Molecular Mechanisms Underlying Vertebrate Adaptive Evolution: A Systematic Review
Abstract
1. Introduction
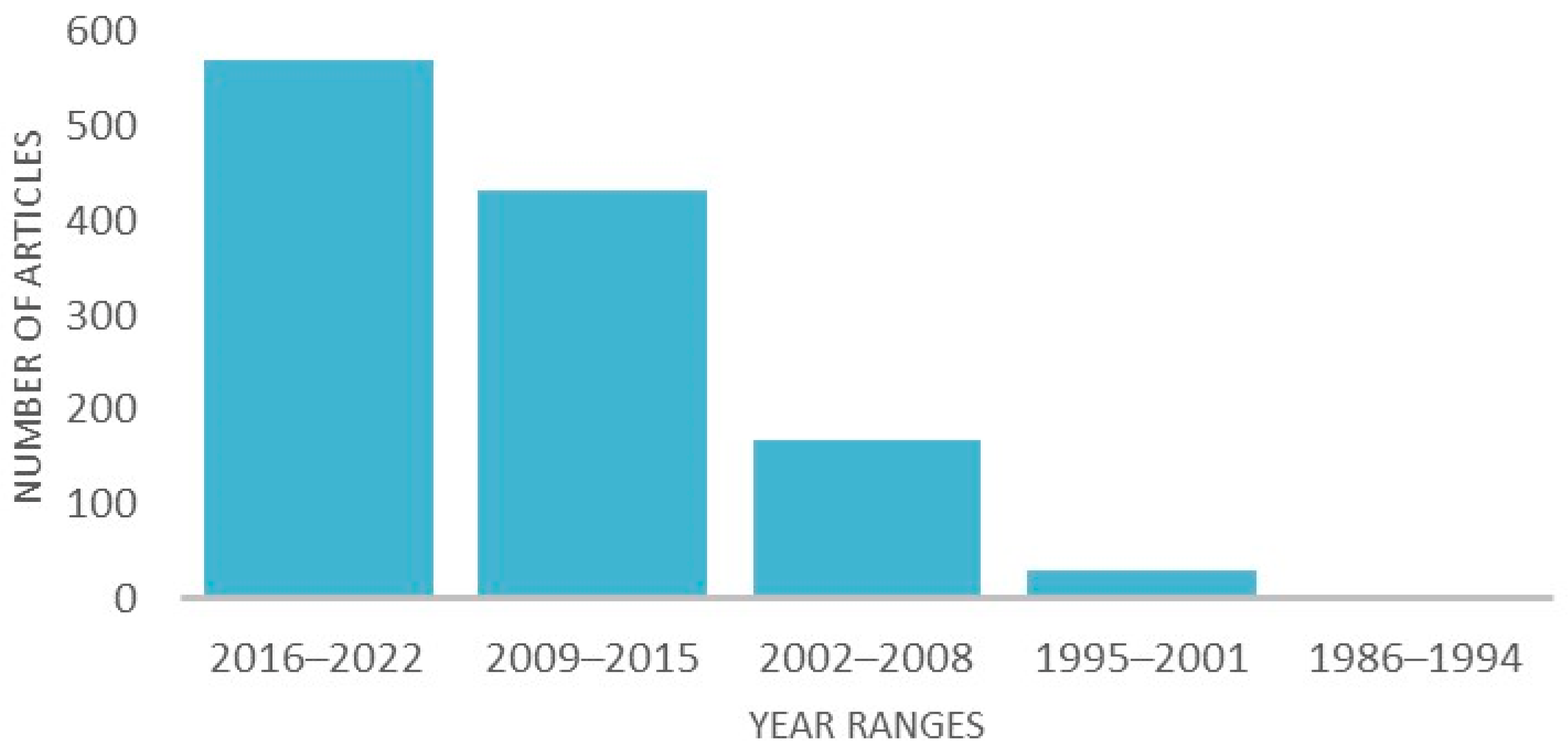
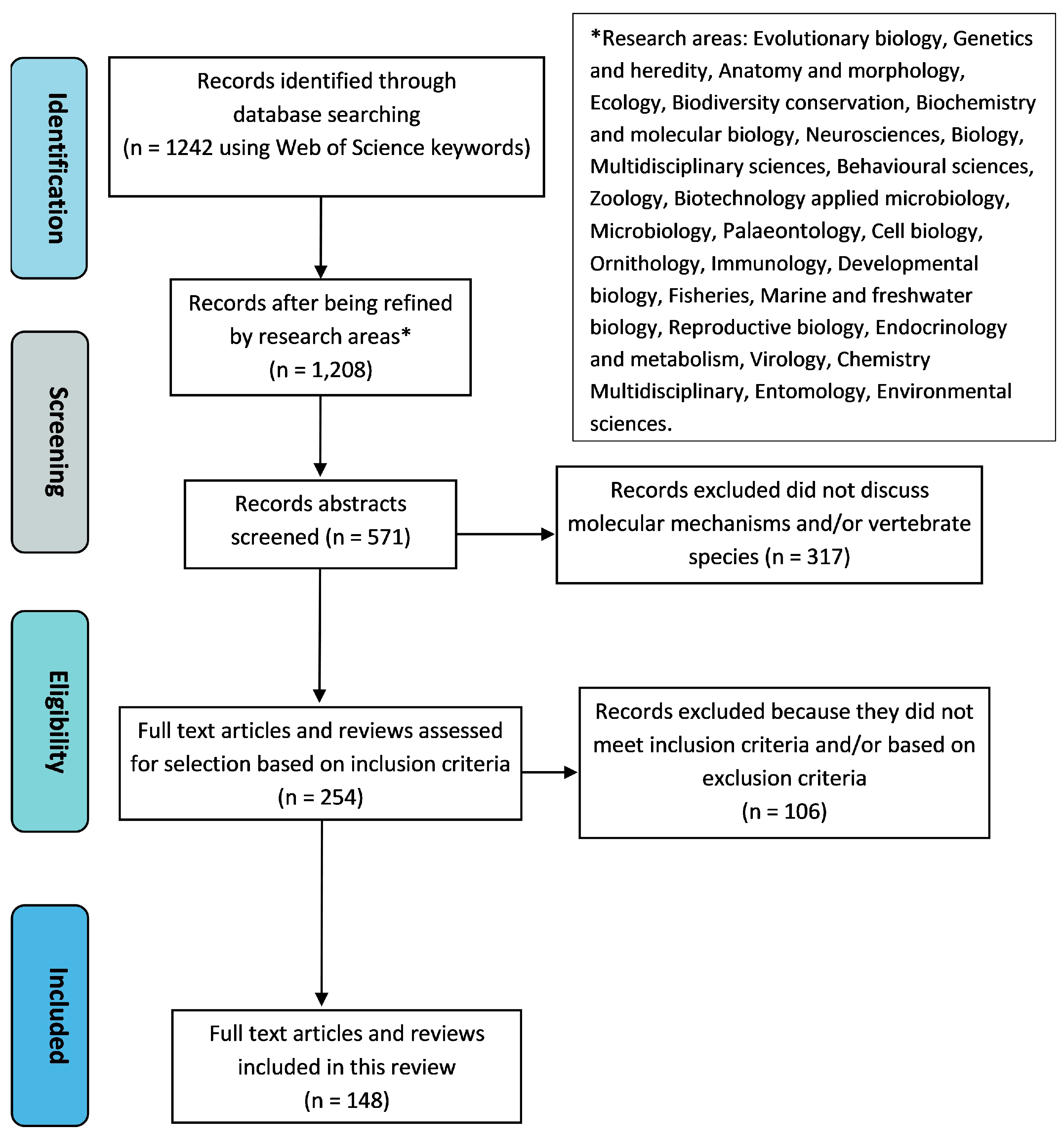
2. Materials and Methods: Evidence Acquisition
2.1. Study Selection
2.2. Eligibility Criteria
- I.
- We included research articles or reviews that investigated or discussed molecular mechanisms involved in adaptive evolution and/or the evolution of a trait that was shown to increase fitness.
- II.
- The following articles were excluded:
- i.
- Research articles or reviews that described observations suggesting the adaptive evolution of a trait (i.e., signatures of selection) but neither offered detailed information about the underlying mechanisms for adaptive evolution nor were supported by other articles that did investigate the underlying mechanism.
- ii.
- Research articles that deemed their results inconclusive or in need of validation, and/or found no evidence to support the adaptive molecular mechanisms they were investigating.
- iii.
- Research articles or reviews that described the evolution of a trait that had not been classified as adaptive.
