Arabidopsis ANAC102, Chloroplastic or Nucleocytosolic Localization?
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Growth Conditions and Treatments
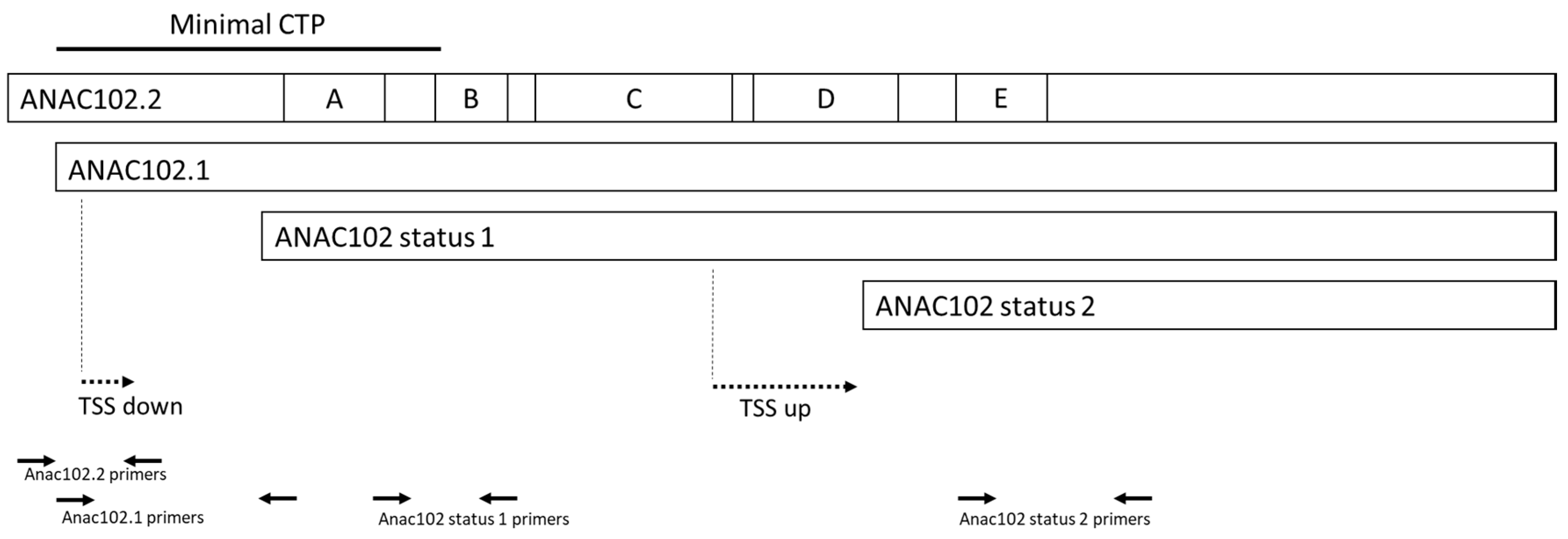
2.2. RNA Preparation, cDNA Cloning and qRT-PCR Assays
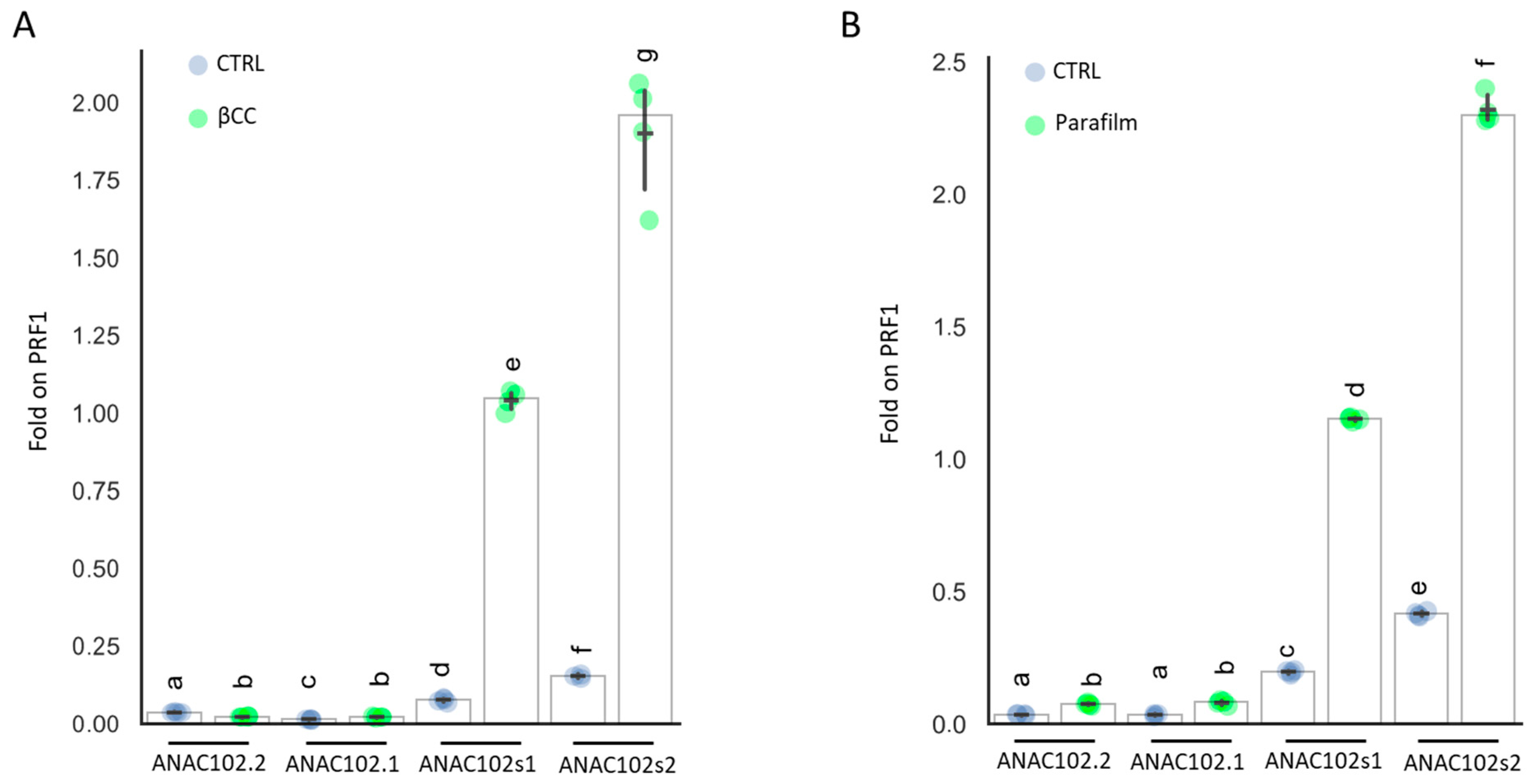
2.3. Data Plotting and Statistical Analysis
2.4. Protein Sequence Retrieval and Alignment
3. Results
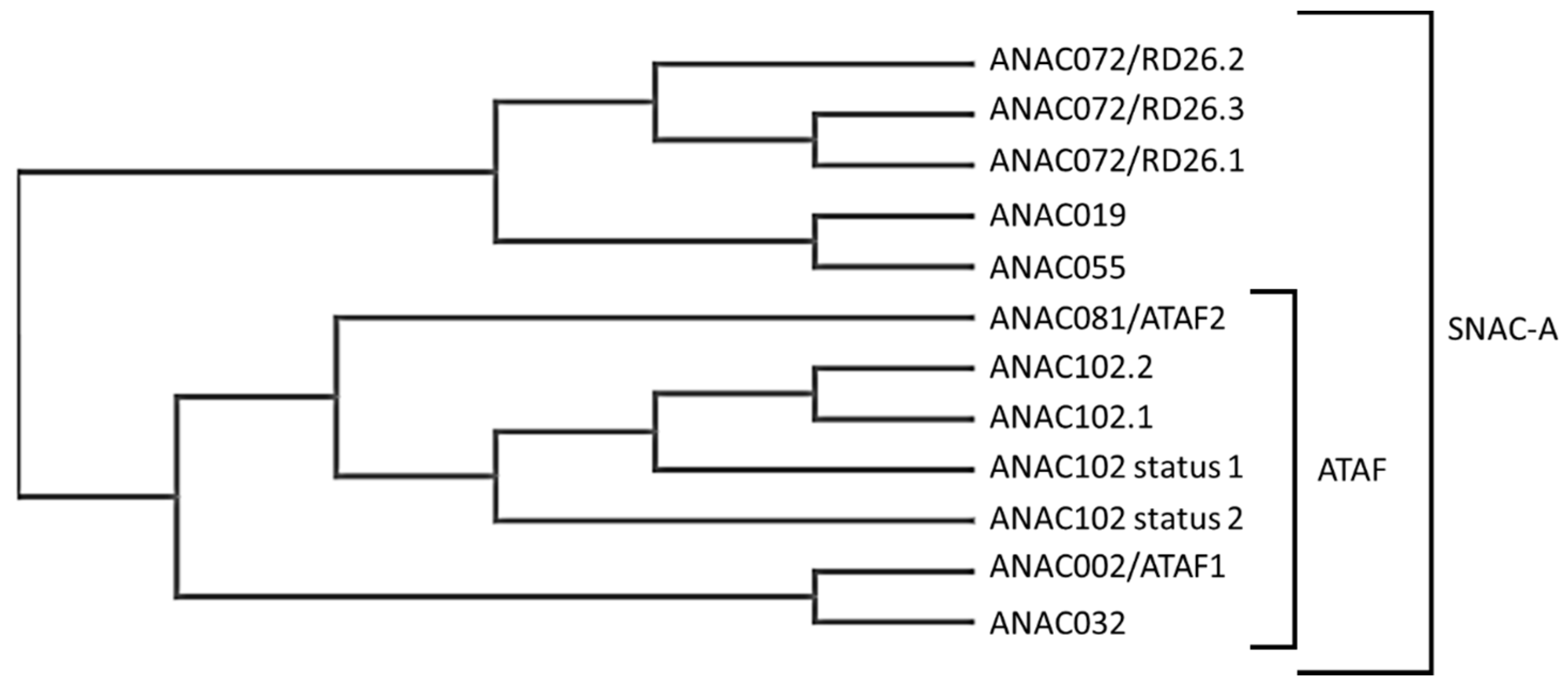
3.1. ANAC102, Chief or Teammate?
3.2. ANAC102 Is Involved in Several Pathways
3.3. ANAC102 Chloroplast or Nuclear?
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lodeyro, A.F.; Krapp, A.R.; Carrillo, N. Photosynthesis and chloroplast redox signaling in the age of global warming: Stress tolerance, acclimation, and developmental plasticity. J. Exp. Bot. 2021, 72, 5919–5937. [Google Scholar] [CrossRef]
- Loro, G.; Wagner, S.; Doccula, F.G.; Behera, S.; Weinl, S.; Kudla, J.; Schwarzländer, M.; Costa, A.; Zottini, M. Chloroplast-Specific in Vivo Ca2+ Imaging Using Yellow Cameleon Fluorescent Protein Sensors Reveals Organelle-Autonomous Ca2+ Signatures in the Stroma. Plant Physiol. 2016, 171, 2317–2330. [Google Scholar] [CrossRef] [PubMed]
- Estavillo, G.M.; Crisp, P.A.; Pornsiriwong, W.; Wirtz, M.; Collinge, D.; Carrie, C.; Giraud, E.; Whelan, J.; David, P.; Javot, H.; et al. Evidence for a SAL1-PAP Chloroplast Retrograde Pathway That Functions in Drought and High Light Signaling in Arabidopsis. Plant Cell 2011, 23, 3992–4012. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.-Z.; Bock, R. GUN control in retrograde signaling: How GENOMES UNCOUPLED proteins adjust nuclear gene expression to plastid biogenesis. Plant Cell 2021, 33, 457–474. [Google Scholar] [CrossRef]
