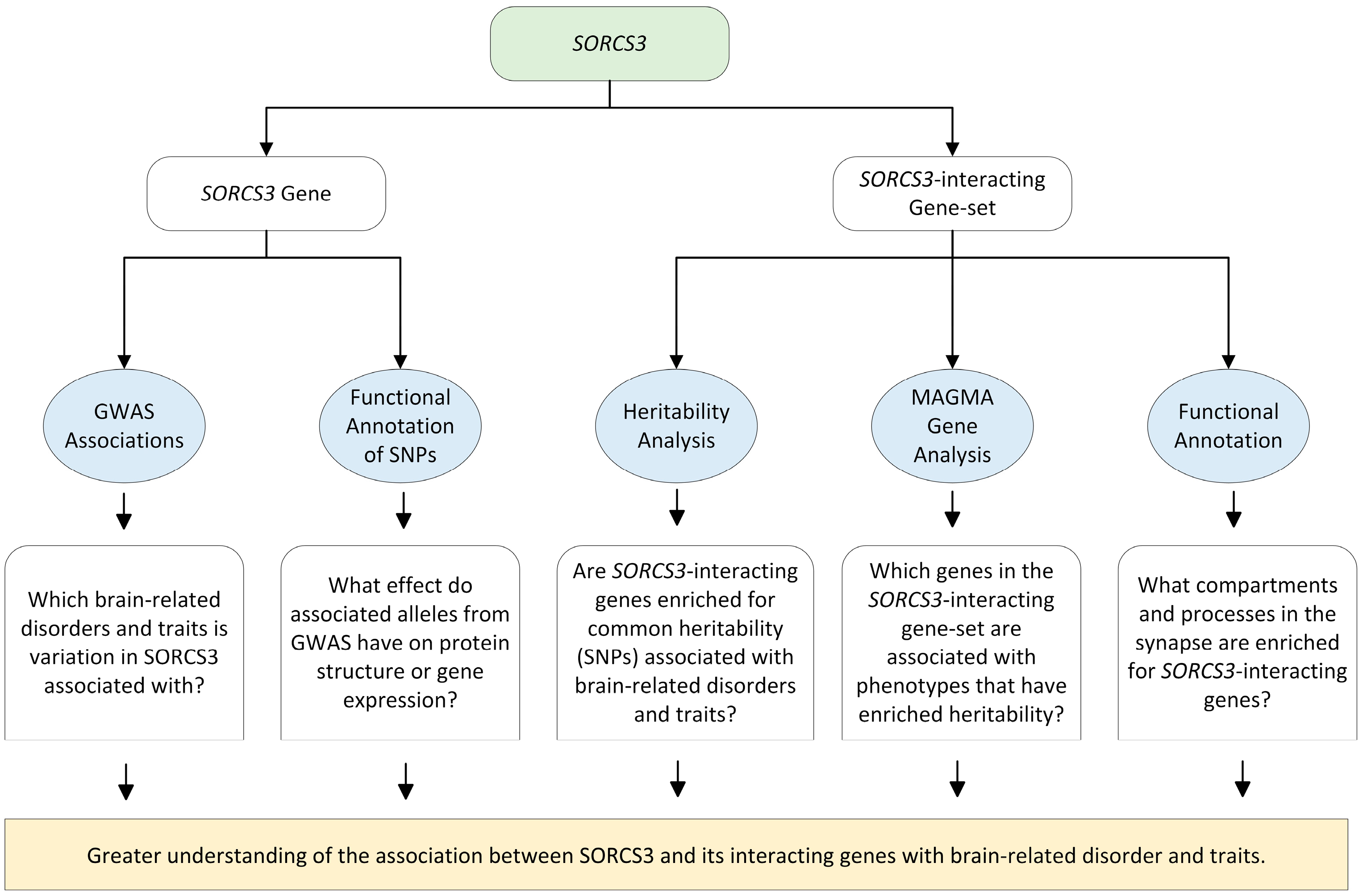
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Sourcing of GWAS Results, LD Information and eQTL Data
2.3. GWAS Data
2.4. SORCS3 Gene-Set
2.5. Stratified Linkage Disequilibrium Score Regression (sLDSC) Analysis
2.6. MAGMA Gene-Set Analysis
2.7. Functional Annotation
3. Results
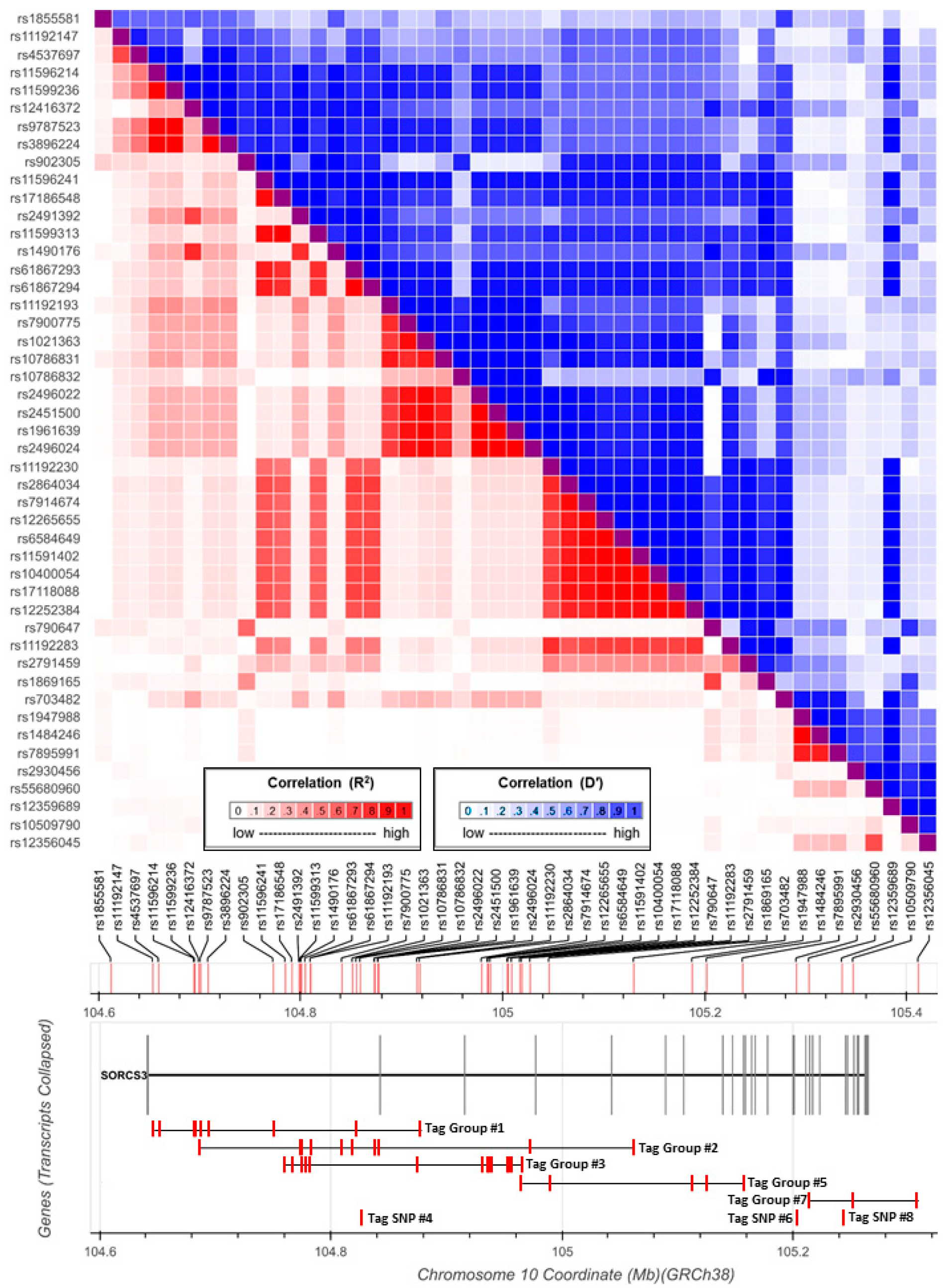
3.1. Association Signals at SORCS3 in GWAS Data
3.2. Functional Annotation of Associated SNPS at SORCS3
3.3. Development of SORCS3 Protein Interaction Gene-Set
3.4. Heritability Analysis of SORCS3 Gene-Set
3.5. Individual Gene Analysis Using MAGMA
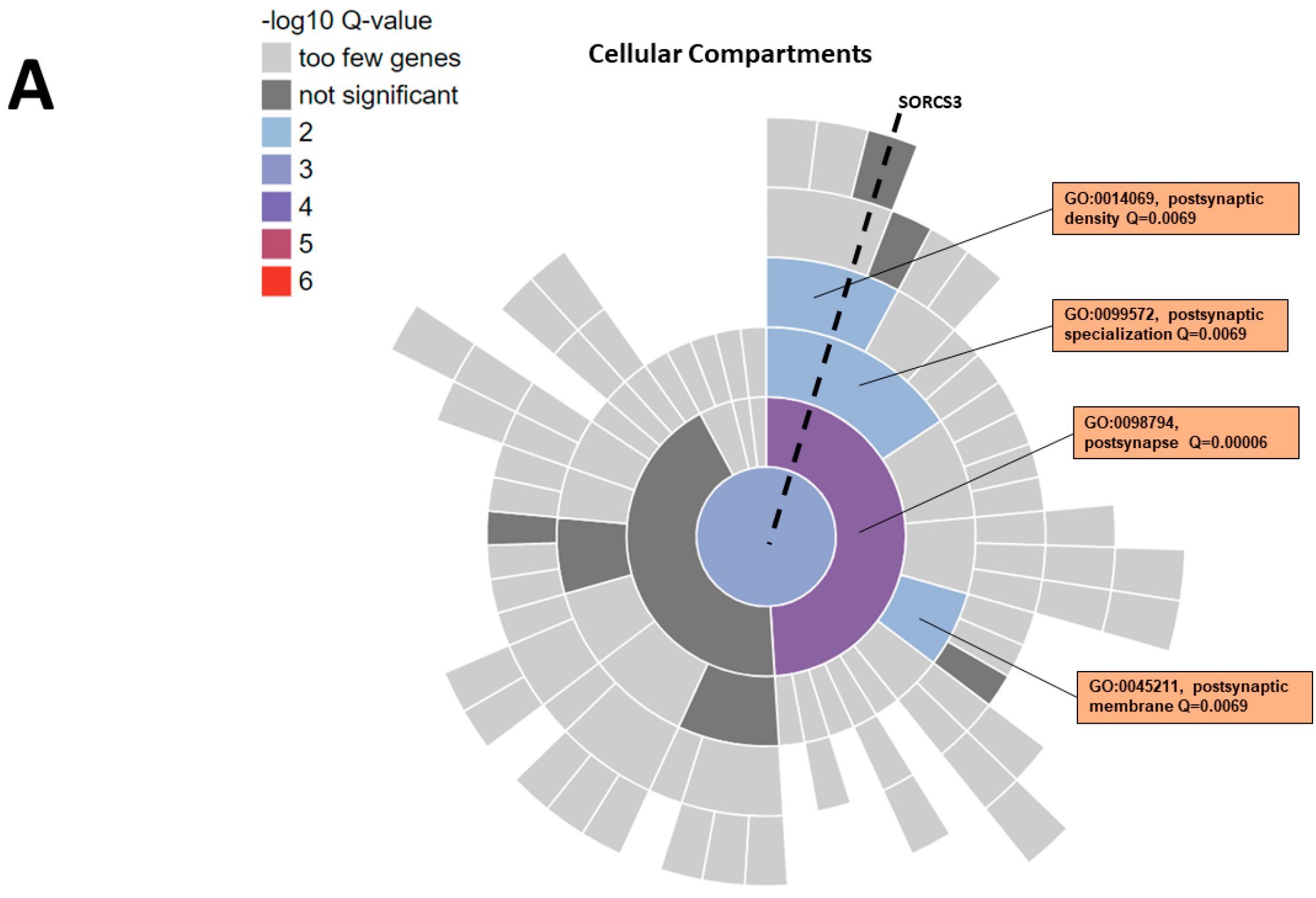
3.6. Functional Annotation of SORCS3 Gene-Set
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Malik, A.R.; Willnow, T.E. VPS10P Domain Receptors: Sorting Out Brain Health and Disease. Trends Neurosci. 2020, 43, 870–885. [Google Scholar] [CrossRef] [PubMed]
- Reitz, C.; Tosto, G.; Vardarajan, B.; Rogaeva, E.; Ghani, M.; Rogers, R.S.; Conrad, C.; Haines, J.L.; Pericak-Vance, M.A.; Fallin, M.D.; et al. Independent and epistatic effects of variants in VPS10-d receptors on Alzheimer disease risk and processing of the amyloid precursor protein (APP). Transl. Psychiatry 2013, 3, e256. [Google Scholar] [CrossRef] [PubMed]
- Marcusson, E.G.; Horazdovsky, B.F.; Cereghino, J.L.; Gharakhanian, E.; Emr, S.D. The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10 gene. Cell 1994, 77, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Hermey, G. The Vps10p-domain receptor family. Cell. Mol. Life Sci. 2009, 66, 2677–2689. [Google Scholar] [CrossRef]
- Schmidt, V.; Willnow, T.E. Protein sorting gone wrong–VPS10P domain receptors in cardiovascular and metabolic diseases. Atherosclerosis 2016, 245, 194–199. [Google Scholar] [CrossRef]
- Willnow, T.E.; Petersen, C.M.; Nykjaer, A. VPS10P-domain receptors—Regulators of neuronal viability and function. Nat. Rev. Neurosci. 2008, 9, 899–909. [Google Scholar] [CrossRef]
- Nielsen, M.S.; Madsen, P.; Christensen, E.I.; Nykjær, A.; Gliemann, J.; Kasper, D.; Pohlmann, R.; Petersen, C.M. The sortilin cytoplasmic tail conveys Golgi–endosome transport and binds the VHS domain of the GGA2 sorting protein. EMBO J. 2001, 20, 2180–2190. [Google Scholar] [CrossRef] [PubMed]
- Subkhangulova, A.; Malik, A.R.; Hermey, G.; Popp, O.; Dittmar, G.; Rathjen, T.; Poy, M.N.; Stumpf, A.; Beed, P.S.; Schmitz, D.; et al. SORCS1 and SORCS3 control energy balance and orexigenic peptide production. EMBO Rep. 2018, 19. [Google Scholar] [CrossRef]
- Christiansen, G.B.; Andersen, K.H.; Riis, S.; Nykjaer, A.; Bolcho, U.; Jensen, M.S.; Holm, M.M. The sorting receptor SorCS3 is a stronger regulator of glutamate receptor functions compared to GABAergic mechanisms in the hippocampus. Hippocampus 2017, 27, 235–248. [Google Scholar] [CrossRef]