2.3. Limitations
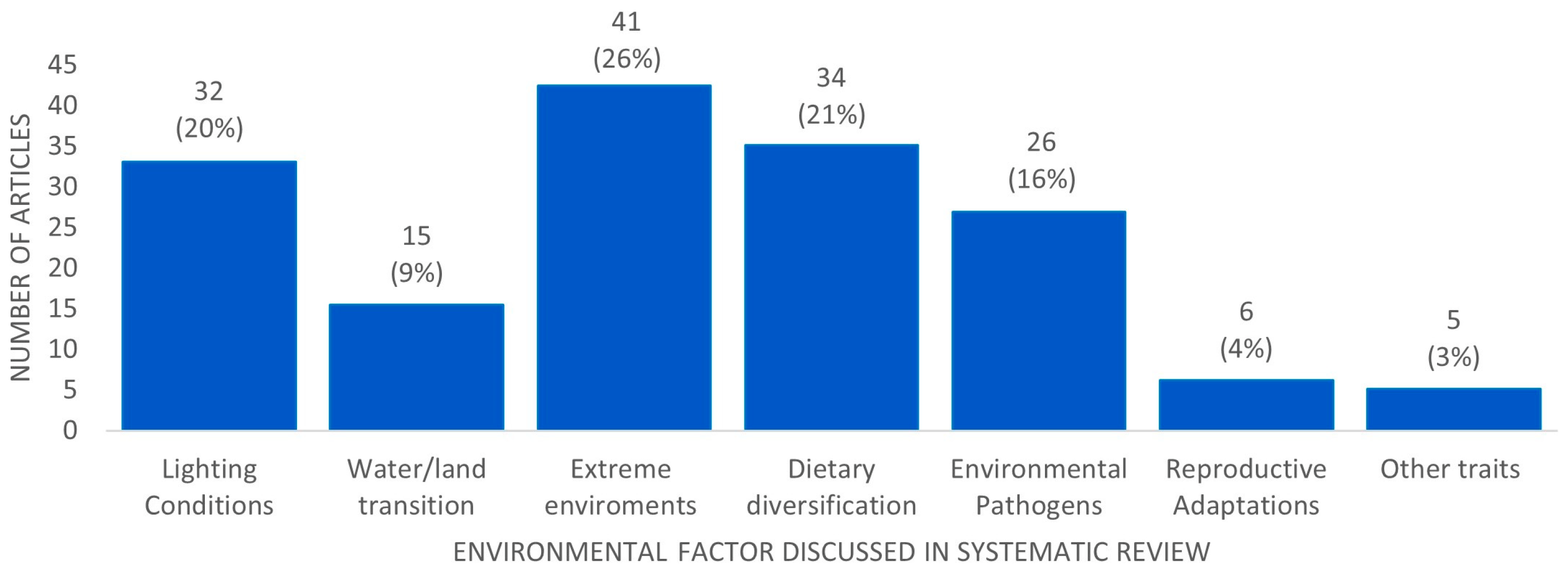
3. General Aspects of the Molecular Mechanisms Underlying Adaptive Evolution in Vertebrates
4. Adaptive Evolution in Response to Variation in Lighting Conditions
5. Adaptations for Colonization of Aquatic and Terrestrial Environments
6. Adapting to Extreme Environmental Conditions
6.1. Hypoxic Environments
6.2. Extreme Aquatic Environments
6.3. Temperature Extremes
7. Adaptations to Dietary Changes
7.1. Metabolic Adaptations
7.2. Non-Metabolic Adaptations
7.3. Anatomical and Behavioural Adaptations
8. Environmental Pathogens
9. Reproductive Adaptations
10. Other Environmental Factors
10.1. Changes in Circadian Rhythm
10.2. Morphological Adaptations
11. Limitations of the Study
12. Future Research
13. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Beldade, P.; Mateus, A.R.; Keller, R.A. Evolution and molecular mechanisms of adaptive developmental plasticity. Mol. Ecol. 2011, 20, 1347–1363. [Google Scholar] [CrossRef] [PubMed]
- Olson-Manning, C.F.; Wagner, M.R.; Mitchell-Olds, T. Adaptive evolution: Evaluating empirical support for theoretical predictions. Nat. Rev. Genet. 2012, 13, 867–877. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Chen, L.; Wang, W. Genomic insights into ruminant evolution: From past to future prospects. Zool. Res. 2019, 40, 476–487. [Google Scholar] [CrossRef]
- Slodkowicz, G.; Goldman, N. Integrated structural and evolutionary analysis reveals common mechanisms underlying adaptive evolution in mammals. Proc. Natl. Acad. Sci. USA 2020, 117, 5977–5986. [Google Scholar] [CrossRef]
- Hao, Y.; Qu, Y.; Song, G.; Lei, F. Genomic Insights into the Adaptive Convergent Evolution. Curr. Genom. 2019, 20, 81–89. [Google Scholar] [CrossRef]
- Carleton, K.L.; Escobar-Camacho, D.; Stieb, S.M.; Cortesi, F.; Marshall, N.J. Seeing the rainbow: Mechanisms underlying spectral sensitivity in teleost fishes. J. Exp. Biol. 2020, 223, jeb193334. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, F.A.; Corrêa Guimarães, E.M.; Carvalho, N.; Ferreira, A.; Schneider, C.H.; Carvalho-Zilse, G.A.; Feldberg, E.; Gross, M. Transposable DNA Elements in Amazonian Fish: From Genome Enlargement to Genetic Adaptation to Stressful Environments. Cytogenet. Genome Res. 2020, 160, 148–155. [Google Scholar] [CrossRef]
- Perry, B.W.; Card, D.C.; McGlothlin, J.W.; Pasquesi, G.I.M.; Adams, R.H.; Schield, D.R.; Hales, N.R.; Corbin, A.B.; Demuth, J.P.; Hoffmann, F.G.; et al. Molecular Adaptations for Sensing and Securing Prey and Insight into Amniote Genome Diversity from the Garter Snake Genome. Genome Biol. Evol. 2018, 10, 2110–2129. [Google Scholar] [CrossRef]
- Luo, J.; Wang, Y.; Yuan, J.; Zhao, Z.; Lu, J. MicroRNA duplication accelerates the recruitment of new targets during vertebrate evolution. RNA 2018, 24, 787–802. [Google Scholar] [CrossRef]
- Penso-Dolfin, L.; Haerty, W.; Hindle, A.; Di Palma, F. microRNA profiling in the Weddell seal suggests novel regulatory mechanisms contributing to diving adaptation. BMC Genom. 2020, 21, 303. [Google Scholar] [CrossRef]
- Li, K.; Sun, X.; Chen, M.; Sun, Y.; Tian, R.; Wang, Z. Evolutionary changes of Hox genes and relevant regulatory factors provide novel insights into mammalian morphological modifications. Integr. Zool. 2018, 13, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.B.; Zhang, Y.; Wang, K. Perspectives on studying molecular adaptations of amphibians in the genomic era. Zool. Res. 2020, 41, 351–364. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Fan, S.; Zhang, Y.; Xu, M.; Zhang, H.; Yang, Y.; Xia, W.; Liu, C.; Zhu, W.; Wang, H.; et al. The seahorse genome and the evolution of its specialized morphology. Nature 2016, 540, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Bloch, N.I. The evolution of opsins and color vision: Connecting genotype to a complex phenotype. Acta Biol. Colomb. 2016, 21, 481–494. [Google Scholar] [CrossRef]
- Bian, C.; Huang, Y.; Li, J.; You, X.; Yi, Y.; Ge, W.; Shi, Q. Divergence, evolution and adaptation in ray-finned fish genomes. Sci. China Life Sci. 2019, 62, 1003–1018. [Google Scholar] [CrossRef]
- Katti, C.; Stacey-Solis, M.; Coronel-Rojas, N.A.; Davies, W.I.L. The Diversity and Adaptive Evolution of Visual Photopigments in Reptiles. Front. Ecol. Evol. 2019, 7, 352. [Google Scholar] [CrossRef]
- Musilova, Z.; Salzburger, W.; Cortesi, F. The Visual Opsin Gene Repertoires of Teleost Fishes: Evolution, Ecology, and Function. Annu. Rev. Cell Dev. Biol. 2021, 37, 441–468. [Google Scholar] [CrossRef]
- Torres-Dowdall, J.; Pierotti, M.E.R.; Härer, A.; Karagic, N.; Woltering, J.M.; Henning, F.; Elmer, K.R.; Meter, A. Rapid and Parallel Adaptive Evolution of the Visual System of Neotropical Midas Cichlid Fishes. Mol. Biol. Evol. 2017, 34, 2469–2485. [Google Scholar] [CrossRef]
- Lin, J.J.; Wang, F.Y.; Li, W.H.; Wang, T.Y. The rises and falls of opsin genes in 59 ray-finned fish genomes and their implications for environmental adaptation. Sci. Rep. 2017, 7, 15568. [Google Scholar] [CrossRef]
- Carleton, K.L.; Dalton, B.E.; Escobar-Camacho, D.; Nandamuri, S.P. Proximate and ultimate causes of variable visual sensitivities: Insights from cichlid fish radiations. Genesis 2016, 54, 299–325. [Google Scholar] [CrossRef]
- Hauser, F.E.; Ilves, K.L.; Schott, R.K.; Alvi, E.; López-Fernández, H.; Chang, B.S.W. Evolution, inactivation and loss of short wavelength-sensitive opsin genes during the diversification of Neotropical cichlids. Mol. Ecol. 2021, 30, 1688–1703. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Fan, Y.; Yue, Y.; Li, P.; Yan, J.; Zhou, K. Molecular Evolution of Visual Opsin Genes during the Behavioral Shifts between Different Photic Environments in Geckos. Asian Herpetol. Res. 2021, 12, 280–288. [Google Scholar] [CrossRef]
- Dungan, S.Z.; Kosyakov, A.; Chang, B.S. Spectral Tuning of Killer Whale (Orcinus orca) Rhodopsin: Evidence for Positive Selection and Functional Adaptation in a Cetacean Visual Pigment. Mol. Biol. Evol. 2016, 33, 323–336. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Shao, F.; Tao, W.; Liu, Z.; Long, J.; Wang, X.; Zhang, S.; Zhao, Q.; Carleton, K.L.; Kocher, T.D. Chromosome-level assembly of southern catfish (Silurus meridionalis) provides insights into visual adaptation to nocturnal and benthic lifestyles. Mol. Ecol. Resour. 2021, 21, 1575–1592. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, Y.; Zhang, W.; Du, X.; Liu, J. Benthic visual adaptation by fine-tuning light sensitivity in Japanese flounder (Paralichthys olivaceus). Front. Mar. Sci. 2022, 9, 1019660. [Google Scholar] [CrossRef]
- Ito, R.K.; Harada, S.; Tabata, R.; Watanabe, K. Molecular evolution and convergence of the rhodopsin gene in Gymnogobius, a goby group having diverged into coastal to freshwater habitats. J. Evol. Biol. 2021, 35, 333–346. [Google Scholar] [CrossRef]
- Ishengoma, E.; Agaba, M.; Cavener, D.R. Evolutionary analysis of vision genes identifies potential drivers of visual differences between giraffe and okapi. PeerJ 2017, 5, e3145. [Google Scholar] [CrossRef]
- Wu, Y.; Hadly, E.A.; Teng, W.; Hao, Y.; Liang, W.; Liu, Y.; Wang, H. Retinal transcriptome sequencing sheds light on the adaptation to nocturnal and diurnal lifestyles in raptors. Sci. Rep. 2016, 6, 33578. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, H.; Hadly, E.A. Invasion of Ancestral Mammals into Dim-light Environments Inferred from Adaptive Evolution of the Phototransduction Genes. Sci. Rep. 2017, 7, 46542. [Google Scholar] [CrossRef]
- Luehrmann, M.; Stieb, S.M.; Carleton, K.L.; Pietzker, A.; Cheney, K.L.; Marshall, N.J. Short-term colour vision plasticity on the reef: Changes in opsin expression under varying light conditions differ between ecologically distinct fish species. J. Exp. Biol. 2018, 221, jeb175281. [Google Scholar] [CrossRef]
- Rennison, D.J.; Owens, G.L.; Heckman, N.; Schluter, D.; Veen, T. Rapid adaptive evolution of colour vision in the threespine stickleback radiation. Proc. Biol. Sci. 2016, 283, 20160242. [Google Scholar] [CrossRef]
- Mehta, T.K.; Koch, C.; Nash, W.; Knaack, S.A.; Sudhakar, P.; Olbei, M.; Bastkowski, S.; Penso-Dolfin, L.; Korcsmaros, T.; Haerty, W.; et al. Evolution of regulatory networks associated with traits under selection in cichlids. Genome Biol. 2021, 22, 25. [Google Scholar] [CrossRef] [PubMed]
- Mehta, T.K.; Penso-Dolfin, L.; Nash, W.; Roy, S.; Di-Palma, F.; Haerty, W. Evolution of miRNA-Binding Sites and Regulatory Networks in Cichlids. Mol. Biol. Evol. 2022, 39, 146. [Google Scholar] [CrossRef] [PubMed]
- Wilwert, E.; Etienne, R.S.; van de Zande, L.; Maan, M.E. Contribution of opsins and chromophores to cone pigment variation across populations of Lake Victoria cichlids. J. Fish Biol. 2022, 101, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Mohun, S.M.; Davies, W.I.L. The Evolution of Amphibian Photoreception. Front. Ecol. Evol. 2019, 7, 321. [Google Scholar] [CrossRef]