- Wang, L.; Kim, C.; Xu, X.; Piskurewicz, U.; Dogra, V.; Singh, S.; Mahler, H.; Apel, K. Singlet oxygen- and EXECUTER1-mediated signaling is initiated in grana margins and depends on the protease FtsH2. Proc. Natl. Acad. Sci. USA 2016, 113, E3792–E3800. [Google Scholar] [CrossRef] [PubMed]
- D’Alessandro, S.; Ksas, B.; Havaux, M. Decoding β-Cyclocitral-Mediated Retrograde Signaling Reveals the Role of a Detoxification Response in Plant Tolerance to Photooxidative Stress. Plant Cell 2018, 30, 2495–2511. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, H.; Ma, T.; Li, J.; Yuan, J.; Xu, Y.-C.; Sun, R.; Zhang, X.; Jing, Y.; Guo, Y.-L.; et al. Arabidopsis EXECUTER1 interacts with WRKY transcription factors to mediate plastid-to-nucleus singlet oxygen signaling. Plant Cell 2022, koac330. [Google Scholar] [CrossRef]
- Fode, B.; Siemsen, T.; Thurow, C.; Weigel, R.; Gatz, C. The Arabidopsis GRAS Protein SCL14 Interacts with Class II TGA Transcription Factors and Is Essential for the Activation of Stress-Inducible Promoters. Plant Cell 2008, 20, 3122–3135. [Google Scholar] [CrossRef]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P.; et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 2003, 10, 239–247. [Google Scholar] [CrossRef]
- Jensen, M.K.; Kjaersgaard, T.; Nielsen, M.M.; Galberg, P.; Petersen, K.; O’Shea, C.; Skriver, K. The Arabidopsis thaliana NAC transcription factor family: Structure–function relationships and determinants of ANAC019 stress signalling. Biochem. J. 2010, 426, 183–196. [Google