- Westergaard, U.; Kirkegaard, K.; Sørensen, E.; Jacobsen, C.; Nielsen, M.; Petersen, C.; Madsen, P. SorCS3 does not require propeptide cleavage to bind nerve growth factor. FEBS Lett. 2005, 579, 1172–1176. [Google Scholar] [CrossRef]
- Hermey, G.; Sjøgaard, S.S.; Petersen, C.M.; Nykjær, A.; Gliemann, J. Tumour necrosis factor α-converting enzyme mediates ectodomain shedding of Vps10p-domain receptor family members. Biochem. J. 2006, 395, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Hermey, G.; Mahlke, C.; Gutzmann, J.J.; Schreiber, J.; Blüthgen, N.; Kuhl, D. Genome-wide profiling of the activity-dependent hippocampal transcriptome. PLoS ONE 2013, 8, e76903. [Google Scholar] [CrossRef] [PubMed]
- Oetjen, S.; Mahlke, C.; Hermans-Borgmeyer, I.; Hermey, G. Spatiotemporal expression analysis of the growth factor receptor SorCS3. J. Comp. Neurol. 2014, 522, 3386–3402. [Google Scholar] [CrossRef] [PubMed]
- Breiderhoff, T.; Christiansen, G.B.; Pallesen, L.T.; Vaegter, C.; Nykjaer, A.; Holm, M.M.; Glerup, S.; Willnow, T.E. Sortilin-related receptor SORCS3 is a postsynaptic modulator of synaptic depression and fear extinction. PLoS ONE 2013, 8, e75006. [Google Scholar] [CrossRef] [PubMed]
- Hampe, W.; Rezgaoui, M.; Hermans-Borgmeyer, I.; Schaller, C.H. The genes for the human VPS10 domain-containing receptors are large and contain many small exons. Hum. Genet. 2001, 108, 529–536. [Google Scholar] [CrossRef]
- Alfadhel, M.A.; Albahkali, S.; Almuaysib, A.; Alrfaei, B.M. The SORCS3 gene is mutated in brothers with infantile spasms and intellectual disability. Discov. Med. 2018, 26, 147–153. [Google Scholar] [PubMed]
- Binzer, S.; Stenager, E.; Binzer, M.; Kyvik, K.O.; Hillert, J.; Imrell, K. Genetic analysis of the isolated Faroe Islands reveals SORCS3 as a potential multiple sclerosis risk gene. Mult. Scler. J. 2016, 22, 733–740. [Google Scholar] [CrossRef] [PubMed]
- De Rubeis, S.; He, X.; Goldberg, A.P.; Poultney, C.S.; Samocha, K.; Cicek, A.E.; Kou, Y.; Liu, L.; Fromer, M.; Walker, S.; et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 2014, 515, 209–215. [Google Scholar] [CrossRef]
- Levey, D.F.; Stein, M.B.; Wendt, F.R.; Pathak, G.A.; Zhou, H.; Aslan, M.; Quaden, R.; Harrington, K.M.; Nuñez, Y.Z.; Overstreet, C.; et al. Bi-ancestral depression GWAS in the Million Veteran Program and meta-analysis in >1.2 million individuals highlight new therapeutic directions. Nat. Neurosci. 2021, 24, 954–963. [Google Scholar] [CrossRef]
- Bigdeli, T.B.; Fanous, A.H.; Li, Y.; Rajeevan, N.; Sayward, F.; Genovese, G.; Gupta, R.; Radhakrishnan, K.; Malhotra, A.K.; Sun, N.; et al. Genome-Wide Association Studies of Schizophrenia and Bipolar Disorder in a Diverse Cohort of US Veterans. Schizophr. Bull. 2021, 47, 517–529. [Google Scholar] [CrossRef]