- van Nynatten, A.; Janzen, F.H.; Brochu, K.; Maldonado-Ocampo, J.A.; Crampton, W.G.R.; Chang, B.S.W. To see or not to see: Molecular evolution of the rhodopsin visual pigment in neotropical electric fishes. Proc. Biol. Sci. 2019, 286, 20191182. [Google Scholar] [CrossRef]
- Wu, Y. Widespread nocturnality of living birds stemming from their common ancestor. BMC Evol. Biol. 2019, 19, 189. [Google Scholar] [CrossRef] [PubMed]
- Damsgaard, C.; Lauridsen, H.; Harter, T.S.; Kwan, G.T.; Thomsen, J.S.; Funder, A.M.; Supuran, C.T.; Tresguerres, M.; Matthews, P.G.; Brauner, C.J. A novel acidification mechanism for greatly enhanced oxygen supply to the fish retina. eLife 2020, 9, e58995. [Google Scholar] [CrossRef]
- Partha, R.; Chauhan, B.K.; Ferreira, Z.; Robinson, J.D.; Lathrop, K.; Nischal, K.K.; Chikina, M.; Ckarmk, N.L. Subterranean mammals show convergent regression in ocular genes and enhancers, along with adaptation to tunneling. eLife 2017, 6, e25884. [Google Scholar] [CrossRef]
- Borges, R.; Fonseca, J.; Gomes, C.; Johnson, W.E.; O’Brien, S.J.; Zhang, G.; Gilbers, M.T.P.; Jarvis, E.D.; Atunes, A. Avian Binocularity and Adaptation to Nocturnal Environments: Genomic Insights from a Highly Derived Visual Phenotype. Genome Biol. Evol. 2019, 11, 2244–2255. [Google Scholar] [CrossRef]
- Baldwin, M.W.; Ko, M.C. Functional evolution of vertebrate sensory receptors. Horm. Behav. 2020, 124, 104771. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Shi, L.; Li, X.; Dong, Q.; Sun, H.; Du, Y.; Zhang, Y.; Shao, T.; Cheng, H.; Chen, W.; et al. Genome-wide adaptive evolution to underground stresses in subterranean mammals: Hypoxia adaption, immunity promotion, and sensory specialization. Ecol. Evol. 2020, 10, 7377–7388. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chen, H.; Li, C.; Chen, S.; Xiao, H. Comparative Transcriptomics Reveals the Molecular Genetic Basis of Cave Adaptability in Sinocyclocheilus Fish Species. Front. Ecol. Evol. 2020, 8, 636503. [Google Scholar] [CrossRef]
- Yuan, Z.; Liu, S.; Zhou, T.; Tian, C.; Bao, L.; Dunham, R.; Liu, Z. Comparative genome analysis of 52 fish species suggests differential associations of repetitive elements with their living aquatic environments. BMC Genom. 2018, 19, 141. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Bao, L.; Zhou, T.; Yuan, Z.; Liu, S.; Dunham, R.; Li, Y.; Wang, K.; Xu, X.; Jin, Y.; et al. Genome sequence of walking catfish (Clarias batrachus) provides insights into terrestrial adaptation. BMC Genom. 2018, 19, 952. [Google Scholar] [CrossRef]
- Tian, R.; Yin, D.; Liu, Y.; Seim, I.; Xu, S.; Yang, G. Adaptive Evolution of Energy Metabolism-Related Genes in Hypoxia-Tolerant Mammals. Front. Genet. 2017, 8, 205. [Google Scholar] [CrossRef]
- Sharma, V.; Hecker, N.; Roscito, J.G.; Foerster, L.; Langer, B.E.; Hiller, M. A genomics approach reveals insights into the importance of gene losses for mammalian adaptations. Nat. Commun. 2018, 9, 1215. [Google Scholar] [CrossRef]
- Zhou, X.; Sun, D.; Guang, X.; Ma, S.; Fang, X.; Mariotti, M.; Nielsen, R.; Gladyshev, V.N.; Yang, G. Molecular Footprints of Aquatic Adaptation Including Bone Mass Changes in Cetaceans. Genome Biol. Evol. 2018, 10, 967–975. [Google Scholar] [CrossRef]
- Zhang, X.; Chi, H.; Li, G.; Irwin, D.M.; Zhang, S.; Rossiter, S.J.; Liu, Y. Parallel Independent Losses of G-Type Lysozyme Genes in Hairless Aquatic Mammals. Genome Biol. Evol. 2021, 13, evab201. [Google Scholar] [CrossRef]
- Chikina, M.; Robinson, J.D.; Clark, N.L. Hundreds of Genes Experienced Convergent Shifts in Selective Pressure in Marine Mammals. Mol. Biol. Evol. 2016, 33, 2182–2192. [Google Scholar] [CrossRef]
- Silva, M.C.; Chibucos, M.; Munro, J.B.; Daugherty, S.; Coelho, M.M.; Silva, J.C. Signature of adaptive evolution in olfactory receptor genes in Cory’s Shearwater supports molecular basis for smell in procellariiform seabirds. Sci. Rep. 2020, 10, 543. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Zhou, X.; Yu, Z.; Xu, S.; Seim, I.; Yang, G. Accelerated evolution and diversifying selection drove the adaptation of cetacean bone microstructure. BMC Evol. Biol. 2019, 19, 194. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Liu, Y.; Sun, X.; Lin, Y.; Yin, D.; Xu, S.; Yang, G. Insights into body size variation in cetaceans from the evolution of body-size-related genes. BMC Evol. Biol. 2019, 19, 157. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Shang, S.; Fang, N.; Zhu, Y.; Zhang, J.; Irwin, D.M.; Zhang, S.; Wang, Z. Accelerated Evolution of Limb-Related Gene Hoxd11 in the Common Ancestor of Cetaceans and Ruminants (Cetruminantia). G3 (Bethesda Md.) 2020, 10, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Wei, H.; Bi, J.; Ding, X.; Li, L.; Xu, S.; Yang, G.; Ren, W. Insights into Dietary Switch in Cetaceans: Evidence from Molecular Evolution of Proteinases and Lipases. J. Mol. Evol. 2020, 88, 521–535. [Google Scholar] [CrossRef]
- Mori, S.; Matsunami, M. Signature of positive selection in mitochondrial DNA in Cetartiodactyla. Genes Genet. Syst. 2018, 93, 65–73. [Google Scholar] [CrossRef]
- Tian, R.; Geng, Y.; Yang, Y.; Seim, I.; Yang, G. Oxidative stress drives divergent evolution of the glutathione peroxidase (GPX) gene family in mammals. Integr. Zool. 2021, 16, 696–711. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, S.; Lyu, T.; Shi, L.; Dong, Y.; He, S.; Zhang, H. Comparative Genome Analysis Reveals the Genomic Basis of Semi-Aquatic Adaptation in American Mink (Neovison vison). Animals 2022, 12, 2385. [Google Scholar] [CrossRef]
- Qu, Y.; Chen, C.; Chen, X.; Hao, Y.; She, H.; Wang, M.; Ericson, P.; Lin, H.; Cai, T.; Song, G.; et al. The evolution of ancestral and species-specific adaptations in snowfinches at the Qinghai-Tibet Plateau. Proc. Natl. Acad. Sci. USA 2021, 118, e2012398118. [Google Scholar] [CrossRef]
- Tian, R.; Seim, I.; Ren, W.; Xu, S.; Yang, G. Contraction of the ROS Scavenging Enzyme Glutathione S-Transferase Gene Family in Cetaceans. G3 (Bethesda Md.) 2019, 9, 2303–2315. [Google Scholar] [CrossRef]
- Yang, L.; Wang, Y.; Sun, N.; Chen, J.; He, S. Genomic and functional evidence reveals convergent evolution in fishes on the Tibetan Plateau. Mol. Ecol. 2021, 30, 5752–5764. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Wang, S.; Wang, Y.F.; Li, S.; Wu, S.W.; Yan, R.G.; Zhang, Y.W.; Wan, R.D.; He, Z.; Song, R.D.; et al. Long read genome assemblies complemented by single cell RNA-sequencing reveal genetic and cellular mechanisms underlying the adaptive evolution of yak. Nat. Commun. 2022, 13, 4887. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Ge, D.; Wen, Z.; Xia, L.; Yang, Q. Evolutionary Genetics of Hypoxia and Cold Tolerance in Mammals. J. Mol. Evol. 2018, 86, 618–634. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.D.; Yang, C.P.; Wang, M.S.; Dong, K.Z.; Yan, D.W.; Hao, Z.Q.; Fan, S.; Chu, S.; Shen, Q.; Jiang, L.; et al. Convergent genomic signatures of high-altitude adaptation among domestic mammals. Natl. Sci. Rev. 2020, 7, 952–963. [Google Scholar] [CrossRef]
- Ma, X.; Dai, W.; Kang, J.; Yang, L.; He, S. Comprehensive Transcriptome Analysis of Six Catfish Species from an Altitude Gradient Reveals Adaptive Evolution in Tibetan Fishes. G3 (Bethesda Md.) 2016, 6, 141–148. [Google Scholar] [CrossRef]
- Tian, R.; Losilla, M.; Lu, Y.; Yang, G.; Zakon, H. Molecular evolution of globin genes in Gymnotiform electric fishes: Relation to hypoxia tolerance. BMC Evol. Biol. 2017, 17, 51. [Google Scholar] [CrossRef]
- Bartáková, V.; Bryjová, A.; Nicolas, V.; Lavrenchenko, L.A.; Bryja, J. Mitogenomics of the endemic Ethiopian rats: Looking for footprints of adaptive evolution in sky islands. Mitochondrion 2021, 57, 182–191. [Google Scholar] [CrossRef]
- Xu, Q.; Zhang, C.; Zhang, D.; Jiang, H.; Peng, S.; Liu, Y.; Zhao, K.; Wang, C.; Chen, L. Analysis of the erythropoietin of a Tibetan Plateau schizothoracine fish (Gymnocypris dobula) reveals enhanced cytoprotection function in hypoxic environments. BMC Evol. Biol. 2016, 16, 11. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, Q.; Liu, Y.; Xu, Q. Characterization of EPO H131S as a key mutation site in the hypoxia-adaptive evolution of Gymnocypris dobula. Fish Physiol. Biochem. 2022, 48, 723–733. [Google Scholar] [CrossRef]
- Gibbons, T.C.; Metzger, D.; Healy, T.M.; Schulte, P.M. Gene expression plasticity in response to salinity acclimation in threespine stickleback ecotypes from different salinity habitats. Mol. Ecol. 2017, 26, 2711–2725. [Google Scholar] [CrossRef]
- Verta, J.P.; Jones, F.C. Predominance of cis-regulatory changes in parallel expression divergence of sticklebacks. eLife 2019, 8, e43785. [Google Scholar] [CrossRef] [PubMed]
- Kitano, J.; Ishikawa, A.; Kusakabe, M. Parallel transcriptome evolution in stream threespine sticklebacks. Dev. Growth Differ. 2019, 61, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Hart, J.C.; Ellis, N.A.; Eisen, M.B.; Miller, C.T. Convergent evolution of gene expression in two high-toothed stickleback populations. PLoS Genet. 2018, 14, e1007443. [Google Scholar] [CrossRef] [PubMed]