Scholar] [CrossRef]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Takasaki, H.; Maruyama, K.; Takahashi, F.; Fujita, M.; Yoshida, T.; Nakashima, K.; Myouga, F.; Toyooka, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. SNAC-As, stress-responsive NAC transcription factors, mediate ABA-inducible leaf senescence. Plant J. 2015, 84, 1114–1123. [Google Scholar] [CrossRef] [PubMed]
- Reback, J.; Jbrockmendel; McKinney, W.; Van Den Bossche, J.; Roeschke, M.; Augspurger, T.; Hawkins, S.; Cloud, P.; Gfyoung; Sinhrks; et al. Pandas-Dev/Pandas: Pandas 1.4.3; Zenodo: Geneva, Switzerland, 2022. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Waskom, M.L. seaborn: Statistical data visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
- Vallat, R. Pingouin: Statistics in Python. J. Open Source Softw. 2018, 3, 1026. [Google Scholar] [CrossRef]
- Inzé, A. Protein Subcellular Trafficking during the Oxidative Stress Response in Plants. Ph.D. Thesis, Ghent University, Ghent, Belgium, 2011. [Google Scholar]
- Xin, K.; Pan, T.; Gao, S.; Yan, S. A Transcription Factor Regulates Gene Expression in Chloroplasts. Int. J. Mol. Sci. 2021, 22, 6769. [Google Scholar] [CrossRef]
- Peng, H.; Neff, M.M. Two ATAF transcription factors ANAC102 and ATAF1 contribute to the suppression of cytochrome P450-mediated brassinosteroid catabolism in Arabidopsis. Physiol. Plant. 2021, 172, 1493–1505. [Google Scholar] [CrossRef]
- Vanderauwera, S.; Zimmermann, P.; Rombauts, S.; Vandenabeele, S.; Langebartels, C.; Gruissem, W.; Inzé, D.; Van Breusegem, F. Genome-Wide Analysis of Hydrogen Peroxide-Regulated Gene Expression in Arabidopsis Reveals a High Light-Induced Transcriptional Cluster Involved in Anthocyanin Biosynthesis. Plant Physiol. 2005, 139, 806–821. [Google Scholar] [CrossRef]
