Genetic Modifiers of Non-Penetrance and RNA Expression Levels in PRPF31-Associated Retinitis Pigmentosa in a Danish Cohort
Abstract
1. Introduction and Background
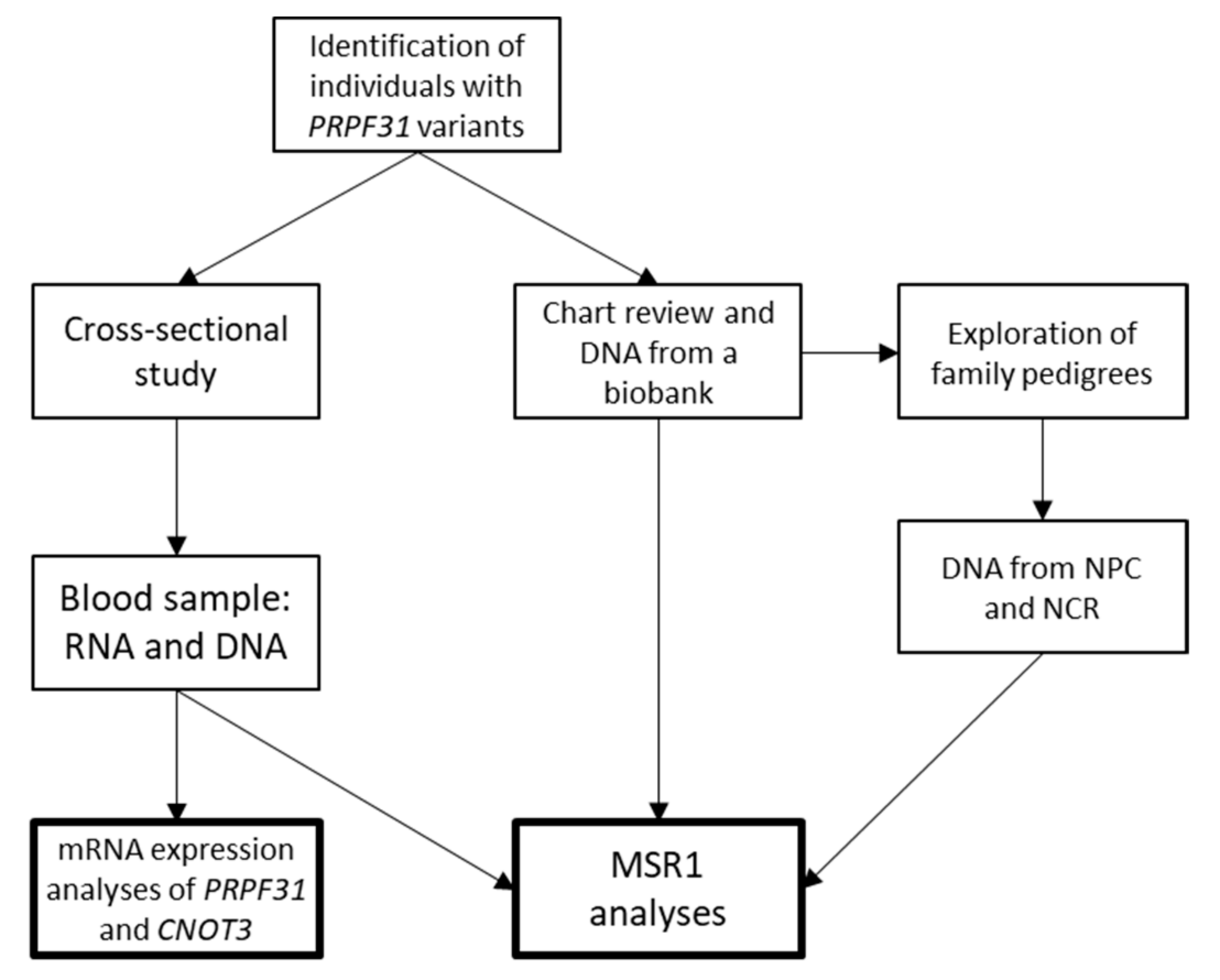
2. Materials and Methods
2.1. Subjects
2.2. Genetic Testing and Classification
2.3. RNA Expression Analysis of PRPF31 and CNOT3
2.4. Copy Number of MSR1
2.5. Statistical Methods
3. Results
3.1. Expression Analyses of PRPF31 and CNOT3
3.2. MSR1 Analyses
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vithana, E.N.; Abu-Safieh, L.; Allen, M.J.; Carey, A.; Papaioannou, M.; Chakarova, C.; Al-Maghtheh, M.; Ebenezer, N.D.; Willis, C.; Moore, A.T.; et al. A Human Homolog of Yeast Pre-MRNA Splicing Gene, PRP31, Underlies Autosomal Dominant Retinitis Pigmentosa on Chromosome 19q13.4 (RP11). Mol. Cell 2001, 8, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Verbakel, S.K.; van Huet, R.A.C.; Boon, C.J.F.; den Hollander, A.I.; Collin, R.W.J.; Klaver, C.C.W.; Hoyng, C.B.; Roepman, R.; Klevering, B.J. Non-Syndromic Retinitis Pigmentosa. Prog. Retin. Eye Res. 2018, 66, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, S.; Di Iorio, E.; Barbaro, V.; Ponzin, D.; Sorrentino, F.S.; Parmeggiani, F. Retinitis Pigmentosa: Genes and Disease Mechanisms. Curr. Genom. 2011, 12, 238–249. [Google Scholar] [CrossRef]
- Haim, M. The Epidemiology of Retinitis Pigmentosa in Denmark. Acta Ophthalmol. Scand. Suppl. 2002, 80, 1–34. [Google Scholar] [CrossRef]
- Hartong, D.T.; Berson, E.L.; Dryja, T.P. Retinitis Pigmentosa. Lancet 2006, 368, 1795–1809. [Google Scholar] [CrossRef]
- Dias, M.F.; Joo, K.; Kemp, J.A.; Fialho, S.L.; da Silva Cunha, A.; Woo, S.J.; Kwon, Y.J. Molecular Genetics and Emerging Therapies for Retinitis Pigmentosa: Basic Research and Clinical Perspectives. Prog. Retin. Eye Res. 2018, 63, 107–131. [Google Scholar] [CrossRef]
- Ruberto, F.P.; Balzano, S.; Namburi, P.; Kimchi, A.; Pescini-Gobert, R.; Obolensky, A.; Banin, E.; Ben-Yosef, T.; Sharon, D.; Rivolta, C. Heterozygous Deletions of Noncoding Parts of the PRPF31 Gene Cause Retinitis Pigmentosa via Reduced Gene Expression. Mol. Vis. 2021, 27, 107–116. [Google Scholar]
- Abu-Safieh, L.; Vithana, E.N.; Mantel, I.; Holder, G.E.; Pelosini, L.; Bird, A.C.; Bhattacharya, S.S. A Large Deletion in the AdRP Gene PRPF31: Evidence That Haploinsufficiency Is the Cause of Disease. Mol. Vis. 2006, 12, 384–388. [Google Scholar]