- Ripke, S.; O’Dushlaine, C.; Chambert, K.; Moran, J.L.; Kähler, A.K.; Akterin, S.; Bergen, S.E.; Collins, A.L.; Crowley, J.J.; Fromer, M.; et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet. 2013, 45, 1150–1159. [Google Scholar] [CrossRef] [PubMed]
- Demontis, D.; Walters, G.B.; Athanasiadis, G.; Walters, R.; Therrien, K.; Nielsen, T.T.; Farajzadeh, L.; Voloudakis, G.; Bendl, J.; Zeng, B.; et al. Genome-wide analyses of ADHD identify 27 risk loci, refine the genetic architecture and implicate several cognitive domains. Nat. Genet. 2023. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Cao, H.; Baranova, A.; Huang, H.; Li, S.; Cai, L.; Rao, S.; Dai, M.; Xie, M.; Dou, Y.; et al. Multi-trait analysis for genome-wide association study of five psychiatric disorders. Transl. Psychiatry 2020, 10, 209. [Google Scholar] [CrossRef]
- Blue, E.E.; Thornton, T.A.; Kooperberg, C.; Liu, S.; Wactawski-Wende, J.; Manson, J.; Kuller, L.; Hayden, K.; Reiner, A.P. Non-coding variants in MYH11, FZD3, and SORCS3 are associated with dementia in women. Alzheimer’s Dement. J. Alzheimer’s Assoc. 2021, 17, 215–225. [Google Scholar] [CrossRef]
- Eszlari, N.; Millinghoffer, A.; Petschner, P.; Gonda, X.; Baksa, D.; Pulay, A.; Réthelyi, J.; Elliott, R.; Anderson, I.; Deakin, J. Genome-wide gene-based tests replicate the association of the SORCS3 gene with neuroticism. Eur. Neuropsychopharmacol. 2017, 27, S579–S580. [Google Scholar] [CrossRef]
- Sütöri, S.; Eszlari, N.; Baksa, D.; Petschner, P.; Gal, Z.; Gonda, X.; Bagdy, G.; Juhasz, G.P. 205 Lifetime depression is associated with SORCS3 gene polymorphisms: Replicating results from a large GWAS study in an independent population. Eur. Neuropsychopharmacol. 2019, 29, S157–S159. [Google Scholar] [CrossRef]
- Lee, J.J.; Wedow, R.; Okbay, A.; Kong, E.; Maghzian, O.; Zacher, M.; Nguyen-Viet, T.A.; Bowers, P.; Sidorenko, J.; Karlsson Linner, R.; et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet. 2018, 50, 1112–1121. [Google Scholar] [CrossRef]
- Savage, J.E.; Jansen, P.R.; Stringer, S.; Watanabe, K.; Bryois, J.; de Leeuw, C.A.; Nagel, M.; Awasthi, S.; Barr, P.B.; Coleman, J.R.I.; et al. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat. Genet. 2018, 50, 912–919. [Google Scholar] [CrossRef]
- Turley, P.; Walters, R.K.; Maghzian, O.; Okbay, A.; Lee, J.J.; Fontana, M.A.; Nguyen-Viet, T.A.; Wedow, R.; Zacher, M.; Furlotte, N.A.; et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat. Genet. 2018, 50, 229–237. [Google Scholar] [CrossRef]
- Buniello, A.; MacArthur, J.A.L.; Cerezo, M.; Harris, L.W.; Hayhurst, J.; Malangone, C.; McMahon, A.; Morales, J.; Mountjoy, E.; Sollis, E.; et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019, 47, D1005–D1012. [Google Scholar] [CrossRef]
- Machiela, M.J.; Chanock, S.J. LDlink: A web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 2015, 31, 3555–3557. [Google Scholar] [CrossRef] [PubMed]
