Glucosinolate Biosynthetic Genes of Cabbage: Genome-Wide Identification, Evolution, and Expression Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Sources
2.2. Identification of GBGs in Cabbage
2.3. Non-Synonymous/Synonymous Substitution (Ka/Ks) Analysis
2.4. Expression Analysis of GBGs
2.5. Phylogenetic Analysis
3. Results
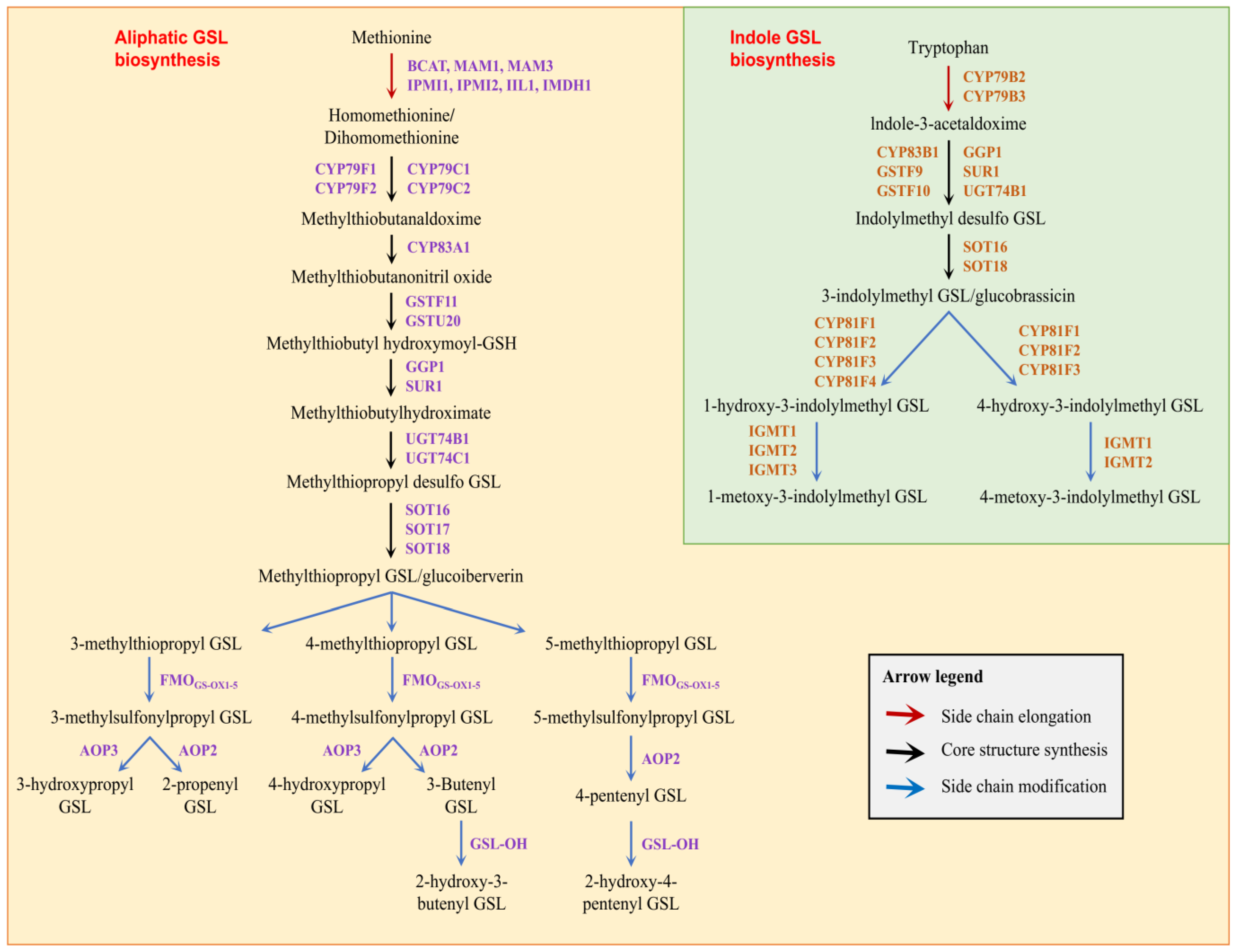
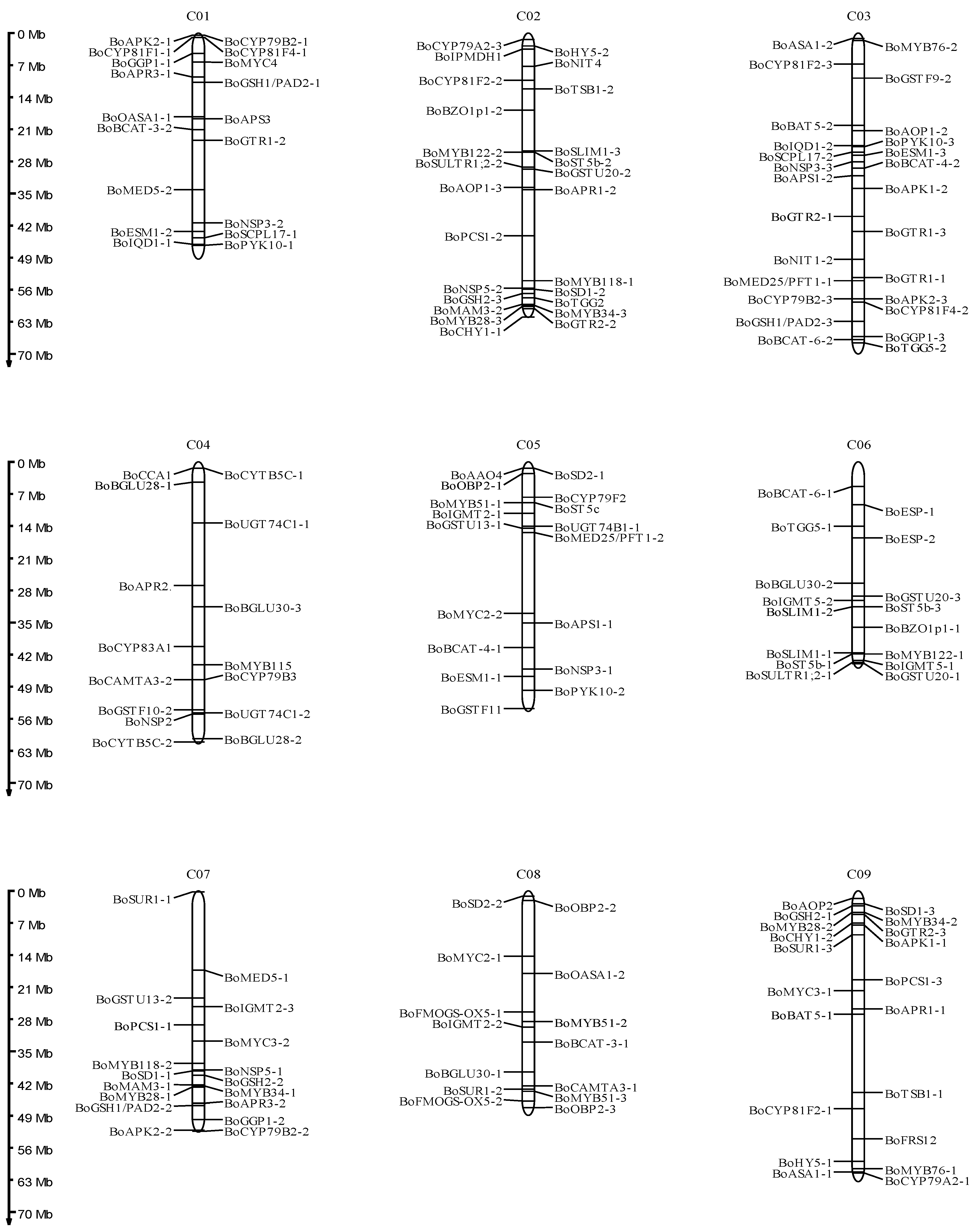
3.1. Identification of Glucosinolate Biosynthetic Genes in B. oleracea
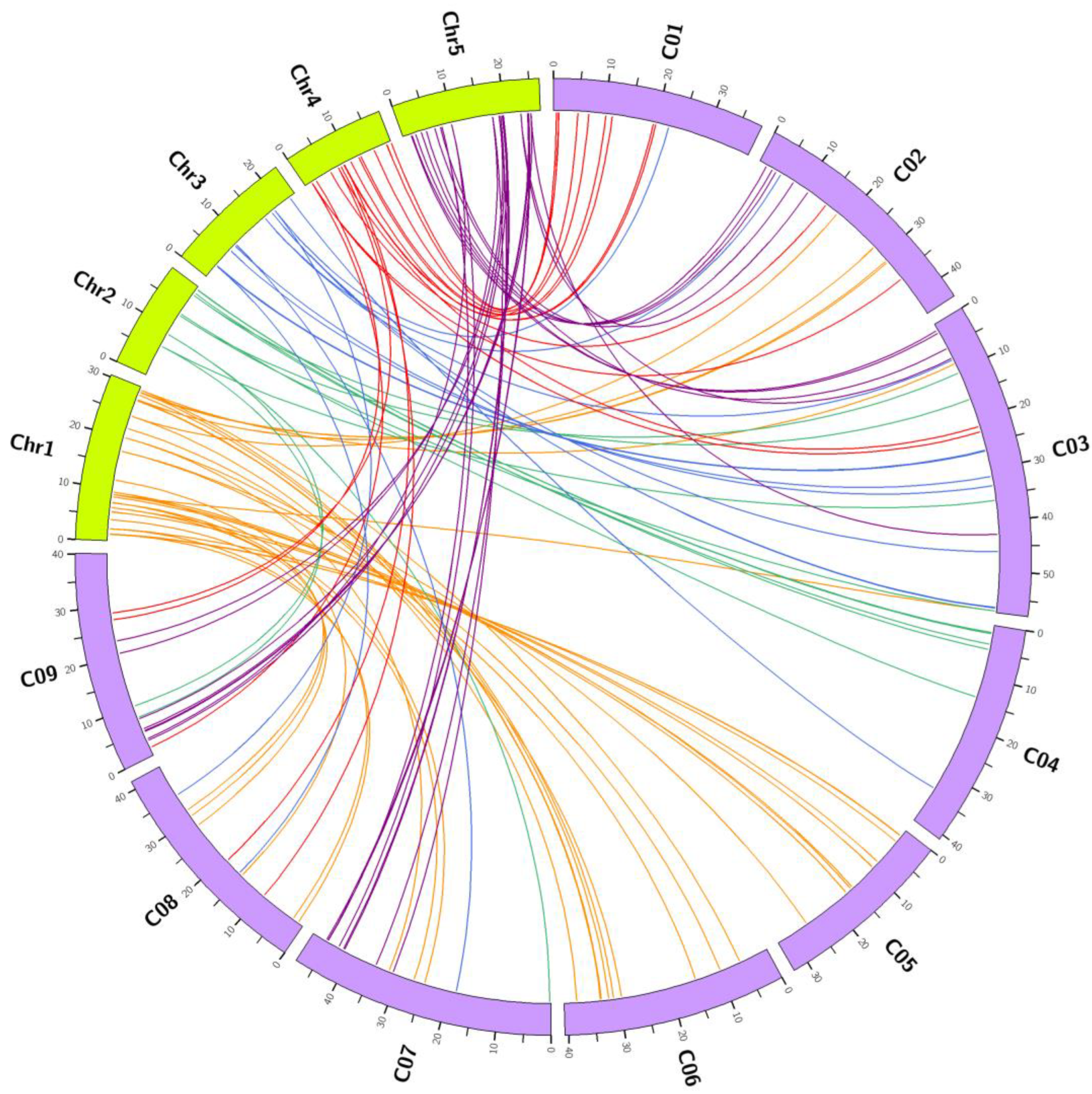
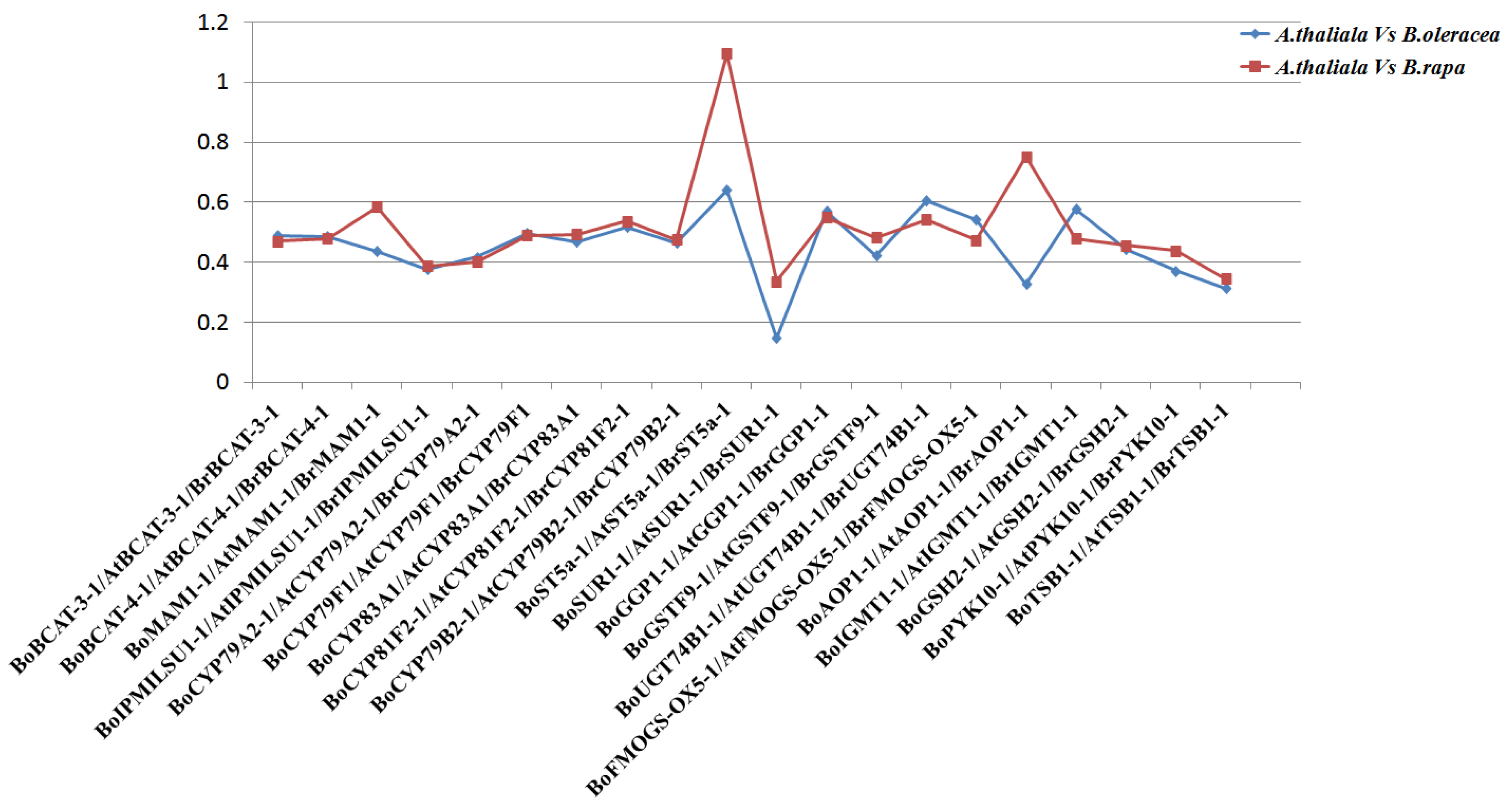
3.2. Comparative Evolutionary Analyses of Orthologous Gene Pairs of Glucosinolate Biosynthetic Genes
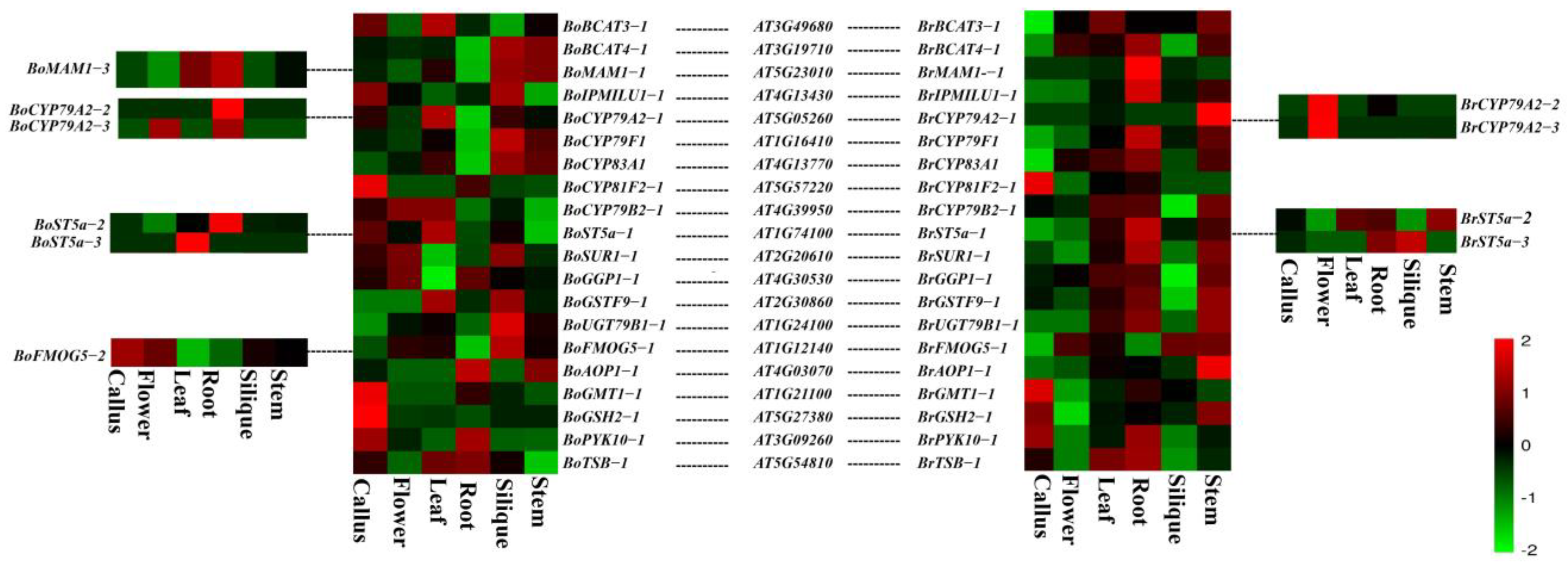
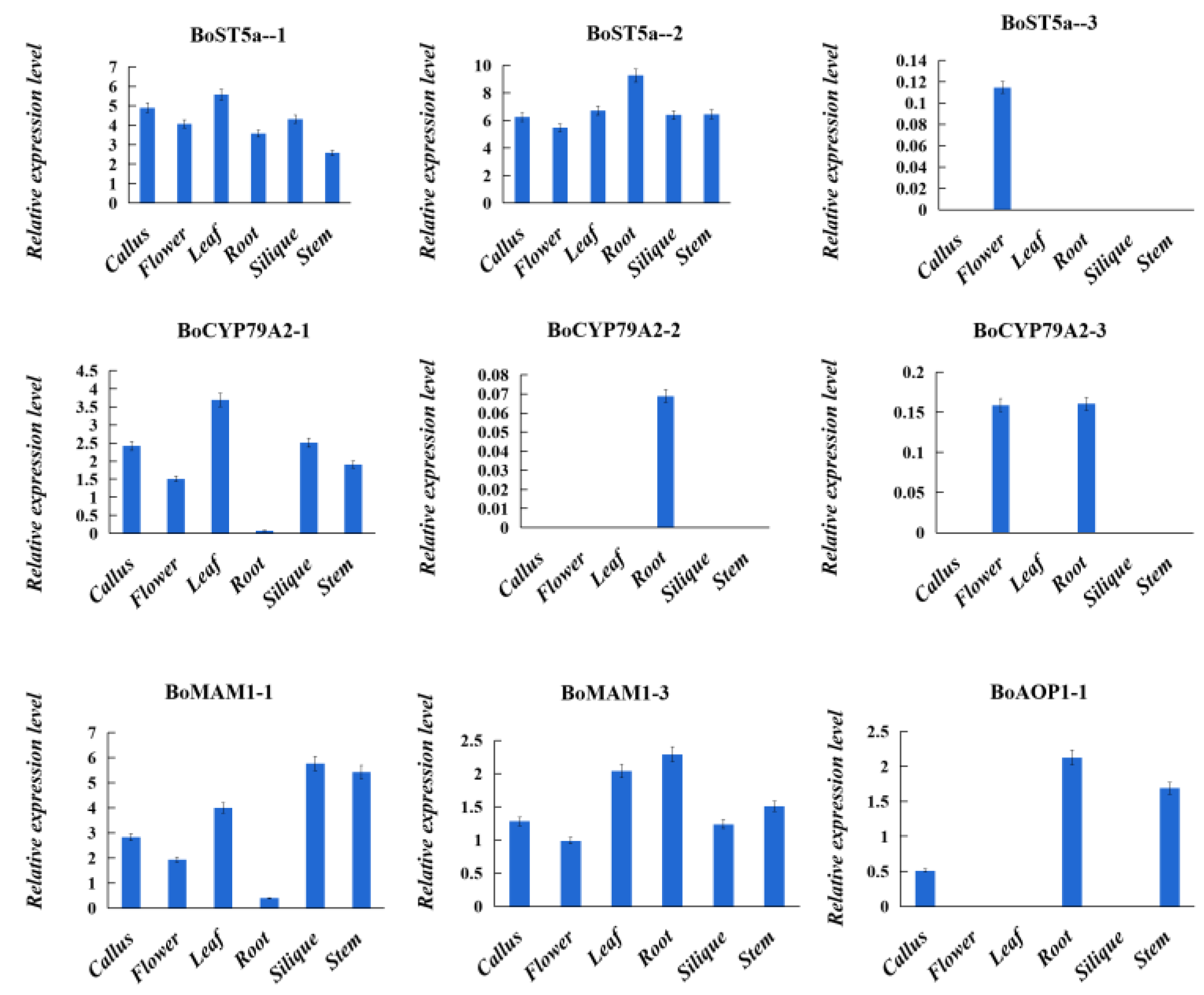
3.3. Expression Analysis of Glucosinolate Biosynthetic Genes Orthologs from B. oleracea, B. rapa, and A. thaliana genomes
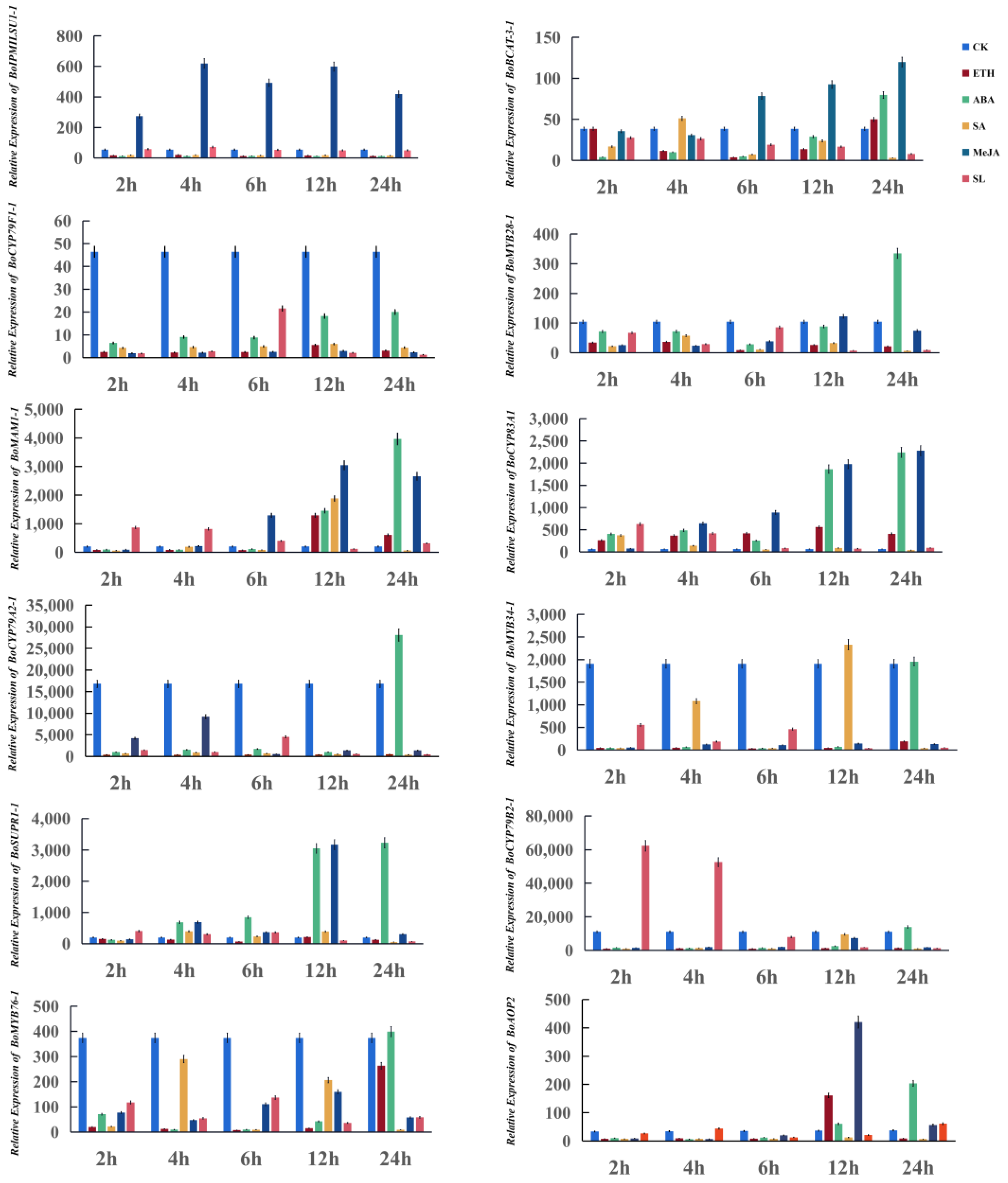
3.4. Twelve Glucosinolate Biosynthetic Genes Respond to Exogenous Phytohormone Treatments
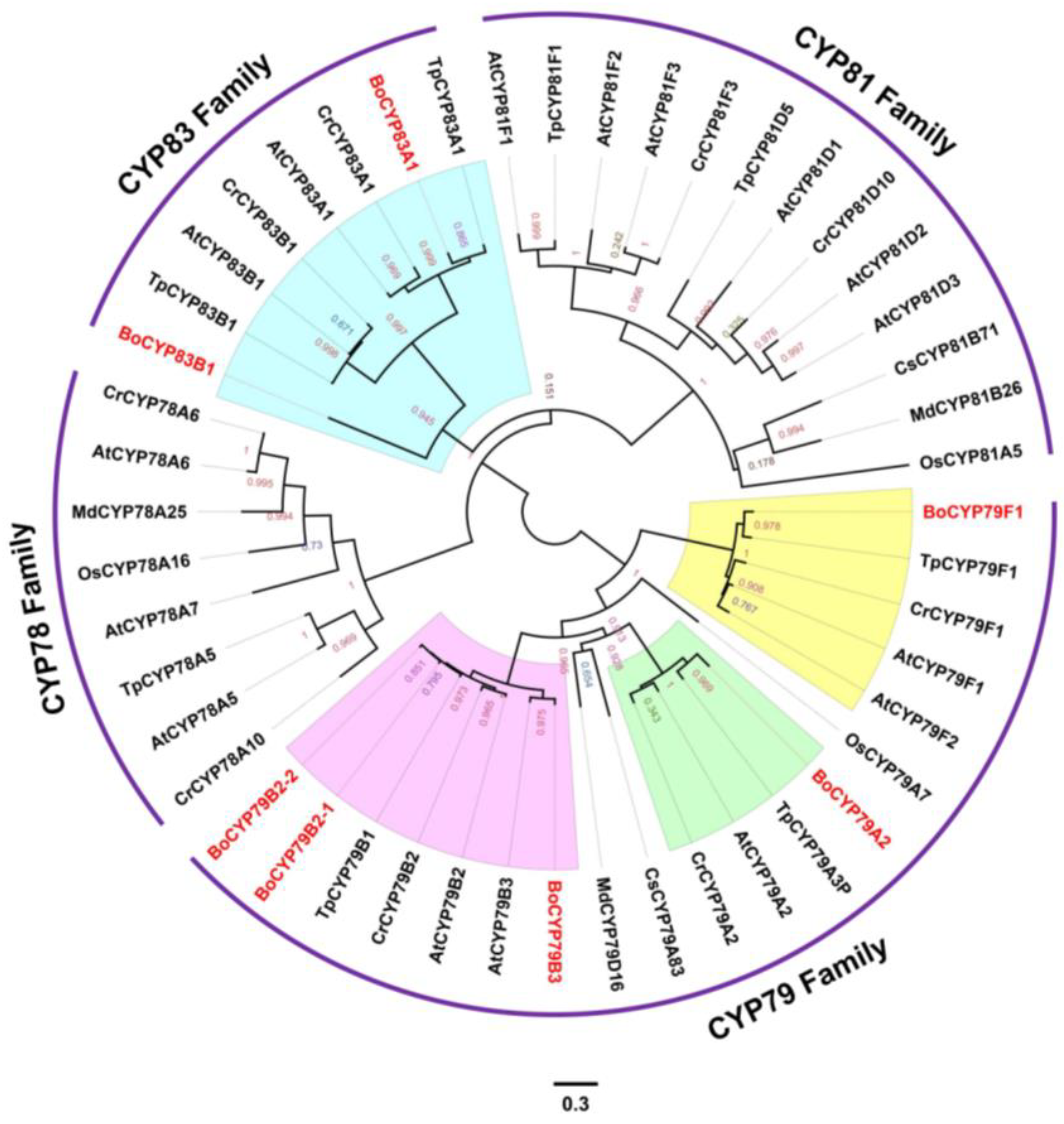
3.5. Evolutionary History of P450 Genes among Seven Plant Species
4. Discussion
4.1. Characterization of Glucosinolate Biosynthetic Genes in B. oleracea
4.2. Exogenous Plant Hormones Regulate the Expression of Glucosinolate Biosynthetic Genes
4.3. Role of CYP450 in Glucosinolate Biosynthesis in Members of Cruciferae
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fahey, J.W.; Zalcmann, A.T.; Talalay, P. The Chemical Diversity and Distribution of Glucosinolates and Isothiocyanates among Plants. Phytochemistry 2001, 56, 5–51. [Google Scholar] [CrossRef] [PubMed]
- Clarke, D.B. Glucosinolates Structures and Analysis in Food. Anal. Methods 2010, 2, 310–325. [Google Scholar] [CrossRef]
- Agerbirk, N.; Olsen, C.E. Glucosinolate Structures in Evolution. Phytochemistry 2012, 77, 16–45. [Google Scholar] [CrossRef] [PubMed]