- Lisbjerg, K.; Bertelsen, M.; Lyng Forman, J.; Grønskov, K.; Prener Holtan, J.; Kessel, L. Disease Progression of Retinitis Pigmentosa Caused by PRPF31 Variants in a Nordic Population: A Retrospective Study with up to 36 Years Follow-Up. Ophthalmic Genet. 2022, 1–8. [Google Scholar] [CrossRef]
- Kiser, K.; Webb-Jones, K.D.; Bowne, S.J.; Sullivan, L.S.; Daiger, S.P.; Birch, D.G. Time Course of Disease Progression of PRPF31-Mediated Retinitis Pigmentosa. Am. J. Ophthalmol. 2019, 200, 76–84. [Google Scholar] [CrossRef]
- Moore, A.T.; Fitzke, F.; Jay, M.; Arden, G.B.; Inglehearn, C.F.; Keen, T.J.; Bhattacharya, S.S.; Bird, A.C. Autosomal Dominant Retinitis Pigmentosa with Apparent Incomplete Penetrance: A Clinical, Electrophysiological, Psychophysical, and Molecular Genetic Study. Br. J. Ophthalmol. 1993, 77, 473–479. [Google Scholar] [CrossRef]
- Russell, S.; Bennett, J.; Wellman, J.A.; Chung, D.C.; Yu, Z.F.; Tillman, A.; Wittes, J.; Pappas, J.; Elci, O.; McCague, S.; et al. Efficacy and Safety of Voretigene Neparvovec (AAV2-HRPE65v2) in Patients with RPE65-Mediated Inherited Retinal Dystrophy: A Randomised, Controlled, Open-Label, Phase 3 Trial. Lancet 2017, 390, 849–860. [Google Scholar] [CrossRef]
- Thompson, D.A.; Iannaccone, A.; Ali, R.R.; Arshavsky, V.Y.; Audo, I.; Bainbridge, J.W.B.; Besirli, C.G.; Birch, D.G.; Branham, K.E.; Cideciyan, A.V.; et al. Advancing Clinical Trials for Inherited Retinal Diseases: Recommendations from the Second Monaciano Symposium. Transl. Vis. Sci. Technol. 2020, 9, 2. [Google Scholar] [CrossRef]
- Rose, A.M.; Luo, R.; Radia, U.K.; Bhattacharya, S.S. Gene of the Month: PRPF31. J. Clin. Pathol. 2017, 70, 729–732. [Google Scholar] [CrossRef]
- Hoskins, A.A.; Friedman, L.J.; Gallagher, S.S.; Crawford, D.J.; Anderson, E.G.; Wombacher, R.; Ramirez, N.; Cornish, V.W.; Gelles, J.; Moore, M.J. Ordered and Dynamic Assembly of Single Spliceosomes. Science 2011, 331, 1289–1295. [Google Scholar] [CrossRef]
- Tanackovic, G.; Ransijn, A.; Thibault, P.; Elela, S.A.; Klinck, R.; Berson, E.L.; Chabot, B.; Rivolta, C. PRPF Mutations Are Associated with Generalized Defects in Spliceosome Formation and Pre-MRNA Splicing in Patients with Retinitis Pigmentosa. Hum. Mol. Genet. 2011, 20, 2116–2130. [Google Scholar] [CrossRef]
- Tanackovic, G.; Rivolta, C. PRPF31 Alternative Splicing and Expression in Human Retina. Ophthalmic Genet. 2009, 30, 76–83. [Google Scholar] [CrossRef]
- Linder, B.; Dill, H.; Hirmer, A.; Brocher, J.; Lee, G.P.; Mathavan, S.; Bolz, H.J.; Winkler, C.; Laggerbauer, B.; Fischer, U. Systemic Splicing Factor Deficiency Causes Tissue-Specific Defects: A Zebrafish Model for Retinitis Pigmentosa. Hum. Mol. Genet. 2011, 20, 368–377. [Google Scholar] [CrossRef]