- Ward, L.D.; Kellis, M. HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012, 40, D930–D934. [Google Scholar] [CrossRef] [PubMed]
- Consortium, G.T. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Trubetskoy, V.; Pardinas, A.F.; Qi, T.; Panagiotaropoulou, G.; Awasthi, S.; Bigdeli, T.B.; Bryois, J.; Chen, C.Y.; Dennison, C.A.; Hall, L.S.; et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 2022, 604, 502–508. [Google Scholar] [CrossRef]
- Mullins, N.; Forstner, A.J.; O’Connell, K.S.; Coombes, B.; Coleman, J.R.I.; Qiao, Z.; Als, T.D.; Bigdeli, T.B.; Borte, S.; Bryois, J.; et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat. Genet. 2021, 53, 817–829. [Google Scholar] [CrossRef] [PubMed]
- Howard, D.M.; Adams, M.J.; Clarke, T.K.; Hafferty, J.D.; Gibson, J.; Shirali, M.; Coleman, J.R.I.; Hagenaars, S.P.; Ward, J.; Wigmore, E.M.; et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci. 2019, 22, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Grove, J.; Ripke, S.; Als, T.D.; Mattheisen, M.; Walters, R.K.; Won, H.; Pallesen, J.; Agerbo, E.; Andreassen, O.A.; Anney, R.; et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 2019, 51, 431–444. [Google Scholar] [CrossRef] [PubMed]
- Warrier, V.; Kwong, A.S.F.; Luo, M.; Dalvie, S.; Croft, J.; Sallis, H.M.; Baldwin, J.; Munafo, M.R.; Nievergelt, C.M.; Grant, A.J.; et al. Gene-environment correlations and causal effects of childhood maltreatment on physical and mental health: A genetically informed approach. Lancet Psychiatry 2021, 8, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Nagel, M.; Watanabe, K.; Stringer, S.; Posthuma, D.; van der Sluis, S. Item-level analyses reveal genetic heterogeneity in neuroticism. Nat. Commun. 2018, 9, 905. [Google Scholar] [CrossRef] [PubMed]
- Karlsson Linner, R.; Biroli, P.; Kong, E.; Meddens, S.F.W.; Wedow, R.; Fontana, M.A.; Lebreton, M.; Tino, S.P.; Abdellaoui, A.; Hammerschlag, A.R.; et al. Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat. Genet. 2019, 51, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Baselmans, B.M.L.; Jansen, R.; Ip, H.F.; van Dongen, J.; Abdellaoui, A.; van de Weijer, M.P.; Bao, Y.; Smart, M.; Kumari, M.; Willemsen, G.; et al. Multivariate genome-wide analyses of the well-being spectrum. Nat. Genet. 2019, 51, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.; Tunbridge, E.M.; Sandor, C.; Lyall, L.M.; Ferguson, A.; Strawbridge, R.J.; Lyall, D.M.; Cullen, B.; Graham, N.; Johnston, K.J.A.; et al. The genomic basis of mood instability: Identification of 46 loci in 363,705 UK Biobank participants, genetic correlation with psychiatric disorders, and association with gene expression and function. Mol. Psychiatry 2020, 25, 3091–3099. [Google Scholar] [CrossRef] [PubMed]