- Christianson, J.A.; Wilson, I.W.; Llewellyn, D.J.; Dennis, E.S. The Low-Oxygen-Induced NAC Domain Transcription Factor ANAC102 Affects Viability of Arabidopsis Seeds following Low-Oxygen Treatment. Plant Physiol. 2009, 149, 1724–1738. [Google Scholar] [CrossRef]
- Roach, T.; Stöggl, W.; Baur, T.; Kranner, I. Distress and eustress of reactive electrophiles and relevance to light stress acclimation via stimulation of thiol/disulphide-based redox defences. Free Radic. Biol. Med. 2018, 122, 65–73. [Google Scholar] [CrossRef]
- D’Alessandro, S.; Beaugelin, I.; Havaux, M. Tanned or Sunburned: How Excessive Light Triggers Plant Cell Death. Mol. Plant 2020, 13, 1545–1555. [Google Scholar] [CrossRef]
- Inzé, A.; Vanderauwera, S.; Hoeberichts, F.A.; Vandorpe, M.; Van Gaever, T.; Van Breusegem, F. A subcellular localization compendium of hydrogen peroxide-induced proteins. Plant Cell Environ. 2012, 35, 308–320. [Google Scholar] [CrossRef]
- Bhakta, S.; Negi, S.; Tak, H.; Singh, S.; Ganapathi, T.R. MusaATAF2 like protein, a stress-related transcription factor, induces leaf senescence by regulating chlorophyll catabolism and H2O2 accumulation. Physiol. Plant. 2022, 174, e13593. [Google Scholar] [CrossRef] [PubMed]
- Ushijima, T.; Hanada, K.; Gotoh, E.; Yamori, W.; Kodama, Y.; Tanaka, H.; Kusano, M.; Fukushima, A.; Tokizawa, M.; Yamamoto, Y.Y.; et al. Light Controls Protein Localization through Phytochrome-Mediated Alternative Promoter Selection. Cell 2017, 171, 1316–1325.e12. [Google Scholar] [CrossRef] [PubMed]