- Yusuf, I.H.; Birtel, J.; Shanks, M.E.; Clouston, P.; Downes, S.M.; Charbel Issa, P.; Maclaren, R.E. Clinical Characterization of Retinitis Pigmentosa Associated with Variants in SNRNP200. JAMA Ophthalmol. 2019, 137, 1295–1300. [Google Scholar] [CrossRef]
- Rose, A.M.; Bhattacharya, S.S. Variant Haploinsufficiency and Phenotypic Non-Penetrance in PRPF31-Associated Retinitis Pigmentosa. Clin. Genet. 2016, 90, 118–126. [Google Scholar] [CrossRef]
- Wheway, G.; Douglas, A.; Baralle, D.; Guillot, E. Mutation Spectrum of PRPF31, Genotype-Phenotype Correlation in Retinitis Pigmentosa, and Opportunities for Therapy. Exp. Eye Res. 2020, 192, 107950. [Google Scholar] [CrossRef] [PubMed]
- Roshandel, D.; Thompson, J.A.; Jeffery, R.C.H.; Zhang, D.; Lamey, T.M.; McLaren, T.L.; De Roach, J.N.; McLenachan, S.; Mackey, D.A.; Chen, F.K. Clinical Evidence for the Importance of the Wild-Type Prpf31 Allele in the Phenotypic Expression of Rp11. Genes 2021, 12, 915. [Google Scholar] [CrossRef] [PubMed]
- McGee, T.L.; Devoto, M.; Ott, J.; Berson, E.L.; Dryja, T.P. Evidence That the Penetrance of Mutations at the RP11 Locus Causing Dominant Retinitis Pigmentosa Is Influenced by a Gene Linked to the Homologous RP11 Allele. Am. J. Hum. Genet. 1997, 61, 1059–1066. [Google Scholar] [CrossRef] [PubMed]
- Rose, A.M.; Shah, A.Z.; Venturini, G.; Krishna, A.; Chakravarti, A.; Rivolta, C.; Bhattacharya, S.S. Transcriptional Regulation of PRPF31 Gene Expression by MSR1 Repeat Elements Causes Incomplete Penetrance in Retinitis Pigmentosa. Sci. Rep. 2016, 6, 19450. [Google Scholar] [CrossRef] [PubMed]
- Venturini, G.; Rose, A.M.; Shah, A.Z.; Bhattacharya, S.S.; Rivolta, C. CNOT3 Is a Modifier of PRPF31 Mutations in Retinitis Pigmentosa with Incomplete Penetrance. PLoS Genet. 2012, 8, e1003040. [Google Scholar] [CrossRef]
- McLenachan, S.; Zhang, D.; Grainok, J.; Zhang, X.; Huang, Z.; Chen, S.C.; Zaw, K.; Lima, A.; Jennings, L.; Roshandel, D.; et al. Determinants of Disease Penetrance in Prpf31-Associated Retinopathy. Genes 2021, 12, 1542. [Google Scholar] [CrossRef]
- Jespersgaard, C.; Fang, M.; Bertelsen, M.; Dang, X.; Jensen, H.; Chen, Y.; Bech, N.; Dai, L.; Rosenberg, T.; Zhang, J.; et al. Molecular Genetic Analysis Using Targeted NGS Analysis of 677 Individuals with Retinal Dystrophy. Sci. Rep. 2019, 9, 1219. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://Www.R-Project.Org/ (accessed on 1 April 2019).