- Roberts Kingman, G.A.; Vyas, D.N.; Jones, F.C.; Brady, S.D.; Chen, H.I.; Reid, K.; Milhaven, M.; Bertino, T.S.; Aguirre, W.E.; Heins, D.C.; et al. Predicting future from past: The genomic basis of recurrent and rapid stickleback evolution. Sci. Adv. 2021, 7, eabg5285. [Google Scholar] [CrossRef]
- Deng, Z.; Hui, L.; He, C.; Shou, C.; Han, Z. Heat shock protein 70 (Hsp70) and heat shock transcription factor (Hsf) gene families in Cynoglossus semilaevis: Genome-wide identification and correlation analysis in response to low salinity stress. Mar. Freshw. Resh. 2021, 72, 1132–1141. [Google Scholar] [CrossRef]
- Barth, J.; Berg, P.R.; Jonsson, P.R.; Bonanomi, S.; Corell, H.; Hemmer-Hansen, J.; Jakobsen, K.S.; Johannesson, K.; Jorde, P.E.; Knutsen, H.; et al. Genome architecture enables local adaptation of Atlantic cod despite high connectivity. Mol. Ecol. 2017, 26, 4452–4466. [Google Scholar] [CrossRef]
- Ahmad, S.F.; Singchat, W.; Panthum, T.; Srikulnath, K. Impact of Repetitive DNA Elements on Snake Genome Biology Evolution. Cells 2021, 10, 1707. [Google Scholar] [CrossRef]
- Indjeian, V.B.; Kingman, G.A.; Jones, F.C.; Guenther, C.A.; Grimwood, J.; Schmutz, J.; Myers, R.M.; Kingsley, D.M. Evolving New Skeletal Traits by cis-Regulatory Changes in Bone Morphogenetic Proteins. Cell 2016, 164, 45–56. [Google Scholar] [CrossRef]
- Peichel, C.L.; Marques, D.A. The genetic and molecular architecture of phenotypic diversity in sticklebacks. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372, 20150486. [Google Scholar] [CrossRef]
- Xie, P.; Yi, S.K.; Yao, H.; Chi, W.; Guo, Y.; Ma, X.F.; Wang, F.P. Comparative transcriptome analysis reveals potential evolutionary differences in adaptation of temperature and body shape among four Percidae species. PLoS ONE 2019, 14, e0215933. [Google Scholar] [CrossRef]
- Reid, K.; Bell, M.A.; Veeramah, K.R. Threespine Stickleback: A Model System for Evolutionary Genomics. Annu. Rev. Genom. Hum. Genet. 2021, 22, 357–383. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.T.; Wang, G.; Thompson, A.C.; Wucherpfennig, J.I.; Reimchen, T.E.; MacColl, A.D.C. DNA fragility in the parallel evolution of pelvic reduction in stickleback fish. Science 2018, 363, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Archambeault, S.L.; Bärtschi, L.R.; Merminod, A.D.; Peichel, C.L. Adaptation via pleiotropy and linkage: Association mapping reveals a complex genetic architecture within the stickleback Eda locus. Evol. Lett. 2020, 4, 282–301. [Google Scholar] [CrossRef] [PubMed]
- Howes, T.R.; Summers, B.R.; Kingsley, D.M. Dorsal spine evolution in threespine sticklebacks via a splicing change in MSX2A. BMC Biol. 2017, 15, 115. [Google Scholar] [CrossRef] [PubMed]
- Greenway, R.; Barts, N.; Henpita, C.; Brown, A.P.; Arias Rodriguez, L.; Rodríguez Peña, C.M.; Arndt, S.; Lau, G.Y.; Murphy, M.P.; Wu, L.; et al. Convergent evolution of conserved mitochondrial pathways underlies repeated adaptation to extreme environments. Proc. Natl. Acad. Sci. USA 2020, 117, 16424–16430. [Google Scholar] [CrossRef]
- Dong, C.; Duan, X.; Younis, L.M.; Zhang, M.; Ma, X.; Chen, B.; Li, X.; Xu, P. Mitogenomic Perspectives on the Adaptation to Extreme Alkaline Environment of Amur ide (Leuciscus waleckii). Mar. Biotechnol. 2020, 22, 220–232. [Google Scholar] [CrossRef]
- Wang, S.; Kuang, Y.; Liang, L.; Sun, B.; Zhao, X.; Zhang, L.; Chang, Y. Resequencing and SNP discovery of Amur ide (Leuciscus waleckii) provides insights into local adaptations to extreme environments. Sci. Rep. 2021, 11, 5064. [Google Scholar] [CrossRef]
- Xu, J.; Li, J.T.; Jiang, Y.; Peng, W.; Yao, Z.; Chen, B.; Jiang, L.; Feng, J.; Ji, P.; Liu, G.; et al. Genomic Basis of Adaptive Evolution: The Survival of Amur Ide (Leuciscus waleckii) in an Extremely Alkaline Environment. Mol. Biol. Evol. 2017, 34, 145–159. [Google Scholar] [CrossRef]
- Jia, J.; Qin, J.; Yuan, X.; Liao, Z.; Huang, J.; Wang, B.; Sun, C.; Li, W. Microarray and metabolome analysis of hepatic response to fasting and subsequent refeeding in zebrafish (Danio rerio). BMC Genom. 2019, 20, 919. [Google Scholar] [CrossRef]
- Chebii, V.J.; Oyola, S.O.; Kotze, A.; Domelevo Entfellner, J.B.; Musembi Mutuku, J.; Agaba, M. Genome-Wide Analysis of Nubian Ibex Reveals Candidate Positively Selected Genes That Contribute to Its Adaptation to the Desert Environment. Animals 2020, 10, 2181. [Google Scholar] [CrossRef]
- Elayadeth-Meethal, M.; Thazhathu Veettil, A.; Asaf, M.; Pramod, S.; Maloney, S.K.; Martin, G.B.; Rivero, M.J.; Sejian, V.; Naseef, P.P.; Kuruniyan, M.S.; et al. Comparative Expression Profiling and Sequence Characterization of ATP1A1 Gene Associated with Heat Tolerance in Tropically Adapted Cattle. Animals 2021, 11, 2368. [Google Scholar] [CrossRef] [PubMed]
- Lighten, J.; Incarnato, D.; Ward, B.J.; van Oosterhout, C.; Bradbury, I.; Hanson, M.; Bentzen, P. Adaptive phenotypic response to climate enabled by epigenetics in a K-strategy species, the fish Leucoraja ocellata (Rajidae). R. Soc. Open Sci. 2016, 3, 160299. [Google Scholar] [CrossRef]
- Zhuang, X.; Yang, C.; Murphy, K.R.; Cheng, C.C. Molecular mechanism and history of non-sense to sense evolution of antifreeze glycoprotein gene in northern gadids. Proc. Natl. Acad. Sci. USA 2019, 116, 4400–4405. [Google Scholar] [CrossRef] [PubMed]
- Irwin, D.M. Duplication and diversification of insulin genes in ray-finned fish. Zool. Res. 2019, 40, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Yamagishi, G.; Yoshida, A.; Kobayashi, A.; Park, M.K. Molecular characterization of insulin from squamate reptiles reveals sequence diversity and possible adaptive evolution. Gen. Comp. Endocrinol. 2016, 225, 197–211. [Google Scholar] [CrossRef]
- Chen, Y.H.; Zhao, H. Evolution of digestive enzymes and dietary diversification in birds. PeerJ 2019, 7, e6840. [Google Scholar] [CrossRef]
- Janiak, M.C.; Burrell, A.S.; Orkin, J.D.; Disotell, T.R. Duplication and parallel evolution of the pancreatic ribonuclease gene (RNASE1) in folivorous non-colobine primates, the howler monkeys (Alouatta spp.). Sci. Rep. 2019, 9, 20366. [Google Scholar] [CrossRef]
- Song, J.; Zheng, H.; Xue, J.; Liu, J.; Sun, Q.; Yang, W. GPR15-C10ORF99 functional pairing initiates colonic Treg homing in amniotes. EMBO Rep. 2022, 23, e53246. [Google Scholar] [CrossRef]
- Endo, Y.; Kamei, K.I.; Inoue-Murayama, M. Genetic signatures of lipid metabolism evolution in Cetacea since the divergence from terrestrial ancestor. J. Evol. Biol. 2018, 31, 1655–1665. [Google Scholar] [CrossRef]
- Gao, X.; Li, Y.; Adetula, A.A.; Wu, Y.; Chen, H. Analysis of new retrogenes provides insight into dog adaptive evolution. Ecol. Evol. 2019, 9, 11185–11197. [Google Scholar] [CrossRef]
- Shan, L.; Wu, Q.; Wang, L.; Zhang, L.; Wei, F. Lineage-specific evolution of bitter taste receptor genes in the giant and red pandas implies dietary adaptation. Integr. Zool. 2018, 13, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Shen, F.; Jie, X.; Zhang, L.; Yan, G.; Wu, H.; Huang, Y.; Hou, R.; Yue, B.; Zhang, X. Comparative Transcriptomics and Methylomics Reveal Adaptive Responses of Digestive and Metabolic Genes to Dietary Shift in Giant and Red Pandas. Genes 2022, 13, 1446. [Google Scholar] [CrossRef] [PubMed]
- Rinker, D.C.; Specian, N.K.; Zhao, S.; Gibbons, J.G. Polar bear evolution is marked by rapid changes in gene copy number in response to dietary shift. Proc. Natl. Acad. Sci. USA 2019, 116, 13446–13451. [Google Scholar] [CrossRef] [PubMed]
- Botero-Castro, F.; Tilak, M.K.; Justy, F.; Catzeflis, F.; Delsuc, F.; Douzery, E. In Cold Blood: Compositional Bias and Positive Selection Drive the High Evolutionary Rate of Vampire Bats Mitochondrial Genomes. Genome Biol. Evol. 2018, 10, 2218–2239. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.J.; Xia, J.M.; Wang, Q.; Yu, J.L.; Song, Z.; Zhao, H. Diet and Adaptive Evolution of Alanine-Glyoxylate Aminotransferase Mitochondrial Targeting in Birds. Mol. Biol. Evol. 2020, 37, 786–798. [Google Scholar] [CrossRef]
- McGlothlin, J.W.; Kobiela, M.E.; Feldman, C.R.; Castoe, T.A.; Geffeney, S.L.; Hanifin, C.T.; Toledo, G.; Vonk, F.J.; Richardson, M.K.; Brodie, E.D.; et al. Historical Contingency in a Multigene Family Facilitates Adaptive Evolution of Toxin Resistance. Curr. Biol. 2016, 26, 1616–1621. [Google Scholar] [CrossRef]
- Feldman, C.R.; Durso, A.M.; Hanifin, C.T.; Pfrender, M.E.; Ducey, P.K.; Stokes, A.N.; Barnett, K.E.; Brodie, E.D., III; Brodie, E.D., Jr. Is there more than one way to skin a newt? Convergent toxin resistance in snakes is not due to a common genetic mechanism. Heredity 2017, 119, 468. [Google Scholar] [CrossRef]
- Gendreau, K.L.; Hornsby, A.D.; Hague, M.T.J.; McGlothlin, J.W. Gene Conversion Facilitates the Adaptive Evolution of Self-Resistance in Highly Toxic Newts. Mol. Biol. Evol. 2021, 38, 4077–4094. [Google Scholar] [CrossRef]