- Jansen, I.E.; Savage, J.E.; Watanabe, K.; Bryois, J.; Williams, D.M.; Steinberg, S.; Sealock, J.; Karlsson, I.K.; Hagg, S.; Athanasiu, L.; et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 2019, 51, 404–413. [Google Scholar] [CrossRef] [PubMed]
- Malik, R.; Rannikmae, K.; Traylor, M.; Georgakis, M.K.; Sargurupremraj, M.; Markus, H.S.; Hopewell, J.C.; Debette, S.; Sudlow, C.L.M.; Dichgans, M.; et al. Genome-wide meta-analysis identifies 3 novel loci associated with stroke. Ann. Neurol. 2018, 84, 934–939. [Google Scholar] [CrossRef]
- Schunkert, H.; Konig, I.R.; Kathiresan, S.; Reilly, M.P.; Assimes, T.L.; Holm, H.; Preuss, M.; Stewart, A.F.; Barbalic, M.; Gieger, C.; et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 2011, 43, 333–338. [Google Scholar] [CrossRef]
- Liu, J.Z.; van Sommeren, S.; Huang, H.; Ng, S.C.; Alberts, R.; Takahashi, A.; Ripke, S.; Lee, J.C.; Jostins, L.; Shah, T.; et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 2015, 47, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, A.; Taliun, D.; Thurner, M.; Robertson, N.R.; Torres, J.M.; Rayner, N.W.; Payne, A.J.; Steinthorsdottir, V.; Scott, R.A.; Grarup, N.; et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 2018, 50, 1505–1513. [Google Scholar] [CrossRef]
- Bulik-Sullivan, B.K.; Loh, P.R.; Finucane, H.K.; Ripke, S.; Yang, J.; Schizophrenia Working Group of the Psychiatric Genomics Consortium; Patterson, N.; Daly, M.J.; Price, A.L.; Neale, B.M. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015, 47, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Finucane, H.K.; Bulik-Sullivan, B.; Gusev, A.; Trynka, G.; Reshef, Y.; Loh, P.R.; Anttila, V.; Xu, H.; Zang, C.; Farh, K.; et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 2015, 47, 1228–1235. [Google Scholar] [CrossRef]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef]
- Finucane, H.K.; Reshef, Y.A.; Anttila, V.; Slowikowski, K.; Gusev, A.; Byrnes, A.; Gazal, S.; Loh, P.R.; Lareau, C.; Shoresh, N.; et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet. 2018, 50, 621–629. [Google Scholar] [CrossRef] [PubMed]
- de Leeuw, C.A.; Mooij, J.M.; Heskes, T.; Posthuma, D. MAGMA: Generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 2015, 11, e1004219. [Google Scholar] [CrossRef] [PubMed]
- Herwig, R.; Hardt, C.; Lienhard, M.; Kamburov, A. Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat. Protoc. 2016, 11, 1889–1907. [Google Scholar] [CrossRef] [PubMed]
- Kamran, M.; Bibi, F.; Ur Rehman, A.; Morris, D.W. Major Depressive Disorder: Existing Hypotheses about Pathophysiological Mechanisms and New Genetic Findings. Genes 2022, 13, 646. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Suzuki, H.; Maegawa, S.; Endo, H.; Sugano, S.; Hashimoto, K.; Yasuda, K.; Inoue, K. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 2003, 22, 905–912. [Google Scholar] [CrossRef] [PubMed]