- Shumbe, L.; D’Alessandro, S.; Shao, N.; Chevalier, A.; Ksas, B.; Bock, R.; Havaux, M. METHYLENE BLUE SENSITIVITY 1 (MBS1) is required for acclimation of Arabidopsis to singlet oxygen and acts downstream of β-cyclocitral. Plant Cell Environ. 2017, 40, 216–226. [Google Scholar] [CrossRef] [PubMed]
- Waszczak, C.; Kerchev, P.I.; Mühlenbock, P.; Hoeberichts, F.A.; Van Der Kelen, K.; Mhamdi, A.; Willems, P.; Denecker, J.; Kumpf, R.P.; Noctor, G.; et al. SHORT-ROOT Deficiency Alleviates the Cell Death Phenotype of the Arabidopsis catalase2 Mutant under Photorespiration-Promoting Conditions. Plant Cell 2016, 28, 1844–1859. [Google Scholar] [CrossRef]
- Kerchev, P.; Mühlenbock, P.; Denecker, J.; Morreel, K.; Hoeberichts, F.A.; Van Der Kelen, K.; Vandorpe, M.; Nguyen, L.; Audenaert, D.; Van Breusegem, F. Activation of auxin signalling counteracts photorespiratory H2O2-dependent cell death. Plant Cell Environ. 2013, 38, 253–265. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cresta, A.; D’Alessandro, S. Arabidopsis ANAC102, Chloroplastic or Nucleocytosolic Localization? Genes 2023, 14, 438. https://doi.org/10.3390/genes14020438
Cresta A, D’Alessandro S. Arabidopsis ANAC102, Chloroplastic or Nucleocytosolic Localization? Genes. 2023; 14(2):438. https://doi.org/10.3390/genes14020438
Chicago/Turabian StyleCresta, Alessandro, and Stefano D’Alessandro. 2023. "Arabidopsis ANAC102, Chloroplastic or Nucleocytosolic Localization?" Genes 14, no. 2: 438. https://doi.org/10.3390/genes14020438
APA StyleCresta, A., & D’Alessandro, S. (2023). Arabidopsis ANAC102, Chloroplastic or Nucleocytosolic Localization? Genes, 14(2), 438. https://doi.org/10.3390/genes14020438