- Wang, L.; Sun, L.; Wan, Q.H.; Fang, S.G. Comparative Genomics Provides Insights into Adaptive Evolution in Tactile-Foraging Birds. Genes 2022, 13, 678. [Google Scholar] [CrossRef]
- Cleves, P.A.; Hart, J.C.; Agoglia, R.M.; Jimenez, M.T.; Erickson, P.A.; Gai, L.; Miller, C.T. An intronic enhancer of Bmp6 underlies evolved tooth gain in sticklebacks. PLoS Genet. 2018, 14, e1007449. [Google Scholar] [CrossRef]
- Erickson, P.A.; Baek, J.; Hart, J.C.; Cleves, P.A.; Miller, C.T. Genetic Dissection of a Supergene Implicates Tfap2a in Craniofacial Evolution of Threespine Sticklebacks. Genetics 2018, 209, 591–605. [Google Scholar] [CrossRef] [PubMed]
- Karagic, N.; Schneider, R.F.; Meyer, A.; Hulsey, C.D. A Genomic Cluster Containing Novel and Conserved Genes is Associated with Cichlid Fish Dental Developmental Convergence. Mol. Biol. Evol. 2020, 37, 3165–3174. [Google Scholar] [CrossRef] [PubMed]
- Machado, J.P.; Philip, S.; Maldonado, E.; O’Brien, S.J.; Johnson, W.E.; Antunes, A. Positive Selection Linked with Generation of Novel Mammalian Dentition Patterns. Genome Biol. Evol. 2016, 8, 2748–2759. [Google Scholar] [CrossRef] [PubMed]
- Mu, Y.; Tian, R.; Xiao, L.; Sun, D.; Zhang, Z.; Xu, S.; Yang, G. Molecular Evolution of Tooth-Related Genes Provides New Insights into Dietary Adaptations of Mammals. J. Mol. Evol. 2021, 89, 458–471. [Google Scholar] [CrossRef] [PubMed]
- Conith, A.J.; Albertson, R.C. The cichlid oral and pharyngeal jaws are evolutionarily and genetically coupled. Nat. Commun. 2021, 12, 5477. [Google Scholar] [CrossRef]
- He, S.; Li, L.; Lv, L.Y.; Cai, W.J.; Dou, Y.Q.; Li, J.; Tang, S.L.; Chen, X.; Zhang, Z.; Xu, J.; et al. Mandarin fish (Sinipercidae) genomes provide insights into innate predatory feeding. Commun. Biol. 2020, 3, 361. [Google Scholar] [CrossRef]
- O’Gorman, M.; Thakur, S.; Imrie, G.; Moran, R.L.; Choy, S.; Sifuentes-Romero, I.; Bilandžija, H.; Renner, K.J.; Duboué, E.; Rohner, N.; et al. Pleiotropic function of the oca2 gene underlies the evolution of sleep loss and albinism in cavefish. Curr. Biol. 2021, 31, 3694–3701. [Google Scholar] [CrossRef]
- Xu, S.; Sun, X.; Niu, X.; Zhang, Z.; Tian, R.; Ren, W.; Zhou, K.; Yang, G. Genetic basis of brain size evolution in cetaceans: Insights from adaptive evolution of seven primary microcephaly (MCPH) genes. BMC Evol. Biol. 2017, 17, 206. [Google Scholar] [CrossRef]
- Huang, Y.; Chain, F.J.; Panchal, M.; Eizaguirre, C.; Kalbe, M.; Lenz, T.L.; Samonte, I.E.; Stoll, M.; Bornberg-Bauer, E.; Reusch, T.B.; et al. Transcriptome profiling of immne tissues reveals habitat-specific gene expression between lake and river sticklebacks. Mol. Ecol. 2016, 25, 943–958. [Google Scholar] [CrossRef]
- Ahmad, H.I.; Zhou, J.; Ahmad, M.J.; Afzal, G.; Jiang, H.; Zhang, X.; Elokil, A.A.; Khan, M.A.; Li, L.; Li, H.; et al. Adaptive selection in the evolution of programmed cell death-1 and its ligands in vertebrates. Aging 2020, 12, 3516–3557. [Google Scholar] [CrossRef]
- Xu, D.; Pavlidis, P.; Thamadilok, S.; Redwood, E.; Fox, S.; Blekhman, R. Recent evolution of the salivary mucin MUC7. Sci. Rep. 2016, 6, 31791. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Rong, X.; Li, G.; Wang, Y.; Chen, B.; Ren, W.; Yang, G.; Xu, S. Genomic organization and adaptive evolution of IGHC genes in marine mammals. Mol. Immunol. 2018, 99, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Shang, S.; Wu, X.; Zhong, H.; Zhao, C.; Wei, Q.; Zhang, H.; Xia, T.; Chen, Y.; Zhang, H.; et al. Genomic analysis and adaptive evolution of the RIG-I-like and NOD-like receptors in reptiles. Int. J. Biol. Macromol. 2019, 134, 1045–1051. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Wei, Y.; Zhu, Y.; Xia, Y.; Irwin, D.M.; Liu, Y. Adaptive Evolution of C-Type Lysozyme in Vampire Bats. J. Mol. Evol. 2019, 87, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.; Seim, I.; Zhang, Z.; Yang, Y.; Ren, W.; Xu, S.; Yang, G. Distinct evolution of toll-like receptor signaling pathway genes in cetaceans. Genes Genom. 2019, 41, 1417–1430. [Google Scholar] [CrossRef]
- Palomar, G.; Dudek, K.; Wielstra, B.; Jockusch, E.L.; Vinkler, M.; Arntzen, J.W.; Ficetola, G.F.; Matsunami, M.; Waldman, B.; Těšický, M.; et al. Molecular Evolution of Antigen-Processing Genes in Salamanders: Do They Coevolve with MHC Class I Genes? Genome Biol. Evol. 2021, 13, 259. [Google Scholar] [CrossRef]
- Ametrano, A.; Picchietti, S.; Guerra, L.; Giacomelli, S.; Oreste, U.; Coscia, M.R. Comparative Analysis of the pIgR Gene from the Antarctic Teleost Trematomus bernacchii Reveals Distinctive Features of Cold-Adapted Notothenioidei. Int. J. Mol. Sci. 2022, 23, 7783. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, T.; Chen, J.; Wu, L.; Wu, X.; Zhang, W.; Luo, J.; Xia, J.; Meng, Z.; Liu, X. Whole-genome sequencing of brown-marbled grouper (Epinephelus fuscoguttatus) provides insights into adaptive evolution and growth differences. Mol. Ecol. Resour. 2022, 22, 711–723. [Google Scholar] [CrossRef]
- Wang, S.; Joka, F.R.; Wang, X.; Bai, B. Analysis of the Phylogeny and Evolutionary Selection Pressure of the Mx Gene in 10 Wild Birds. Pak. J. Zool. 2019, 51, 1299–1307. [Google Scholar] [CrossRef]
- Yamada, E.; Yoshikawa, R.; Nakano, Y.; Misawa, N.; Kobayashi, T.; Ren, F.; Izumi, T.; Miyazawa, T.; Koyanagi, Y.; Sato, K. A naturally occurring bovine APOBEC3 confers resistance to bovine lentiviruses: Implication for the co-evolution of bovids and their lentiviruses. Sci. Rep. 2016, 6, 33988. [Google Scholar] [CrossRef]
- Yang, Y.; Wu, L.N.; Chen, J.F.; Wu, X.; Xia, J.H.; Meng, Z.N.; Liu, X.C.; Lin, H.R. Whole-genome sequencing of leopard coral grouper (Plectropomus leopardus) and exploration of regulation mechanism of skin color and adaptive evolution. Zool. Res. 2020, 41, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, S.K.; Gupta, A.; Saxena, R.; Prasoodanan, V.P.K.; Sharma, A.K.; Mittal, P.; Roy, A.; Shafer, A.; Vijay, N.; Sharma, V.K. Genome Sequence of Peacock Reveals the Peculiar Case of a Glittering Bird. Front. Genet. 2018, 9, 392. [Google Scholar] [CrossRef] [PubMed]
- Solbakken, M.H.; Voje, K.L.; Jakobsen, K.S.; Jentoft, S. Linking species habitat and past palaeoclimatic events to evolution of the teleost innate immune system. Proc. Biol. Sci. 2017, 284, 20162810. [Google Scholar] [CrossRef] [PubMed]
- Bagheri, M.; Zahmatkesh, A. Evolution and species-specific conservation of toll-like receptors in terrestrial vertebrates. Int. Rev. Immunol. 2018, 37, 217–228. [Google Scholar] [CrossRef]
- Shang, S.; Zhong, H.; Wu, X.; Wei, Q.; Zhang, H.; Chen, J.; Chen, Y.; Tang, X.; Zhang, H. Genomic evidence of gene duplication and adaptive evolution of Toll like receptors (TLR2 and TLR4) in reptiles. Int. J. Biol. Macromol. 2018, 109, 698–703. [Google Scholar] [CrossRef]
- Tian, R.; Chen, M.; Chai, S.; Rong, X.; Chen, B.; Ren, W.; Xu, S.; Yang, G. Divergent Selection of Pattern Recognition Receptors in Mammals with Different Ecological Characteristics. J. Mol. Evol. 2018, 86, 138–149. [Google Scholar] [CrossRef]
- Neves, F.; Águeda-Pinto, A.; Pinheiro, A.; Abrantes, J.; Esteves, P.J. Strong selection of the TLR2 coding region among the Lagomorpha suggests an evolutionary history that differs from other mammals. Immunogenetics 2019, 71, 437–443. [Google Scholar] [CrossRef]
- Voogdt, C.G.; Bouwman, L.I.; Kik, M.J.; Wagenaar, J.A.; van Putten, J.P. Reptile Toll-like receptor 5 unveils adaptive evolution of bacterial flagellin recognition. Sci. Rep. 2016, 6, 19046. [Google Scholar] [CrossRef]
- Liu, G.; Zhang, H.; Sun, G.; Zhao, C.; Shang, S.; Gao, X.; Xia, T.; Yang, X. Characterization of the peripheral blood transcriptome and adaptive evolution of the MHC I and TLR gene families in the wolf (Canis lupus). BMC Genom. 2017, 18, 584. [Google Scholar] [CrossRef]
- Velová, H.; Gutowska-Ding, M.W.; Burt, D.W.; Vinkler, M. Toll-Like Receptor Evolution in Birds: Gene Duplication, Pseudogenization, and Diversifying Selection. Mol. Biol. Evol. 2018, 35, 2170–2184. [Google Scholar] [CrossRef]
- Xu, S.; Tian, R.; Lin, Y.; Yu, Z.; Zhang, Z.; Niu, X.; Wang, X.; Yan, G. Widespread positive selection on cetacean TLR extracellular domain. Mol. Immunol. 2019, 106, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Schad, J.; Voigt, C.C. Adaptive evolution of virus-sensing toll-like receptor 8 in bats. Immunogenetics 2016, 68, 783–795. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, S.; Yang, Y.; Fu, Q.; Abebe, A.; Liu, Z. Identification of NF-κB related genes in channel catfish and their expression profiles in mucosal tissues after columnaris bacterial infection. Dev. Comp. Immunol. 2017, 70, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Cheng, Y.; Liu, Z.; Tao, W.; Zheng, S.; Wang, D. Bioinformatic analyses of zona pellucida genes in vertebrates and their expression in Nile tilapia. Fish Physiol. Biochem. 2018, 44, 435–449. [Google Scholar] [CrossRef]