- Underwood, J.G.; Boutz, P.L.; Dougherty, J.D.; Stoilov, P.; Black, D.L. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol. Cell. Biol. 2005, 25, 10005–10016. [Google Scholar] [CrossRef]
- O’Leary, A.; Fernàndez-Castillo, N.; Gan, G.; Yang, Y.; Yotova, A.Y.; Kranz, T.M.; Grünewald, L.; Freudenberg, F.; Antón-Galindo, E.; Cabana-Domínguez, J.; et al. Behavioural and functional evidence revealing the role of RBFOX1 variation in multiple psychiatric disorders and traits. Mol. Psychiatry 2022, 27, 4464–4473. [Google Scholar] [CrossRef]
- Weyn-Vanhentenryck, S.M.; Mele, A.; Yan, Q.; Sun, S.; Farny, N.; Zhang, Z.; Xue, C.; Herre, M.; Silver, P.A.; Zhang, M.Q.; et al. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep. 2014, 6, 1139–1152. [Google Scholar] [CrossRef]
- Vuong, C.K.; Wei, W.; Lee, J.A.; Lin, C.H.; Damianov, A.; de la Torre-Ubieta, L.; Halabi, R.; Otis, K.O.; Martin, K.C.; O’Dell, T.J.; et al. Rbfox1 Regulates Synaptic Transmission through the Inhibitory Neuron-Specific vSNARE Vamp1. Neuron 2018, 98, 127–141.e7. [Google Scholar] [CrossRef]
- Gehman, L.T.; Stoilov, P.; Maguire, J.; Damianov, A.; Lin, C.-H.; Shiue, L.; Ares, M.; Mody, I.; Black, D.L. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat. Genet. 2011, 43, 706–711. [Google Scholar] [CrossRef]
- Hamada, N.; Ito, H.; Nishijo, T.; Iwamoto, I.; Morishita, R.; Tabata, H.; Momiyama, T.; Nagata, K.-I. Essential role of the nuclear isoform of RBFOX1, a candidate gene for autism spectrum disorders, in the brain development. Sci. Rep. 2016, 6, 1–19. [Google Scholar] [CrossRef]
- Lee, J.-A.; Damianov, A.; Lin, C.-H.; Fontes, M.; Parikshak, N.N.; Anderson, E.S.; Geschwind, D.H.; Black, D.L.; Martin, K.C. Cytoplasmic Rbfox1 regulates the expression of synaptic and autism-related genes. Neuron 2016, 89, 113–128. [Google Scholar] [CrossRef] [PubMed]
- Fogel, B.L.; Wexler, E.; Wahnich, A.; Friedrich, T.; Vijayendran, C.; Gao, F.; Parikshak, N.; Konopka, G.; Geschwind, D.H. RBFOX1 regulates both splicing and transcriptional networks in human neuronal development. Hum. Mol. Genet. 2012, 21, 4171–4186. [Google Scholar] [CrossRef] [PubMed]
- Bill, B.R.; Lowe, J.K.; DyBuncio, C.T.; Fogel, B.L. Orchestration of neurodevelopmental programs by RBFOX1: Implications for autism spectrum disorder. Int. Rev. Neurobiol. 2013, 113, 251–267. [Google Scholar]
- Fernandez-Castillo, N.; Gan, G.; van Donkelaar, M.M.J.; Vaht, M.; Weber, H.; Retz, W.; Meyer-Lindenberg, A.; Franke, B.; Harro, J.; Reif, A.; et al. RBFOX1, encoding a splicing regulator, is a candidate gene for aggressive behavior. Eur. Neuropsychopharmacol. 2020, 30, 44–55. [Google Scholar] [CrossRef]
