CD99 and the Chicken Alloantigen D Blood System †
Abstract
1. Introduction
2. Materials and Methods
2.1. Genetic Material
2.2. GWAS Analysis
2.3. Identification of Candidate Gene
2.4. SNP Selection and Subsequent Genotyping
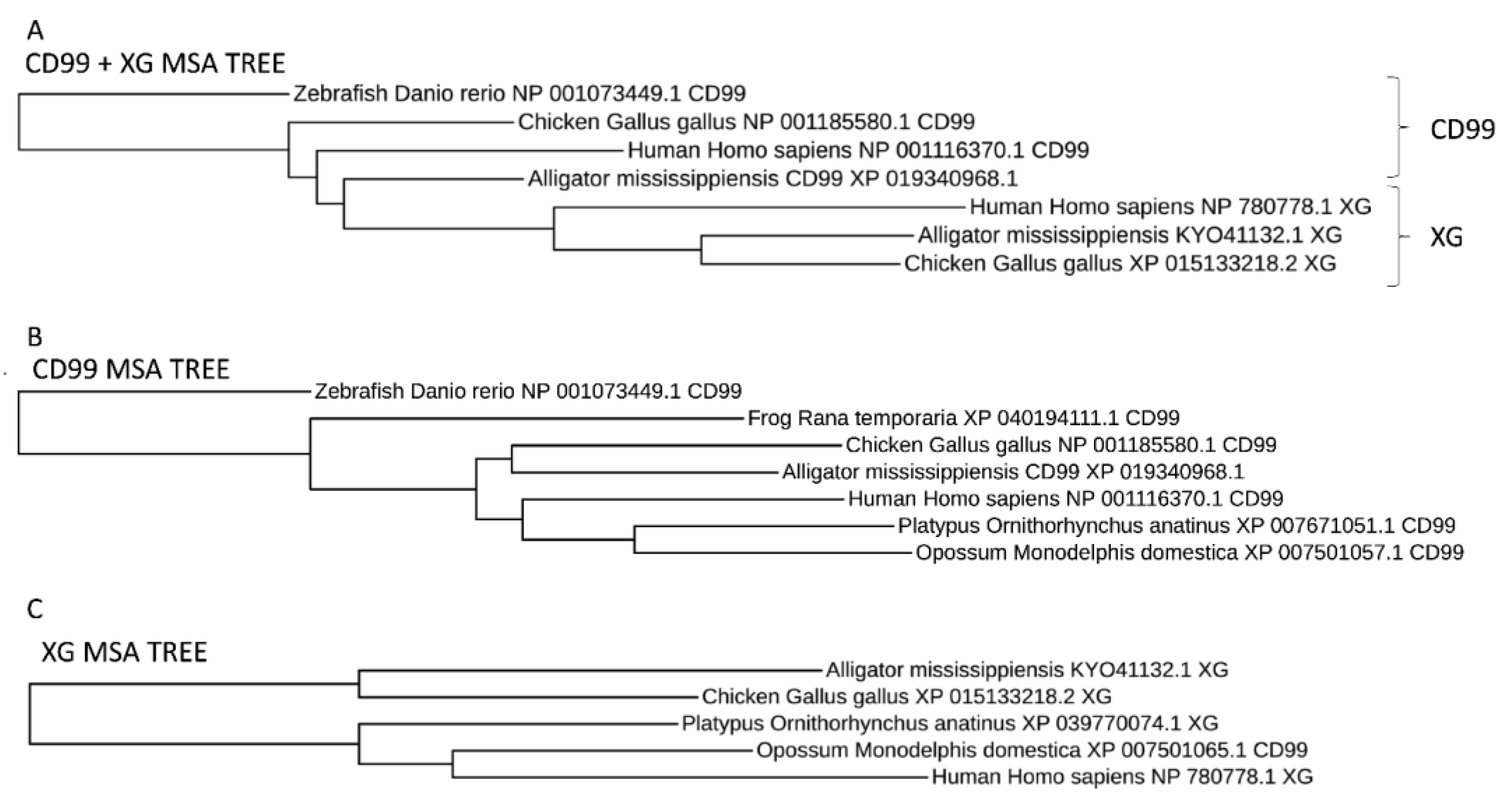
2.5. Sequence Comparison Methods
3. Results and Discussion
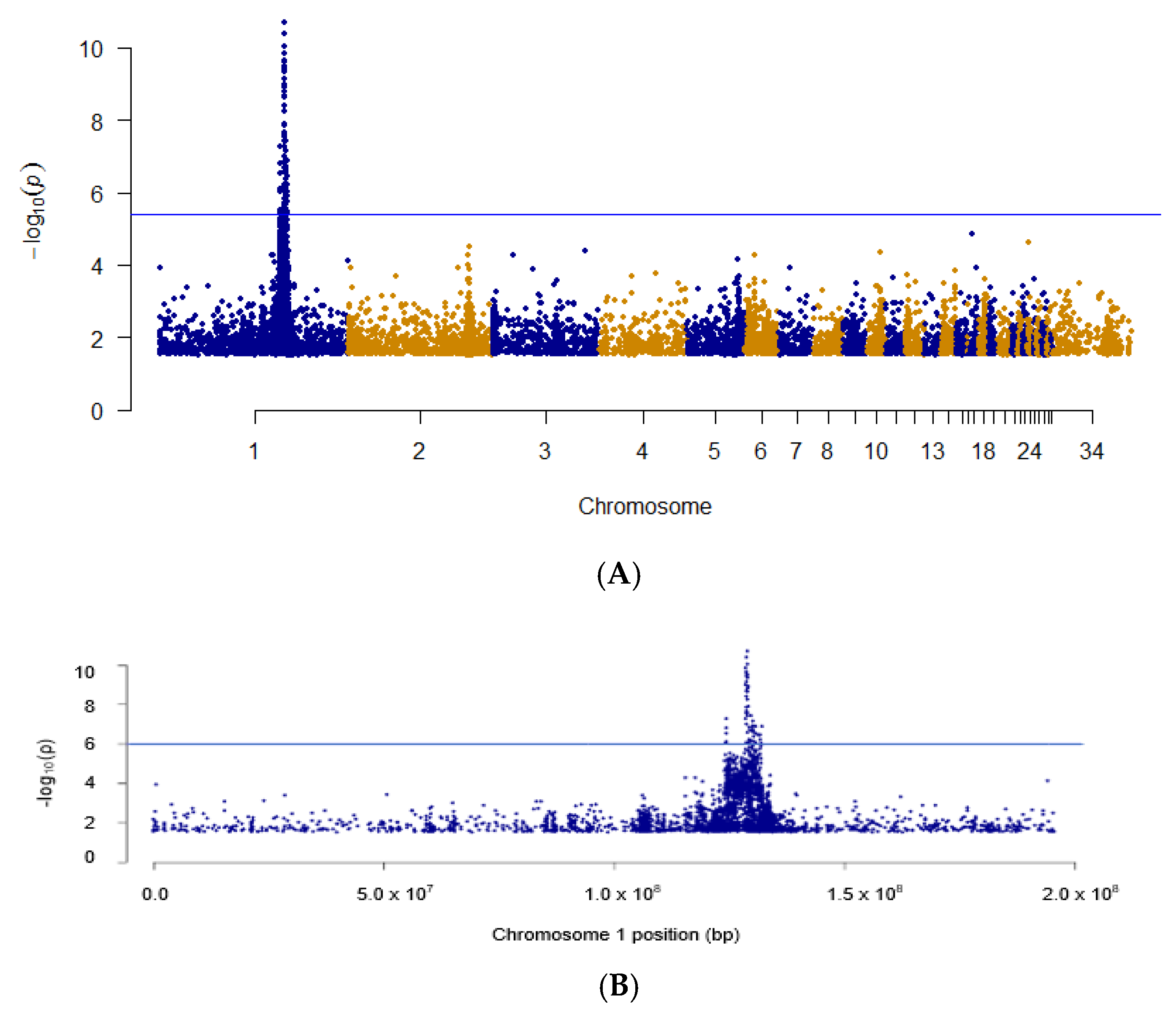
3.1. Identification of D System Candidate Gene
3.2. Haplotype Identification
3.3. Protein Structure and Antigenicity
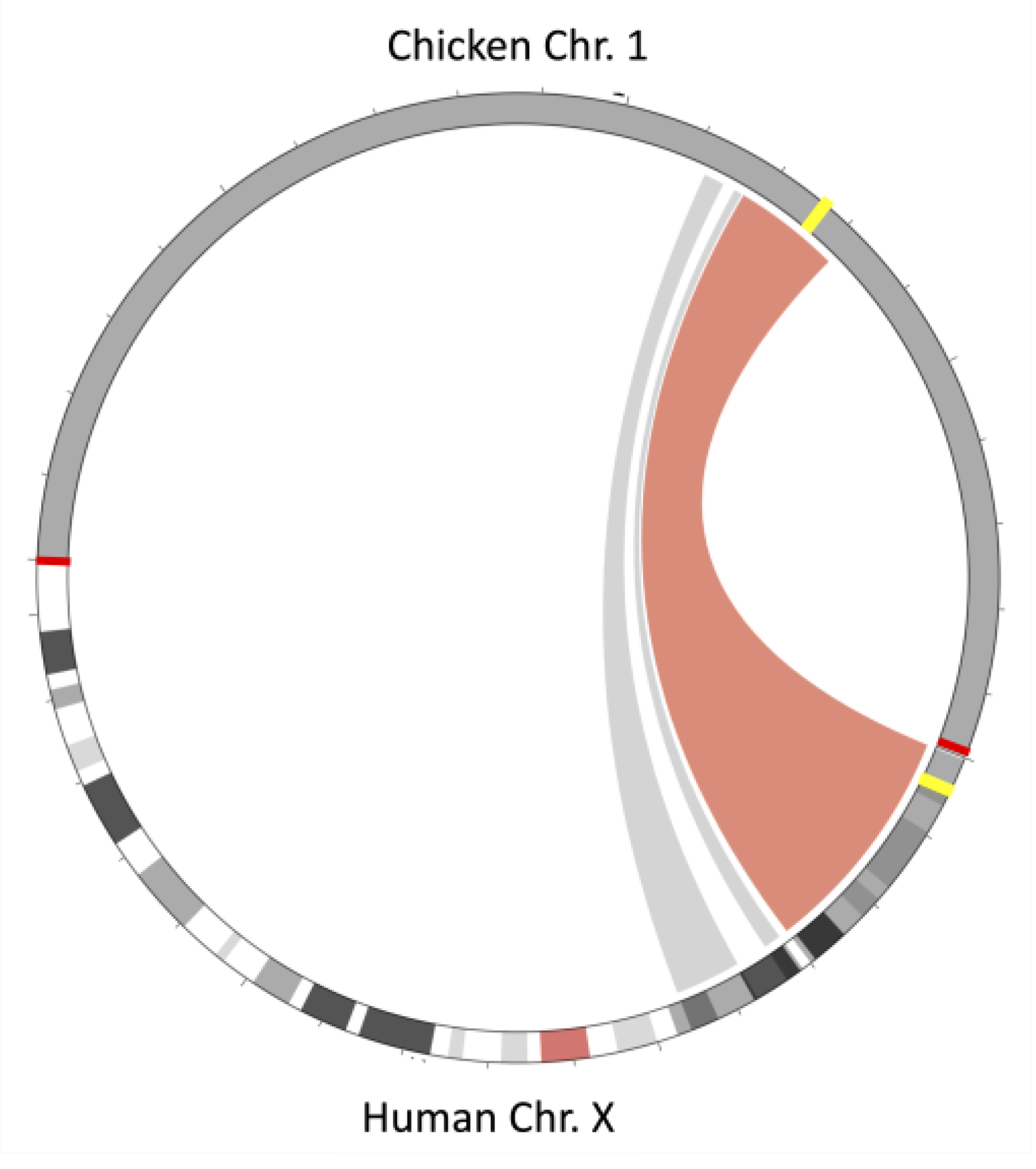
3.4. Synteny Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pasello, M.; Manara, M.C.; Scotlandi, K. CD99 at the crossroads of physiology and pathology. J. Cell Commun. Signal. 2018, 12, 55–68. [Google Scholar] [CrossRef]
- Johnson, N.C. XG: The forgotten blood group system. Immunohematology 2011, 27, 68–71. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, P.A. XG blood group puzzle solved: What’s next? Blood 2018, 132, 243. [Google Scholar] [CrossRef]
- Yeh, C.-C.; Chang, C.-J.; Twu, Y.-C.; Chu, C.-C.; Liu, B.-S.; Huang, J.-T.; Hung, S.-T.; Chan, Y.-S.; Tsai, Y.-J.; Lin, S.-W.; et al. The molecular genetic background leading to the formation of the human erythroid-specific Xg(a)/CD99 blood groups. Blood Adv. 2018, 2, 1854–1864. [Google Scholar] [CrossRef]