- De Winter, J.C.F. Using the Student’s t-Test with Extremely Small Sample Sizes. Pract. Assessment Res. Eval. 2013, 18, 1–12. [Google Scholar]
- Martin-Merida, I.; Sanchez-Alcudia, R.; Jose, P.F.S.; Blanco-Kelly, F.; Perez-Carro, R.; Da Silva, L.R.J.; Almoguera, B.; Garcia-Sandoval, B.; Lopez-Molina, M.I.; Avila-Fernandez, A.; et al. Analysis of the PRPF31 Gene in Spanish Autosomal Dominant Retinitis Pigmentosa Patients: A Novel Genomic Rearrangement. Investig. Ophthalmol. Vis. Sci. 2017, 58, 1045–1053. [Google Scholar] [CrossRef]
- Ali-nasser, T.; Zayit-soudry, S.; Banin, E.; Sharon, D.; Ben-yosef, T.; Ali, T. Autosomal Dominant Retinitis Pigmentosa with Incomplete Penetrance Due to an Intronic Mutation of the PRPF31 Gene. Mol. Vis. 2022, 28, 359–368. [Google Scholar]
- Rio Frio, T.; Civic, N.; Ransijn, A.; Beckmann, J.S.; Rivolta, C. Two Trans-Acting EQTLs Modulate the Penetrance of PRPF31 Mutations. Hum. Mol. Genet. 2008, 17, 3154–3165. [Google Scholar] [CrossRef]
- Vithana, E.N.; Abu-Safieh, L.; Pelosini, L.; Winchester, E.; Hornan, D.; Bird, A.C.; Hunt, D.M.; Bustin, S.A.; Bhattacharya, S.S. Expression of PRPF31 MRNA in Patients with Autosomal Dominant Retinitis Pigmentosa: A Molecular Clue for Incomplete Penetrance? Investig. Ophthalmol. Vis. Sci. 2003, 44, 4204–4209. [Google Scholar] [CrossRef]
- Rivolta, C.; McGee, T.L.; Frio, T.R.; Jensen, R.V.; Berson, E.L.; Dryja, T.P. Variation in Retinitis Pigmentosa-11 (PRPF31 or RP11) Gene Expression between Symptomatic and Asymptomatic Patients with Dominant RP11 Mutations. Hum. Mutat. 2006, 27, 644–653. [Google Scholar] [CrossRef]
- Frio, T.R.; Wade, N.M.; Ransijn, A.; Berson, E.L.; Beckmann, J.S.; Rivolta, C. Premature Termination Codons in PRPF31 Cause Retinitis Pigmentosa via Haploinsufficiency Due to Nonsense-Mediated MRNA Decay. J. Clin. Investig. 2008, 118, 1519–1531. [Google Scholar] [CrossRef]
- Villanueva, A.; Willer, J.R.; Bryois, J.; Dermitzakis, E.T.; Katsanis, N.; Davis, E.E. Whole Exome Sequencing of a Dominant Retinitis Pigmentosa Family Identifies a Novel Deletion in PRPF31. Investig. Ophthalmol. Vis. Sci. 2014, 55, 2121–2129. [Google Scholar] [CrossRef]
- Lenartowicz, M.; Moos, T.; Ogórek, M.; Jensen, T.G.; Møller, L.B. Metal-Dependent Regulation of ATP7A and ATP7B in Fibroblast Cultures. Front. Mol. Neurosci. 2016, 9, 68. [Google Scholar] [CrossRef]
- Liu, J.Y.; Dai, X.; Sheng, J.; Cui, X.; Wang, X.; Jiang, X.; Tu, X.; Tang, Z.; Bai, Y.; Liu, M.; et al. Identification and Functional Characterization of a Novel Splicing Mutation in RP Gene PRPF31. Biochem. Biophys. Res. Commun. 2008, 367, 420–426. [Google Scholar] [CrossRef]