- Lal, D.; Trucks, H.; Moller, R.S.; Hjalgrim, H.; Koeleman, B.P.; de Kovel, C.G.; Visscher, F.; Weber, Y.G.; Lerche, H.; Becker, F.; et al. Rare exonic deletions of the RBFOX1 gene increase risk of idiopathic generalized epilepsy. Epilepsia 2013, 54, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.; Chung, D.W.; Dienel, S.; Belch, M.; Fish, K.; Lewis, D. P484. Altered Rbfox1 Signaling Pathway and Cortical Parvalbumin Neuron Dysfunction in Schizophrenia. Biol. Psychiatry 2022, 91, S284. [Google Scholar] [CrossRef]
- Verhoeven, W.M.A.; Egger, J.I.M.; Jongbloed, R.E.; van Putten, M.M.; de Bruin-van Zandwijk, M.; Zwemer, A.S.; Pfundt, R.; Willemsen, M.H. A de novo CTNNB1 Novel Splice Variant in an Adult Female with Severe Intellectual Disability. Int. Med. Case Rep. J. 2020, 13, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Darbro, B.W.; Singh, R.; Zimmerman, M.B.; Mahajan, V.B.; Bassuk, A.G. Autism Linked to Increased Oncogene Mutations but Decreased Cancer Rate. PLoS ONE 2016, 11, e0149041. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Yang, J.; Huang, J.; Chen, Z.; Wu, X.; Zhu, L.; Huang, G.; Long, J.; Su, L. Influence of CTNNB1 rs2953 polymorphism on schizophrenia susceptibility in Chinese Han population through modifying miR-485 binding to CTNNB1. Genes Brain Behav. 2019, 18, e12524. [Google Scholar] [CrossRef]
- Pappas, A.L.; Bey, A.L.; Wang, X.; Rossi, M.; Kim, Y.H.; Yan, H.; Porkka, F.; Duffney, L.J.; Phillips, S.M.; Cao, X.; et al. Deficiency of Shank2 causes mania-like behavior that responds to mood stabilizers. JCI Insight 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, P.; Feng, G. SHANK proteins: Roles at the synapse and in autism spectrum disorder. Nat. Rev. Neurosci. 2017, 18, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Peca, J.; Feliciano, C.; Ting, J.T.; Wang, W.; Wells, M.F.; Venkatraman, T.N.; Lascola, C.D.; Fu, Z.; Feng, G. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature 2011, 472, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Alsufiani, H.M.; Alkhanbashi, A.S.; Laswad, N.A.B.; Bakhadher, K.K.; Alghamdi, S.A.; Tayeb, H.O.; Tarazi, F.I. Zinc deficiency and supplementation in autism spectrum disorder and Phelan-McDermid syndrome. J. Neurosci. Res. 2022, 100, 970–978. [Google Scholar] [CrossRef]
- Park, S.M.; Park, H.R.; Lee, J.H. MAPK3 at the Autism-Linked Human 16p11.2 Locus Influences Precise Synaptic Target Selection at Drosophila Larval Neuromuscular Junctions. Mol. Cells 2017, 40, 151–161. [Google Scholar] [CrossRef]
- Deane, A.R.; Potemkin, N.; Ward, R.D. Mitogen-activated protein kinase (MAPK) signalling corresponds with distinct behavioural profiles in a rat model of maternal immune activation. Behav. Brain Res. 2021, 396, 112876. [Google Scholar] [CrossRef]
- Tucker, R.P. Teneurins: Domain architecture, evolutionary origins, and patterns of expression. Front. Neurosci. 2018, 12, 938. [Google Scholar] [CrossRef]