- Killingbeck, E.E.; Wilburn, D.B.; Merrihew, G.E.; MacCoss, M.J.; Swanson, W.J. Proteomics support the threespine stickleback egg coat as a protective oocyte envelope. Mol. Reprod. Dev. 2021, 88, 500–515. [Google Scholar] [CrossRef]
- Vicens, A.; Treviño, C.L. Positive Selection in the Evolution of Mammalian CRISPs. J. Mol. Evol. 2018, 86, 635–645. [Google Scholar] [CrossRef]
- Isakov, N. Histocompatibility and Reproduction: Lessons from the Anglerfish. Life 2022, 12, 113. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, Y.; Liang, R.; Cui, C.; Zhu, Y.; Zhang, F.; Zhang, J.; Chen, X. Comparative transcriptome analysis provides insights into the molecular mechanisms of high-frequency hearing differences between the sexes of Odorrana tormota. BMC Genom. 2022, 23, 296. [Google Scholar] [CrossRef]
- Zhang, C.; Tong, C.; Ludwig, A.; Tang, Y.; Liu, S.; Zhang, R.; Feng, C.; Li, G.; Peng, Z.; Zhao, K. Adaptive Evolution of the Eda Gene and Scales Loss in Schizothoracine Fishes in Response to Uplift of the Tibetan Plateau. Int. J. Mol. Sci. 2018, 19, 2953. [Google Scholar] [CrossRef]
- Li, B.; Wang, H.; Li, A.; An, C.; Zhu, L.; Liu, S.; Zhuang, Z. The Landscape of DNA Methylation Generates Insight into Epigenetic Regulation of Differences Between Slow-Twitch and Fast-Twitch Muscles in Pseudocaranx dentex. Front. Mar. Sci. 2022, 9, 916373. [Google Scholar] [CrossRef]
- Lu, L.; Li, Y.; Liu, Z.; Liang, F.; Guo, F.; Yang, S.; Wang, D.; He, Y.; Xiong, J.; Li, D. Functional constraints on adaptive evolution of protein ubiquitination sites. Sci. Rep. 2017, 7, 39949. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Xing, C.; Lu, W.; Liu, Z.; Wang, X.; Cheng, J.; Zhang, Q. Rapid evolution of piRNA pathway and its transposon targets in Japanese flounder (Paralichthys olivaceus). Comp. Biochem. Physiol. Part D Genom. Proteom. 2019, 31, 100609. [Google Scholar] [CrossRef] [PubMed]
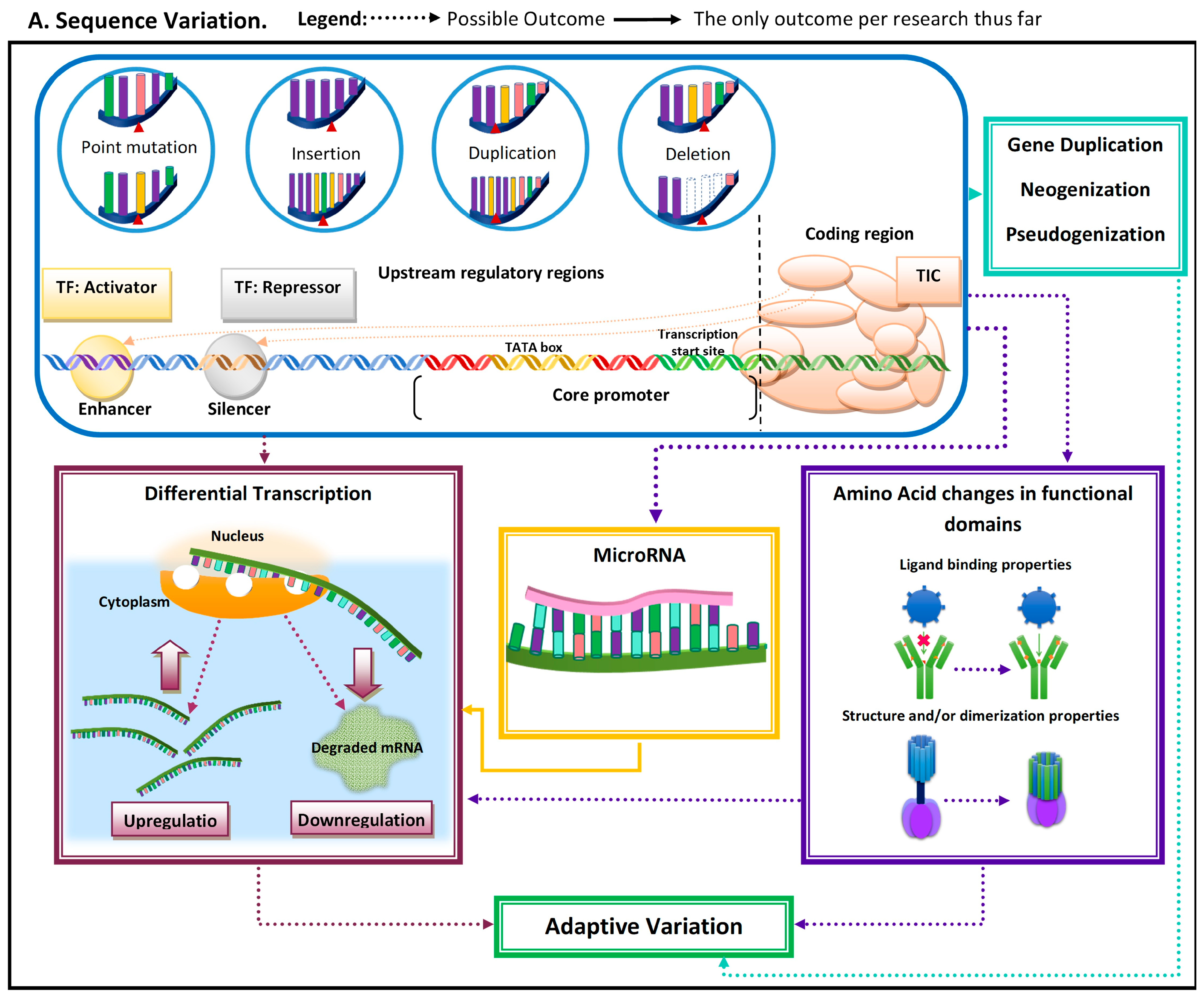
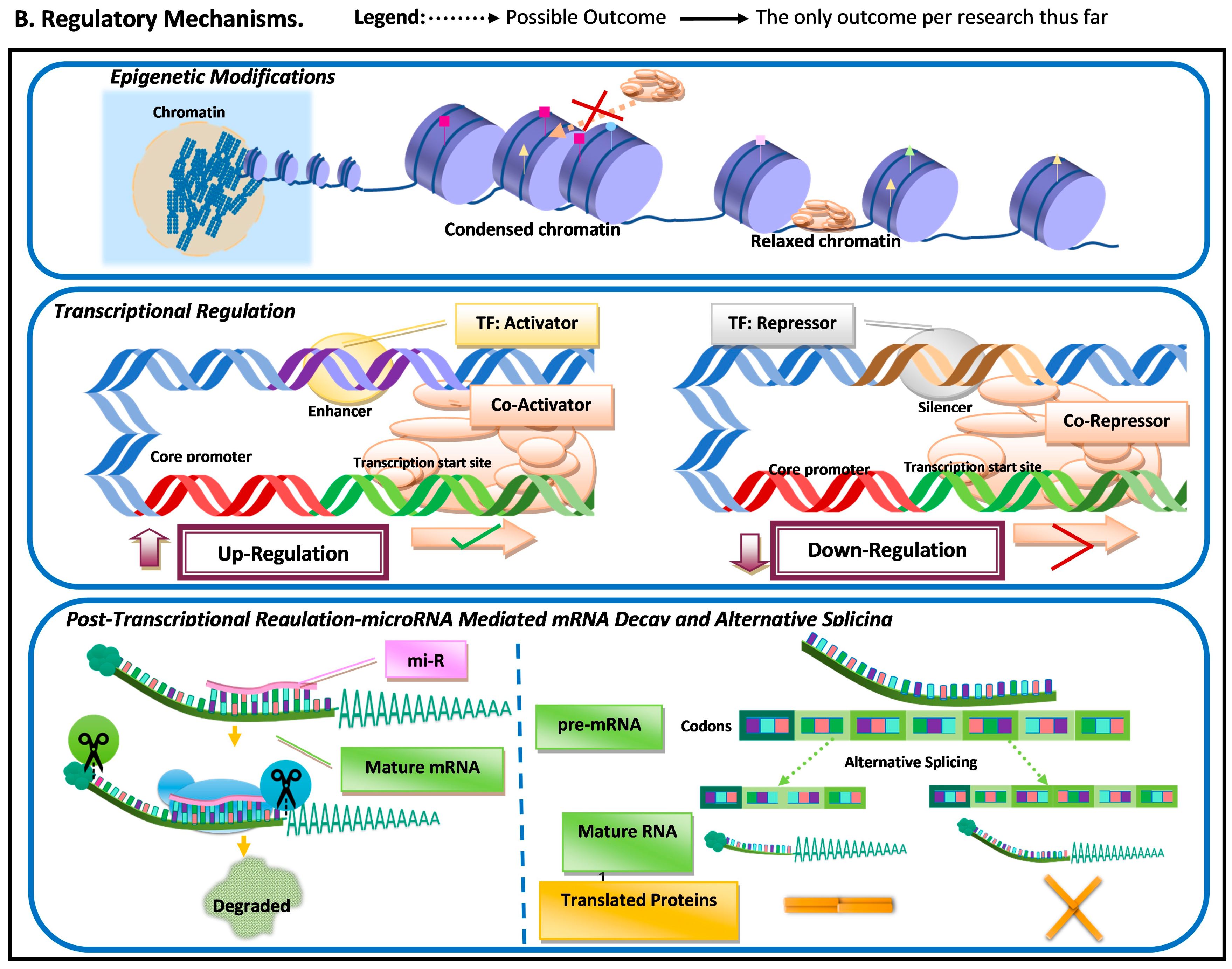
| Concept | Brief Description |
|---|---|
| Point mutation | Change in DNA sequence by a single nucleotide; it can be an insertion, duplication, or deletion. |
| Insertion | DNA sequence variation by insertion of one or more nucleotides. |
| Duplication | Modification in DNA sequence by duplication of a stretch of nucleotides (i.e., one or more). |
| Deletion | Change in DNA sequence by excision of one or more nucleotides. |
| Upstream regulatory regions | Regions upstream (before) from the core promoter to which transcription factors (TF) or coactivators can cis-regulate. Transcription factors can activate or increase transcription when bound to enhancers, while repressors can decrease transcription when bound to silencers. |
| Core promoter | Regulatory region where the transcription initiation complex (TIC) binds, as it has the transcription start site and can include a thymidine adenine-rich region (i.e., TATA box) where the TIC is recruited. |
| Coding region | DNA sequence that codes for the pre-mRNA, which is later modified to the mRNA and translated into proteins. |
| Upregulation | Regulatory change that leads to an increase in transcription of specific mRNA. |
| Gene duplication | Refers to the duplication of a complete gene. |
| Neogenization | DNA sequence gains a novel gene function and can occur in previously non-coding regions, more commonly in duplicated genes. |
| Pseudogenization | Process by which a gene either loses a function or becomes completely non-functional. |
| Downregulation | Regulatory change which leads to a decrease in transcription of specific messenger RNA (mRNA). |
| microRNAs (miRs) | Small RNA fragments that can hybridize with target mRNA and can lead to its decay, thus downregulating it. |
| Epigenetic modifications | Molecular modifications (e.g., acetylation, methylation, and phosphorylation) to histones, proteins involved in the packing of DNA. These modifications can impact chromatin condensation and/or RNA polymerase activity. |
| Chromatin | A protein–DNA molecular complex that is the natural state of Eukaryotic genetic material. |
| Transcriptional regulation | Processes that impact the rate of transcription, either upregulating or downregulating; these can be cis-regulatory elements (e.g., enhancers or silencers) or components of the transcription initiation complex (TIC) (e.g., co-activators and co-repressors). |
| Enhancer | Upstream regulatory element that can cis-regulate transcription (upregulation) by interacting with activator transcription factors, which in turn can interact with co-activators in the TIC. |
| Silencer | Upstream regulatory element that can cis-regulate transcription by interacting with repressor transcription factors, which in turn can interact with a co-repressor in the TIC, for disassociation from DNA. |
| Post-transcriptional regulation | Regulatory mechanisms that operate upon the mRNA transcript itself or in its maturation process. |
| microRNA-mediated mRNA decay | A mechanism in which mi-R hybridize with target DNA and recruit the Argo complex, which leads to removal of the protective elements of the mRNA: its cap and polyadenylation tail, which leads to degradation and subsequent downregulation of said transcript. |
| Splicing | A process that occurs during mRNA maturation, where segments of the raw transcript (pre-mRNA) are removed. |
| Alternative splicing | Alternative mRNA maturation processes that result in variations in the final mature RNA transcript and subsequent variation in the translated amino acid sequence, which in turn can result in functional variation in protein properties (e.g., dimerization, folding, and ligand affinity). |
| Adaptive Evolution in Visual Systems “in Light of” Varying Lighting Conditions | |||
| Abbreviation | Full Name | Gene Ontology | Mechanisms |
| CYP27C1 | Cytochrome P450 Family 27 | Cytochrome enzyme that catalyzes desaturation from Vitamin A1, converting it to Vitamin A2. | D |
| GRK1 | G-Protein-Coupled Receptor Kinase | G-Protein receptor kinase, phosphorylates rhodopsin and initiates deactivation. | S |
| LUM | Lumican | Proteoglycan that, in retinae, regulates organization of collagen fibers. | S |
| LWS | Long-Wavelength-Sensitive Opsin | G-protein-coupled receptor involved in the phototransduction process, sensitive to “reds” and “greens” between 501 and 573 nm. | S |
| RH1 | Rhodopsin | G-protein-coupled receptor involved in the phototransduction process expressed in rods, absorbs in 447–525 nm. | S, L |
| RH2 | Rhodopsin | G-protein-coupled receptor involved in the phototransduction process expressed in rods, absorbs in 452–537 nm. | D, S, L |
| RHO | Rhodopsin | G-protein-coupled receptor involved in the phototransduction activity. | D |
| SWS1 | Short-Wavelength-Sensitive Opsin | G-protein-coupled receptor involved in the phototransduction process, sensitive to “violet”, including ultraviolet between 347 and 383 nm. | D, L |
| SWS2 | Short-Wavelength-Sensitive Opsin 2 | G-protein-coupled receptor involved in the phototransduction process, sensitive to “blue-violet” between 397 and 482 nm. | D, S, L |
| VHA | Vacuolar-Type H+ -ATPase | Mitochondrial protein that participates in the excretion of H+ from endothelial cells into the lumen. | S |
| Adaptations for colonization of aquatic and terrestrial environments | |||
| ACAN | Aggrecan | Integral part of extracellular membrane within cartilaginous tissue. | S |
| AMPD3 | Adenosine Monophosphate Deaminase 3 | Highly regulated enzyme that hydrolytically deaminates adenosine monophosphate into inosine monophosphate within the adenylate catabolic pathway. | L |
| ATP8 | Mitochondrially Encoded ATP Synthetase Membrane Subunit 8 | ATP synthetase that produces ATP from ADP when a protein gradient is present. | S |
| CALHM1 | Calcium Homeostasis Modulator 1 | Calcium–ion channel involved in sweet, bitter, and umami taste transduction. | L |
| DSC1 | Desmocollin 1 | Calcium-dependent glycoprotein found primarily in epidermal cells constituting adhesive proteins within desmosome cell–cell junctions. | L |
| DSG4 | Desmocollin 4 | Transmembrane component of desmosomes. | L |
| ELK1 | ETS Transcription Factor ELK1 | Transcription factor that regulates expression by binding to the promoter of the serum response factor gene. | - |
| FSHR | Follicle-Stimulating Hormone Receptor | Receptor for the follicle-stimulating hormone and functions in gonad development. | S |
| GNAT3 | G Protein Subunit α Transducin 3 | G protein subunit involved in bitter, sweet, and umami taste transduction. | L |
| GSDMA | Gasdermin A | Precursor protein of a cell-pore-forming protein. | L |
| HOX | Homeobox Domain Genes | Family of transcription factor genes involved in body plan along animal bilateral axis. | - |
| HOXD11 | Homeobox D11 | Transcription factor part of a family involved in limb and genital development. | S |
| HOXD12 | Homeobox D12 | Transcription factor part of a family involved in limb and genital development. | S |