- Hare, J.D. Ecological Role of Volatiles Produced by Plants in Response to Damage by Herbivorous Insects. Annu. Rev. Entomol. 2011, 56, 161–180. [Google Scholar] [CrossRef] [PubMed]
- Sarai, Q.M.; Paula, G.I.; Francisco, Q.M.; Débora, V.; Diego, A.M. The Role of Brassica Bioactives on Human Health: Are We Studying It the Right Way? Molecules 2020, 25, 1591. [Google Scholar]
- Mawson, R.; Heaney, R.K.; Zdunczyk, Z.; Kozlowska, H. Rapeseed Meal-glucosinolates and Their Antinutritional Effects Part 6. Taint in End-products. Food/Nahrung 2010, 39, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Herr, I.; Büchler, M.W. Dietary Constituents of Broccoli and Other Cruciferous Vegetables: Implications for Prevention and Therapy of Cancer. Cancer Treat. Rev. 2010, 36, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Abuyusuf, M.; Robin, A.; Lee, J. Glucosinolate Profiling and Expression Analysis of Glucosinolate Biosynthesis Genes Differentiate White Mold Resistant and Susceptible Cabbage Lines. Int. J. Mol. Sci. 2018, 19, 4037. [Google Scholar] [CrossRef] [PubMed]
- Aires, A.; Carvalho, R.; Barbosa, M.D.C. Suppressing Potato Cyst Nematode, Globodera Rostochiensis with Extracts of Brassicacea Plants. Am. J. Pot. Res. 2009, 86, 327–333. [Google Scholar] [CrossRef]
- Salem, A.Z.; Medhat, D.; Fathy, S.A.; Mohamed, M.R.; ElKhayat, Z.; ElDaly, S.M. Indole Glucosinolates Exhibit Anti-inflammatory Effects on Ehrlich Ascites Carcinoma CellsThrough Modulation of Inflammatory Markers and MiRNAs. Mol. Biol. Rep. 2021, 48, 10. [Google Scholar] [CrossRef] [PubMed]
- Punetha, H.; Saif, N.S.; Dinesh, P. Glucosinolates in Oilseed Brassica: Neutraceuticals with Tremendous Health Benefits. Int. J. Biol. Sci. 2020, 9, 26–32. [Google Scholar] [CrossRef]