- Rose, A.M.; Shah, A.Z.; Venturini, G.; Rivolta, C.; Rose, G.E.; Bhattacharya, S.S. Dominant PRPF31 Mutations Are Hypostatic to a Recessive CNOT3 Polymorphism in Retinitis Pigmentosa: A Novel Phenomenon of “Linked Trans -Acting Epistasis”. Ann. Hum. Genet. 2014, 78, 62–71. [Google Scholar] [CrossRef]
- Yang, C.; Georgiou, M.; Atkinson, R.; Collin, J.; Al-Aama, J.; Nagaraja-Grellscheid, S.; Johnson, C.; Ali, R.; Armstrong, L.; Mozaffari-Jovin, S.; et al. Pre-MRNA Processing Factors and Retinitis Pigmentosa: RNA Splicing and Beyond. Front. Cell Dev. Biol. 2021, 9, 2062. [Google Scholar] [CrossRef]
- Yin, J.; Brocher, J.; Fischer, U.; Winkler, C. Mutant Prpf31 Causes Pre-MRNA Splicing Defects and Rod Photoreceptor Cell Degeneration in a Zebrafish Model for Retinitis Pigmentosa. Mol. Neurodegener. 2011, 6, 56. [Google Scholar] [CrossRef] [PubMed]
- Valdés-Sánchez, L.; Calado, S.M.; de la Cerda, B.; Aramburu, A.; Belén García-Delgado, A.; Massalini, S.; Montero-Sánchez, A.; Bhatia, V.; Rodríguez-Bocanegra, E.; Diez-Lloret, A.; et al. Retinal Pigment Epithelium Degeneration Caused by Aggregation of PRPF31 and the Role of HSP70 Family of Proteins. Mol. Med. 2020, 26, 1–22. [Google Scholar] [CrossRef] [PubMed]




| RP Group | Non-Penetrant Carriers | Fold Change | p-Value | |
|---|---|---|---|---|
| PRPF31, relative normalized mean ± SD | 1.14 ± 0.50 | 1.25 ± 0.55 | 1.1 | 0.66 |
| CNOT3, relative normalized mean ± SD | 1.18 ± 0.46 | 1.51 ± 0.84 | 1.3 | 0.27 |
| MSR1 on Wild-Type Allele | RP | NPC |
|---|---|---|
| 4-copy on WT, n | 0 | 3 |
| 3-copy on WT, n | 26 | 4 |
| MSR1 on PRPF31 Variant Allele | RP | NPC |
| 4-copy on mutant, n | 11 | 2 |
| 3-copy on mutant, n | 15 | 5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lisbjerg, K.; Grønskov, K.; Bertelsen, M.; Møller, L.B.; Kessel, L. Genetic Modifiers of Non-Penetrance and RNA Expression Levels in PRPF31-Associated Retinitis Pigmentosa in a Danish Cohort. Genes 2023, 14, 435. https://doi.org/10.3390/genes14020435
Lisbjerg K, Grønskov K, Bertelsen M, Møller LB, Kessel L. Genetic Modifiers of Non-Penetrance and RNA Expression Levels in PRPF31-Associated Retinitis Pigmentosa in a Danish Cohort. Genes. 2023; 14(2):435. https://doi.org/10.3390/genes14020435
Chicago/Turabian StyleLisbjerg, Kristian, Karen Grønskov, Mette Bertelsen, Lisbeth Birk Møller, and Line Kessel. 2023. "Genetic Modifiers of Non-Penetrance and RNA Expression Levels in PRPF31-Associated Retinitis Pigmentosa in a Danish Cohort" Genes 14, no. 2: 435. https://doi.org/10.3390/genes14020435
APA StyleLisbjerg, K., Grønskov, K., Bertelsen, M., Møller, L. B., & Kessel, L. (2023). Genetic Modifiers of Non-Penetrance and RNA Expression Levels in PRPF31-Associated Retinitis Pigmentosa in a Danish Cohort. Genes, 14(2), 435. https://doi.org/10.3390/genes14020435