- Jackson, V.A.; Meijer, D.H.; Carrasquero, M.; van Bezouwen, L.S.; Lowe, E.D.; Kleanthous, C.; Janssen, B.J.; Seiradake, E. Structures of Teneurin adhesion receptors reveal an ancient fold for cell-cell interaction. Nat. Commun. 2018, 9, 1–9. [Google Scholar] [CrossRef]
- Silva, J.-P.; Lelianova, V.G.; Ermolyuk, Y.S.; Vysokov, N.; Hitchen, P.G.; Berninghausen, O.; Rahman, M.A.; Zangrandi, A.; Fidalgo, S.; Tonevitsky, A.G.; et al. Latrophilin 1 and its endogenous ligand Lasso/teneurin-2 form a high-affinity transsynaptic receptor pair with signaling capabilities. Proc. Natl. Acad. Sci. USA 2011, 108, 12113–12118. [Google Scholar] [CrossRef]
- Del Toro, D.; Carrasquero-Ordaz, M.A.; Chu, A.; Ruff, T.; Shahin, M.; Jackson, V.A.; Chavent, M.; Berbeira-Santana, M.; Seyit-Bremer, G.; Brignani, S.; et al. Structural Basis of Teneurin-Latrophilin Interaction in Repulsive Guidance of Migrating Neurons. Cell 2020, 180, 323–339.e19. [Google Scholar] [CrossRef]
- Li, S.; DeLisi, L.E.; McDonough, S.I. Rare germline variants in individuals diagnosed with schizophrenia within multiplex families. Psychiatry Res. 2021, 303, 114038. [Google Scholar] [CrossRef]
- Yi, X.; Li, M.; He, G.; Du, H.; Li, X.; Cao, D.; Wang, L.; Wu, X.; Yang, F.; Chen, X.; et al. Genetic and functional analysis reveals TENM4 contributes to schizophrenia. iScience 2021, 24, 103063. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.-B.; Xu, Z.-H.; Zhu, J.; Wu, Y.; Zhuang, X.-H.; Chen, Q.-L.; Wu, C.-R.; Hu, J.-T.; Zhou, H.-S.; Xie, W.-H.; et al. Exome Sequencing Identifies TENM4 as a Novel Candidate Gene for Schizophrenia in the SCZD2 Locus at 11q14-21. Front. Genet. 2019, 9, 725. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamran, M.; Laighneach, A.; Bibi, F.; Donohoe, G.; Ahmed, N.; Rehman, A.U.; Morris, D.W. Independent Associated SNPs at SORCS3 and Its Protein Interactors for Multiple Brain-Related Disorders and Traits. Genes 2023, 14, 482. https://doi.org/10.3390/genes14020482
Kamran M, Laighneach A, Bibi F, Donohoe G, Ahmed N, Rehman AU, Morris DW. Independent Associated SNPs at SORCS3 and Its Protein Interactors for Multiple Brain-Related Disorders and Traits. Genes. 2023; 14(2):482. https://doi.org/10.3390/genes14020482
Chicago/Turabian StyleKamran, Muhammad, Aodán Laighneach, Farhana Bibi, Gary Donohoe, Naveed Ahmed, Asim Ur Rehman, and Derek W. Morris. 2023. "Independent Associated SNPs at SORCS3 and Its Protein Interactors for Multiple Brain-Related Disorders and Traits" Genes 14, no. 2: 482. https://doi.org/10.3390/genes14020482
APA StyleKamran, M., Laighneach, A., Bibi, F., Donohoe, G., Ahmed, N., Rehman, A. U., & Morris, D. W. (2023). Independent Associated SNPs at SORCS3 and Its Protein Interactors for Multiple Brain-Related Disorders and Traits. Genes, 14(2), 482. https://doi.org/10.3390/genes14020482