- Lucarini, E.; Micheli, L.; Trallori, E. Effect of Glucoraphanin and Sulforaphane Against Chemotherapy-induced Neuropathic Pain:Kv7 Potassium Channels Modulation by H2S Release in Vivo. Phytother. Res. 2018, 32, 2226–2234. [Google Scholar] [CrossRef] [PubMed]
- Tsaur, I.; Thomas, A.; Taskiran, E.; Rutz, J.; Chun Felix, K.H.; Haferkamp, A.J.; Eva, B.; Roman, A. Concomitant Use of Sulforaphane Enhances Antitumor Efficacy of Sunitinib in Renal Cell Carcinoma In Vitro. Cancers 2022, 14, 4643. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Rutz, J.; Maxeiner, S.; Grein, T.; Thomas, A.; Juengel, E.; Chun Felix, K.H.; Cinatl, J.; Haferkamp, A.; Tsaur, I.; et al. Plant-Derived Sulforaphane Suppresses Growth and Proliferation of Drug-Sensitive and Drug-Resistant Bladder Cancer Cell Lines In Vitro. Cancers 2022, 14, 4682. [Google Scholar] [CrossRef]
- Liu, B.; Mao, Q.Q.; Cao, M. Cruciferous Vegetables Intake and Risk of Prostate Cancer: A Meta-analysis. Int. J. Urol. 2012, 19, 134–141. [Google Scholar] [CrossRef]
- Wu, Q.J.; Yang, Y.; Vogtmann, E. Cruciferous Vegetables Intake and The Risk of Colorectal Cancer:A Meta-analysis of Observational Studies. Ann. Oncol. 2013, 24, 1079–1087. [Google Scholar] [CrossRef]
- Liu, X.; Lü, K. CruciferousVegetables Intakeis Inversely Associated with Risk of Breast Cancer:A Meta-analysis. Breast 2013, 22, 309–313. [Google Scholar] [CrossRef]
- Citi, V.; Piragine, E.; Pagnotta, E. Anticancer Properties of Erucin, an H2S-releasing Isothiocyanate on Human Pancreatic Adenocarcinoma Cells (AsPC-1). Phytother. Res. 2019, 33, 845–855. [Google Scholar] [CrossRef] [PubMed]
- Abbaoui, B.; Lucas, C.; Riedl, K.M. Cruciferous Vegetables, Isothiocyanates and Bladder Cancer Prevention. Mol. Nutr. Food Res. 2018, 62, e1800079. [Google Scholar] [CrossRef] [PubMed]
- Halkier, B.A.; Du, L. The Biosynthesis of Glucosinolates. Trends Plant Sci. 1997, 2, 425–431. [Google Scholar] [CrossRef]