| HOXD13 | Homeobox D13 | Transcription factor part of a family involved in limb and genital development. | S |
| KRT20 | Keratin 20 | Intermediate filament conferring structural integrity to epidermal cells. | L |
| KRT9 | Keratin 9 | Intermediate filament chain expressed in terminally differentiated epidermal cells. | L |
| LHX3 | LIM Homeobox 3 | Transcription factor required for pituitary development and motor neuron specification. | - |
| LYG1 | Lysozyme G1 | Lysozyme activity | L |
| LYG2 | Lysozyme G2 | Lysozyme activity | |
| MB | Myoglobin | Iron- and oxygen-binding protein typically present in skeletal muscle tissue. | E |
| MLL | Lysine Methyltransferase 2A | Currently called KMT2A, co-activator involved in transcriptional regulation of genes during early development and hematopoiesis. | S |
| MMP12 | Matrix Metalloproteinase 12 | Involved in extracellular matrix breakdown | L |
| ORA1 | Olfactory Receptor Class-A-Like-Protein-1 | Olfactory receptor. | E |
| PIT-1 | POU Class 1 Homeobox 1 | Transcription factor involved in regulating expression of multiple pituitary development genes and hormones. | S |
| SULT6B1 | Sulfotransferase Family 6B Member 1 | Sulfotransferase. | E |
| TGM5 | Transglutaminase 5 | Transglutaminase, enzyme that catalyzes crosslinking between glutamine and lysine residues. | L |
| Adapting to extreme environmental conditions (i.e., hypoxia, salinity, and low temperatures) | |||
| ABCA12 | ATP-Binding-Cassette Subfamily A Member 12 | ATP-Binding-Cassette transporter protein. | S |
| AFGP | Antifreeze Glycoprotein | Proteins that can inhibit growth of ice. | S |
| ALDOA | Aldolase | Glycolytic enzyme. | S |
| BMP6 | Bone Morphogenetic Protein 6 | Ligand of the transforming growth factor β, involved in multiple regulatory processes (e.g., iron homeostasis, fat and bone development, and ovulation). | S |
| CA | Carbonic Anhydrase | Family of genes of zinc metalloenzymes that catalyze reversible hydration of carbon dioxide. | E |
| COX1 | Cytochrome C Oxidase I | Mitochondrial protein, component of the cytochrome C oxidase. | S |
| COX3 | Cytochrome C Oxidase III | Mitochondrial protein, components of the cytochrome C oxidase. | S |
| CSAD | Cysteine Sulfinic Acid Decarboxylase | Member of the group 2 decarboxylase. | D |
| CTH | Cystathionine γ-Lyase | Cytoplasmic enzyme involved in the transculturation pathway. | D |
| EDA | Ectodysplasin A | Membrane protein thought to be involved in cell–cell signaling during development. | S |
| ENO | Enolase | Glycolytic enzyme. | S |
| EPAS1 | Endothelial Pas Domain Protein 1 | Transcription factor involved in the regulation of genes that are controlled by oxygen. | S |
| EPO | Erythropoietin | Secreted glycosylated cytokine involved in promoting red blood cell production. | D, S |
| FBP1 | Fructose-Bisphosphatase 1 | Glucogenesis regulatory enzyme. | S |
| FBP2 | Fructose-Bisphosphatase 2 | Glucogenesis regulatory enzyme. | D |
| G6PCB | Glucose-6-Phosphatase | Involved in the gluconeogenesis pathway. | D |
| GDF6 | Growth Differentiation Factor 6 | Ligand of the transforming growth factor β, involved in regulation of genes associated with formation of some bones, joints, limbs, skull, and axial skeleton. | S |
| GLA | Galactosidase α | Glycoprotein involved in termina hydrolyses from glycolipids and glycoproteins. | S |
| GPI | Glucose-6-Phosphate Isomerase | Multiple functions; glycolytic enzyme and neurotrophic factor that promote survival of skeletal motor neurons and sensory neurons intracellularly and extracellularly, respectively. | S |
| GST | Gluthathione-S-transferase | Conjugation of reduced glutathionone, important in detoxification. | E |
| HB | Hemoglobin | Transport of oxygen. | S |
| IGFLR1 | IGF-Like Family Receptor 1 | Possibly a cell membrane receptor for IGF-like proteins. | S |
| KITLG | KIT Ligand | Ligand of the tyrosine-kinase receptor, a pleiotropic factor involved in embryonic development. | S |
| LDHA | Lactate Dehydrogenase A | Involved in the pyruvate fermentation to lactate pathway. | S |
| LDHD | Lactate Dehydrogenase D | Lactate dehydrogenase. | S |
| NDUFA10 | NADH Dehydrogenase 1 α Subcomplex Subunit 10 | Subunit of the first enzyme complex in the electron transport chain in the inner mitochondrial membrane. | S |
| NDUFA9 | NADH Dehydrogenase 1 α Subcomplex Subunit 9 | Subunit of the first enzyme complex in the electron transport chain in the inner mitochondrial membrane. | S |
| NDUFAB1 | NADH Dehydrogenase 1 α/β Subcomplex 1 | Non-catalytic subunit of the NADH complex involved in the mitochondrial inner membrane electron transport. | S |
| NDUFC2 | NADH Ubiquinone Oxidoreductase Subunit C2 | Suspected to be non-catalytic, a subunit of the NADH complex involved in the mitochondrial inner membrane electron transport. | S |
| NDUFV3 | NADH Ubiquinone Oxidoreductase Subunit V3 | Suspected to be non-catalytic, a subunit of the NADH complex involved in the mitochondrial inner membrane electron transport. | S |
| NPR1 | Natriuretic Peptide Receptor 1 | Guanylyl cyclase involved in catalyzing the production of cGMP from GTP. | E |
| PC | Pyruvate Carboxylase | Involved in glucogenesis, lipogenesis, insulin secretion, and glutamate. | S |
| PCK1 | Phosphoenolpyruvate Carboxykinase 1 | Main control point in the glucogenesis. | S |
| PCXB | Pyruvate Carboxylase | Catalyzes the carboxylation of pyruvate to oxaloacetate a process that requires biotin and ATP. | D |
| PGC-1 | PPARG Coactivator 1 α | Transcriptional coactivator involved in regulation of genes involved in energy metabolism. | S |
| PITX1 | Paired-Like Homeodomain 1 | Transcription factor part of a family involved in organ development and bilateral symmetry. | S |
| PKLR | Pyruvate Kinase L/R | Pyruvate kinase that catalyzes the rate-limiting step in glycolysis. | D |
| SLC12A3 | Solute Carrier Family 12 Member 3 | Important in electrolyte homeostasis, cotransporter that mediates sodium and chloride reabsorption. | S |
| SOD | Sodium Peroxide Dismutase 1 | Binds copper and zinc ions. Functions to destroy free radicals in the body. | E |
| TRPM8 | Transient Receptor Potential Cation Channer Subfamily Member 8 | Receptor-activated non-selection cation channel, suggested to be involved in low temperature sensation. | S |
| UCP1 | Uncoupling Protein 1 | Mitochondrial anion carrier protein. | D |
| VIPR1 | Vasoactive Intestinal Polypeptide Receptor 1 | Receptor for the vasoactive intestinal neuropeptide. | S |
| VHL | Von Hippel–Lindau Tumor Suppressor | Involved in ubiquitination of the hypoxia-inducible factor. | S |
| VOM1 | Vitelline Membrane Outer Layer 1 | Vitelline membrane outer layer proteins. | E |
| ZP | Zona Pellucida Glycoproteins | Proteins involved in the composition and fertilization functions, and preimplantation development in the zona pellucida. | E |
| Adaptations to dietary changes | |||
| ACAD9 | Acyl-CoA Dehydrogenase Family Member 9 | Localized in mitochondria and involved in catalyzing the rate limiting step of the β-oxidation of fatty acyl-CoA. | S |
| AGT | Alanine-Glyoxylate Aminotransferase | Also known as AGXT, involved in glyoxylate detoxification. In carnivores, the AGT is needed in the mitochondria, while in herbivores peroxisomes, and omnivores, have AGT in both organelles. | E |
| AMY | α-Amylase | Carbohydrase, specifically amylase. | S |
| AMY1B | Amylase α 1B | Secreted protein that catalyzes the first steps in digestion of starch and glycogen. | L |
| ASPM | Assembly Factor For Spindle Microtubules | Essential in mitotic spindle function in embryonic neuroblasts. | S |
| BARX1 | BARX Homeobox 1 | Homeobox transcription factor, suggested to be involved in teeth and craniofacial mesenchyme of neural crest origin. | S |
| BMP2 | Bone Morphogenetic Protein 2 | Ligand of the transforming growth factor β, involved in bone and cartilage development. | S |
| BMP4 | Bone Morphogenetic Protein 4 | Ligand of the transforming growth factor β, involved in heart development and adipogenesis. | S |
| CDK5RAP2 | Cyclin-Dependent-Kinase 5 Regulatory Subunit-Associated Protein 2 | Plays a role in centriole engagement and microtubule nucleation. | S |
| CEP152 | Centrosomal Protein 152 | Thought to play a role in centrosome function. | S |
| CYP7A1 | Cytochrome P450 Family 7 Subfamily A Member 1 | Monooxygenase with lipase catalytic function. | S |
| DGAT2 | Diacylglycerol O-Acyltransferase 2 | Enzyme involved in catalyzing the final reaction in triglyceride synthesis. | D |
| DLX2 | Distal-Less Homeobox 2 | Homeobox transcription factor, postulated to play a role in forebrain and craniofacial development. | S |
| EDAR | Ectodysplasin A Receptor | Receptor for Ectodyplasin A and can activate multiple cell death pathways. | S |
| EHHADH | Enoyl-CoA Hydratase | Bifunctional enzyme part of the peroxisomal β-oxidation pathway. | E |
| EVE1 | EVE1 | Transcription factor. | S |