- Glick, G.; Chang, T.S.; Jaap, R.G. The bursa of Fabricius and antibody production. Poult. Sci. 1956, 35, 224–234. [Google Scholar] [CrossRef]
- Cooper, M.D.; Raymond, D.A.; Peterson, R.D.; South, M.A.; Good, R.A. The functions of the thymus system and the bursa system in the chicken. J. Exp. Med. 1966, 123, 75–102. [Google Scholar] [CrossRef]
- Weiss, R.A.; Vogt, P.K. 100 years of Rous sarcoma virus. J. Exp. Med. 2011, 208, 2351–2355. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.L., Jr.; Medarova, Z.; Briles, W.E. Immune effects of chicken non-MHC alloantigens. Poult. Sci. 2016, 95, 447–457. [Google Scholar] [CrossRef]
- Schierman, L.W.; Nordskog, A.W. Relationship of blood type to histocompatibility in chickens. Science 1961, 134, 1008–1009. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.M.; Taylor, R.L., Jr. Brief review of the chicken Major Histocompatibility Complex: The genes, their distribution on chromosome 16, and their contributions to disease resistance. Poult. Sci. 2016, 95, 375–392. [Google Scholar] [CrossRef]
- Fulton, J.E.; Drobik-Czwarno, W.; Wolc, A.; McCarron, A.M.; Lund, A.R.; Schmidt, C.J.; Taylor, J.R.L. The Chicken A and E Blood Systems Arise from Genetic Variation in and around the Regulators of Complement Activation Region. J. Immunol. 2022, 209, 1128–1137. [Google Scholar] [CrossRef]
- Briles, R.W.; Gilmour, D.G. (Eds.) Erythrcyte Alloantigen Characterisitics; FASEB: Bethesda, MD, USA, 1980. [Google Scholar]
- Bitgood, J.J.; Kendall, R.L.; Briles, R.W.; Briles, W.E. Erythrocyte alloantigen loci Ea-D and Ea-I map to chromosome 1 in the chicken. Anim. Genet. 1991, 22, 449–454. [Google Scholar] [CrossRef]
- Scott, T.R.; Oduho, G.W.; Glick, B.; Hagan, F.; Briles, W.E.; Yamamoto, Y. Erythrocyte alloantigen diversity and some immunological effects of the B system in related New Hampshire strains. Poult. Sci. 1988, 67, 1210–1217. [Google Scholar] [CrossRef]
- Martin, A.; Dunnington, E.A.; Gross, W.B.; Briles, W.E.; Briles, R.W.; Siegel, P.B. Production traits and alloantigen systems in lines of chickens selected for high or low antibody responses to sheep erythrocytes. Poult. Sci. 1990, 69, 871–878. [Google Scholar] [CrossRef]