- Padilla, G.; Cartea, M.E.; Velasco, P. Variation of Glucosinolates in Vegetable Crops of Brassica rapa. Phytochemistry 2007, 68, 536–545. [Google Scholar] [CrossRef] [PubMed]
- Schonhof, I.; Krumbein, A.; Brückner, B. Genotypic Effects on Glucosinolates and Sensory Properties of Broccoli and Cauliflower. Food/Nahrung 2010, 48, 25–33. [Google Scholar] [CrossRef]
- Augustine, R.; Bisht, N.C. Biofortification of Oilseed Brassica juncea with the Anti-cancer Compound Glucoraphanin by Suppressing GSL-ALK Gene Family. Sci. Rep. 2015, 5, 18005. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, H.; Liu, Z. A Naturally Occurring Variationinthe BrMAM-3 Geneis Associated with Aliphatic Glucosinolate Accumulationin Brassica Rapa Leaves. Hort. Res. 2018, 5, 69. [Google Scholar] [CrossRef]
- Tamara, G.; Bettina, B.; Hans-peter, M. The Transcription Factor Hig1/Myb51 Regulates Indolic Glucosinolate Biosynthesis in Arabidopsis Thaliana. Plant J. 2010, 50, 886–901. [Google Scholar]
- Gigolashvili, T.; Berger, B.; Flügge, U.I. Specific and Coordinated Control of Indolic and Aliphatic Glucosinolate Biosynthesis by r2r3-Myb Transcription Factors in Arabidopsis thaliana. Phytochem.Rev. 2009, 8, 3–13. [Google Scholar] [CrossRef]
- Cai, C.; Yuan, W.; Miao, H. Functional Characterization of BoaMYB51 Sas Central RegulatorsofIndole Glucosinolate Biosynthesis in Brassica oleracea var. Alboglabra bailey. Front. Plant Sci. 2018, 9, 1599. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Hao, G.; Zhou, J. Identification and Expression Pattern Analysis of Bomyb51 Involvedin Indolic Glucosinolate Biosynthesis from Broccoli (Brassica Oleracea var. Italica). Biochem. Bioph. Res. Commun. 2018, 501, 598–604. [Google Scholar] [CrossRef] [PubMed]
- Sonderby, I.E.; Hansen, B.G.; Bjarnholt, N. A Systems Biology Approach Identifies A r2r3 Myb Gene Subfamily with Distinct and Overlapping Functions in Regulation of Aliphatic Glucosinolates. PLoS ONE 2007, 2, e1322. [Google Scholar] [CrossRef] [PubMed]