| IFG-1 | Insulin Growth Factor 1 | Similar to insulin in function and structure, involved in mediating growth and development. | S |
| IRX5A | Iroquis Homeobox 5A | Transcription factor involved in cell development, embryonic skeletal joint development, chondrocyte differentiation, and neuron differentiation. | L |
| LHX7 | LIM Homeobox 7 | Transcription factor with zinc-finger motifs, involved in patterning and differentiation of multiple tissue types. | S |
| LHX8 | LIM Homeobox 8 | Transcription factor with zinc-finger motifs, involved in patterning and differentiation of multiple tissue types. | S |
| LIPF | Lipase F Gastric Type | Gastric lipase involved in digestion of triglycerides. | S |
| MAO | Monoamine Oxidase A | Enzyme involved in the catalysis of the oxidative deamination of amines of three neurotransmitters (i.e., dopamine, norepinephrine, and serotonin). | S |
| MCPH1 | Microcephalin 1 | DNA damage response protein. | L |
| NOX4 | NADPH Oxidase 4 | Catalytic subunit of the NADPH oxidase complex. | L |
| ODAM | Odontogenic Ameloblast-Associated Protein Precursor | Tooth-associated epithelial protein thought to have a role in odontogenesis. | D |
| PAX9 | Paired Box 9 | Paired box domain transcription factor with a critical role in fetal development and cancer growth. | S |
| PGA | Pepsin A | Enzyme that participates in digestion by activating pepsinogen. | S |
| PITX2 | Paired-Like Homeodomain 2 | Homeodomain transcription factor, regulates procollagen lysyl hydroxylase gene expression, as well as basal and hormone regulated activity of prolactin. Involved in development of the eye, tooth, and abdominal organs. | S |
| PLRP2 | Pancreatic Lipase-Related Protein 2 | Lipase for hydrolyzation of galactolipids. | E |
| RNASE1 | Ribonuclease A Family Member 1 Pancreatic | Pancreatic-type of secretory ribonuclease. | E |
| RPFA | Resuscitation-Promoting Factor A | Protein with peptidoglycan hydrolytic activity functions as a factor that stimulates resuscitation of dormant cells. | D |
| RUNX2 | RUNX Family Transcription Factor 2 | Transcription factor with Runt DNA-binding domain, essential for osteoblastic differentiation and skeletal morphogenesis. | S |
| SALL1A | Spalt-Like Transcription Factor 1 | Transcription factor with zinc-finger motifs. | L |
| SCN8A | Sodium Voltage-Gated Channel α Subunit 8 | Forms the ion-pore region of the voltage-gated sodium channel. Involved in depolarization during the formation of an action potential in excitable neurons. | S |
| SCN9A | Sodium Voltage-Gated Channel α Subunit 9 | Ion channel activity and sodium ion binding. | S |
| SCNA4A | Sodium Voltage-Gated Channel α Subunit 4 | Transmembrane glycoprotein, involved in generation and propagation of action potentials in neurons and muscle. | S |
| SCPP5 | Secretory Calcium-Binding Phosphoprotein 5 | Secretory Calcium-Binding Phosphoprotein. | D |
| SHH | Sonic Hedgehog | Protein in the Sonic Hedgehog signaling pathway, essential in patterning in early embryonic development. | S |
| SHOX | Short Stature Homeobox | Homeodomain transcription factor | L |
| SLC2A3 | Solute Carrier Family 2 Member 3 | Enables dehydroascorbic acid transmembrane transporter activity | D |
| TAS2R | Taste 2 Receptor | Transmembrane G-protein-coupled receptor involved in ability to taste glucosinolates and bitter compounds in plants. | L |
| TBX4 | T-Box Transcription Factor 4 | Transcription factor with a T-Box binding domain. It has been suggested to play a role in limb development. | L |
| TFAP2A | Transcription Factor AP 2 α | Transcription factor that works both as a suppressor and activator of multiple genes. | S |
| TST | Thiosulfate Sulfurtransferase | Catalyzes the conversion of thiosulfate and cyanide to thiocyanate and sulfite | D |
| UNK | Unk Zinc Finger | RNA-binding protein involved in establishment and maintenance of the early morphology of cortical neurons in embryonic development. | D |
| WDR62 | WD Repent Domain 62 | Proposed to play a role in cerebral cortical development. | S |
| Environmental Pathogens | |||
| APOBEC | Apolipoprotein B MRNA Editing Enzyme Catalytic Subunit 3G | Catalyzes site-specific deamination of RNA and single-stranded DNA. | S |
| CD14 | Myeloid Cell-Specific Leucine-Rich | Surface antigen preferentially expressed on monocytes/macrophages. | S |
| CD22 | Sialic Acid-Binding Ig-Like Lectin 2 | Carbohydrate binding protein. Mediates B-cell/B-cell interactions. Thought to localize B-cells in lymphoid tissues. | E |
| CD40 | Tumor Necrosis Factor Receptor Superfamily Member 5 | Receptor on antigen-presenting cells of the immune system. | S |
| CD80 | T-Lymphocyte Activation Antigen CD80 | Membrane receptor activated by CD28 involved in costimulatory signal essential for T-lymphocyte activation. | S |
| IFNAR2 | Interferon α and β Receptor Subunit 2 | Type I membrane protein, forms one of the chains of a interferon α and β receptor. | S |
| IGH | Immunoglobulin Heavy Locus | Antigen-binding protein involved in the Lectin-induced complement pathway and NFAT immune response. | S |
| IGHE | Immunoglobulin Heavy Constant Epsilon | Antigen-binding protein involved in Interleukin 4-mediated and cytokine signaling. | S |
| IGHM | Immunoglobulin Heavy Constant Delta | Antigen binding. | S |
| LY96 | Lymphocyte Antigen 96 | Confers responsiveness to lipopolysaccharides when it associates with a Toll-like receptor. | S |
| MHC | Major Histocompatibility Complex class I | Family of genes associated with antigen processing. | E |
| MDA5 | Interferon Induced with Helicase C Domain 1 | Intracellular sensor of viral RNA that triggers innate immune response. | S |
| MUC7 | Mucin 7-Secreted | Salivary mucin thought to play a role in facilitating clearance of bacteria in the mouth. | S |
| MX | MX Dynamin-Like GTPase | Guanosine triphosphate metabolizing protein that participates in antiviral response. | S |
| NLPRP3 | NLR Family Pyrin Domain-Containing 3 | Peptidoglycan binding protein. | E |
| SSC4D | Scavenger Receptor Cysteine-Rich Family Member with 4 Domains | Scavenger receptor activity. | E |
| TAB 1 | TGF β-Activated Kinase 1 | Regulator of the MAP kinase kinase kinase. Thus, mediates various intracellular signaling pathways. | S |
| TICAM1 | Toll-Like Receptor Adaptor Molecule 1 | Adaptor protein with Toll/Interleukin 1-receptor homology domain. Protein kinase binding and obsolete signal transducer activity. | S |
| TLR | Toll-Like Receptors | Family of genes that play a fundamental role in pathogen recognition and activation of innate immunity. | - |
| TLR1 | Toll-Like Receptor 1 | Non-viral pathogen recognition, protein heterodimerization activity, and transmembrane signaling. | E |
| TLR2 | Toll-Like Receptor 2 | Non-viral pathogen recognition, protein heterodimerization activity, and transmembrane signaling. | E, S |
| TLR4 | Toll-Like Receptor 4 | Non-viral pathogen recognition, lipopolysaccharide binding. | S |
| TLR5 | Toll-Like Receptor 5 | Non-viral pathogen recognition, protein heterodimerization activity, and transmembrane signaling. | S |
| TLR7 | Toll-Like Receptor 7 | Transmembrane signaling receptor activity and double-stranded RNA binding. | E, S |
| TLR8 | Toll-Like Receptor 8 | RNA binding and drug binding. | E, S |
| TLR9 | Toll-Like Receptor 9 | Transmembrane signaling receptor activity and siRNA binding. | E |
| TLR22 | Toll-Like Receptor 22 | Transmembrane signaling receptor activity. | E |
| TLR23 | Toll-Like Receptor 23 | Receptor activity. | E |
| TLR25 | Toll-Like Receptor 25 | Transmembrane signaling receptor activity. | E |
| Other Factors | |||
| MSX2 | Homeobox Protein MSX-2 | Transcription factor with a homeobox-binding domain. | E |
| C6AST | Calpastatin | Endogenous calcium-dependent cysteine protease inhibitor. | E |
| PASTN | Pasttristacin | Metalloendopeptidase activity. | E |
| CRISP | Cysteine-Rich Secretory Proteins | Associated with reptilian venom production and mammalian reproduction. | E |
| PER2 | Period Circadian Regulator 2 | Transcription factor and “activator” activity. Primary circadian pacemaker in the mammalian brain. | S |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez Sosa, F.; Pilot, M. Molecular Mechanisms Underlying Vertebrate Adaptive Evolution: A Systematic Review. Genes 2023, 14, 416. https://doi.org/10.3390/genes14020416
Martínez Sosa F, Pilot M. Molecular Mechanisms Underlying Vertebrate Adaptive Evolution: A Systematic Review. Genes. 2023; 14(2):416. https://doi.org/10.3390/genes14020416
Chicago/Turabian StyleMartínez Sosa, Francelly, and Małgorzata Pilot. 2023. "Molecular Mechanisms Underlying Vertebrate Adaptive Evolution: A Systematic Review" Genes 14, no. 2: 416. https://doi.org/10.3390/genes14020416
APA StyleMartínez Sosa, F., & Pilot, M. (2023). Molecular Mechanisms Underlying Vertebrate Adaptive Evolution: A Systematic Review. Genes, 14(2), 416. https://doi.org/10.3390/genes14020416