- Sonderby, I.E.; Burow, M.; Rowe, H.C. A Complex Interplay of Three r2r3Myb Transcription Factors Determinesthe Profile of Aliphatic Glucosinolates in Arabidopsis. Plant Physiol. 2010, 153, 348–363. [Google Scholar] [CrossRef] [PubMed]
- Sergey, M.; Eyal, B.; Hadar, L. The Transcript and Metabolite Networks Affected by the Two Clades of Arabidopsis Glucosinolate Biosynthesis Regulators. Plant Physiol. 2008, 148, 2021–2049. [Google Scholar]
- Schuster, J.; Knill, T.; Reichelt, M.; Gershenzon, J.; Binder, S. Branched Chain Aminotransferase 4 is Part of The Chain Elongation Pathway in The Biosynthesis of Methionnie-derived Glucosinolates in Arabidopsis. Plant Cell. 2006, 18, 2664–2679. [Google Scholar] [CrossRef]
- Miao, H.; Wei, J.; Zhao, Y.; Yan, H.; Sun, B.; Huang, J.; Wang, Q. Glucose Signalling Positively Regulates Aliphatic Glucosinolate Biosynthesis. J. Exp. Bot. 2013, 64, 1097–1109. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Luo, R.; Li, J.; Miao, R.; An, H.; Yan, X.; Pang, Q. Arabidopsis Glutathione-S-Transferases GSTF11 and GSTU20 Function in Aliphatic Glucosinolate Biosynthesis. Front. Plant Sci. 2022, 12, 816233. [Google Scholar] [CrossRef] [PubMed]
- Glutathione S-Transferases in The Biosynthesis of Sulfur-containing Secondary Metabolites in Brassicaceae Plants. Front. Plant Sci. 2018, 9, 1639. [CrossRef] [PubMed]
- Kong, W.; Li, j.; Yu, Q.; Cang, W.; Xu, R.; Wang, Y.; Ji, W. Two Novel Flavin-containing Monooxygenases Involved in Biosynthesis of Aliphatic Glucosinolates. Front. Plant Sci. 2016, 7, 1292. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.F.; Liu, Z.Y.; Liang, J.I.; Wu, J.; Cheng, F.; Wang, X.W. Three Genes Encoding AOP2, A Protein Involved in Aliphatic Glucosinolate Biosynthesis, Are Differentially Expressed in Brassica rapa. J. Exp. Bot. 2015, 66, 20. [Google Scholar] [CrossRef]
- Araki, R.; Hasumi, A.; Nishizawa, O.I. Novel Bioresources for Studies of Brassica Oleracea: Identification of A Kale Myb Transcription Factor Responsible for Glucosinolate Production. Plant Biotechnol. J. 2013, 11, 1017–1027. [Google Scholar] [CrossRef] [PubMed]
- Mitreiter, S.; Gigolashvili, T. Regulation of Glucosinolate Biosynthesis. J. Exp. Bot. 2020, 72, 70–91. [Google Scholar] [CrossRef]
- Harun, S.; Abdullah-Zawawi, M.R.; Goh, H.H. A Comprehensive Gene Inventory for Glucosinolate Biosynthetic Pathway in Arabidopsis thaliana. J. Agric. Food Chem. 2020, 68, 7281–7297. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.J.; Chen, C.M.; Chen, G.J. Progress in Glucosinolates and Its Molecular Mechanism of Biosynthesis. J. South China Agric. Univ. 2019, 40, 59–70. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. Mafft Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. Fasttree: Computing large Minimum Evolution Trees with Profiles Instead of A Distance Matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Kazuki, S.; Fumio, M. Metabolomics for Functional Genomics, Systems Biology, and Biotechnology. Annu. Rev. Plant Biol. 2010, 61, 1. [Google Scholar]
- Zhang, Z. KaKs-calculator 3.0: Calculating Selective Pressure on Coding and Non-coding Sequences. Genom. Proteom. Bioinform. 2022, 20, 536–540. [Google Scholar] [CrossRef]
- Das Bidyadhar. Glucosinolate Biosynthesis: Role of MAM Synthase and Its Perspectives. Biosci. Rep. 2021, 41, BSR20211634. [CrossRef]
- Abrahams, R.S.; Pires, J.C.; Schranz, M.E. Genomic Origin and Diversification of the Glucosinolate MAM Locus. Front. Plant Sci. 2020, 11, 111. [Google Scholar] [CrossRef]
- Zhang, P.L.; Chang, T.; Zhang, Q.Z.; Cai, Y.L.; Zhou, Z.Y.; Hu, M.F.; Zhang, J.Y. Identification of MAM1s in Regulation of 3C Glucosinolates Accumulation in Allopolyploid Brassica juncea. Crit. Rev. Biotechnol. 2020, 6, 409–418. [Google Scholar]
- Meenu, A.R.; Majee, M.; Pradhan, A.K.; Bisht, N.C. Genomic Origin, Expression Differentiation and Regulation of Multiple Genes Encoding CYP83A1, A Key Enzyme for Core Glucosinolate Biosynthesis, from The Allotetraploid Brassica juncea. Planta 2015, 241, 3. [Google Scholar]
- Biao, Z.; Wang, Z.Z.; Yang, Y.; Zhu, Z.J.; Wang, H.S. Isolation and Expression of Glucosinolate Synthesis Genes CYP83A1 and CYP83B1 in Pak Choi ( Brassica rapa L. ssp. chinensis var. communis (N. Tsen & S.H. Lee) Hanelt). IJMS 2012, 13, 5. [Google Scholar]
- Bak, S.; Feyereisen, R. The Involvement of Two P450 Enzymes, CYP83B1 and CYP83A1, in Auxin Homeostasis and Glucosinolate Biosynthesis. Plant Physiol. 2001, 127, 1. [Google Scholar] [CrossRef] [PubMed]
- Kiarash, J.G.; Ali Riahi-Madvar, F.R.; Rambod, P.; Fereshteh, J.B.; Mahshid, G.A. Effect of Salinity Stress on Enzymes’ Activity, Ions Concentration, Oxidative Stress Parameters, Biochemical Traits, Content of Sulforaphane, and CYP79F1 Gene Expression Level in Lepidium draba Plant. J. Plant Growth Regul. 2019, 39, 1075–1094. [Google Scholar]
- Chen, S.X. CYP79F1 and CYP79F2 Have Distinct Functions in The Biosynthesis of Aliphatic Glucosinolates in Arabidopsis. Plant J. 2003, 33, 923–937. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Mukhopadhyay, A.; Gupta, V.; Pental, D.; Pradhan, A.K. CYP79F1 Regulates Synthesis of Propyl Fraction of Aliphatic Glucosinolates in Oilseed Mustard Brassica juncea: Functional Validation through Genetic and Transgenic. PLoS ONE 2016, 11, e0150060. [Google Scholar] [CrossRef]
- Czerniawski, P.; PiślewskaBednarek, M.; Piasecka, A.; Kułak, K.; Bednarek, P. Loss of MYB34 transcription Factor Supports Backward Evolution of Indole Glucosinolate Biosynthesis in A Subclade of Camelineae tribe and Releases Feedback Loop in This Pathway in Arabidopsis. Plant Cell Physiol. 2022, 12, pcac142. [Google Scholar] [CrossRef]
- Henninge, F.; Eriche, G.; Tamarae, G. The Role of MYB34, MYB51 and MYB122 in The Regulation of Camalexin Biosynthesis in Arabidopsis thaliana. Front. Plant Sci. 2015, 6, 654. [Google Scholar]
- Hansen, C.C.; Nelson, D.R.; Møller, B.L.; WerckReichhart, D. Plant Cytochrome P450 Plasticity and Evolution. Mol. Plant. 2021, 14, 1772. [Google Scholar] [CrossRef]
- Dimaano, N.G.; Iwakami, S. Cytochrome P450-mediated Herbicide Metabolism in Plants: Current Understanding and Prospects. Pest Manag. Sci. 2021, 77, 22–32. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, P.; Cao, W.; Yang, L.; Zhang, Y.; Fang, Z.; Zhuang, M.; Lv, H.; Wang, Y.; Cheng, S.; Ji, J. Glucosinolate Biosynthetic Genes of Cabbage: Genome-Wide Identification, Evolution, and Expression Analysis. Genes 2023, 14, 476. https://doi.org/10.3390/genes14020476
Wang P, Cao W, Yang L, Zhang Y, Fang Z, Zhuang M, Lv H, Wang Y, Cheng S, Ji J. Glucosinolate Biosynthetic Genes of Cabbage: Genome-Wide Identification, Evolution, and Expression Analysis. Genes. 2023; 14(2):476. https://doi.org/10.3390/genes14020476
Chicago/Turabian StyleWang, Peng, Wenxue Cao, Limei Yang, Yangyong Zhang, Zhiyuan Fang, Mu Zhuang, Honghao Lv, Yong Wang, Shanhan Cheng, and Jialei Ji. 2023. "Glucosinolate Biosynthetic Genes of Cabbage: Genome-Wide Identification, Evolution, and Expression Analysis" Genes 14, no. 2: 476. https://doi.org/10.3390/genes14020476
APA StyleWang, P., Cao, W., Yang, L., Zhang, Y., Fang, Z., Zhuang, M., Lv, H., Wang, Y., Cheng, S., & Ji, J. (2023). Glucosinolate Biosynthetic Genes of Cabbage: Genome-Wide Identification, Evolution, and Expression Analysis. Genes, 14(2), 476. https://doi.org/10.3390/genes14020476