- Dunnington, E.A.; Briles, R.W.; Briles, W.E.; Gross, W.B.; Siegel, P.B. Allelic frequencies in eight alloantigen systems of chickens selected for high and low antibody response to sheep red blood cells. Poult. Sci. 1984, 63, 1470–1472. [Google Scholar] [CrossRef]
- Crittenden, L.B.; Provencher, L.; Santangelo, L.; Levin, I.; Abplanalp, H.; Briles, R.W.; Briles, W.E.; Dodgson, J.B. Characterization of a Red Jungle Fowl by White Leghorn Backcross Reference Population for Molecular Mapping of the Chicken Genome. Poult. Sci. 1993, 72, 334–348. [Google Scholar] [CrossRef]
- Kranis, A.; Gheyas, A.A.; Boschiero, C.; Turner, F.; Yu, L.; Smith, S.; Talbot, R.; Pirani, A.; Brew, F.; Kaiser, P.; et al. Development of a high density 600K SNP genotyping array for chicken. BMC Genom. 2013, 14, 59. [Google Scholar] [CrossRef]
- Abplanalp, H.; Briles, W.E.; Stone, H. (Eds.) Blood Group Systems Part II; FASEB: Bethesda, MD, USA, 1980. [Google Scholar]
- Hala, K. Inbred Lines of Avian Species; Toivanen, A., Toivanen, P., Eds.; CRC Press: Boca Raton, FL, USA, 1987. [Google Scholar]
- International Chicken Genome Sequencing C. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 2004, 432, 695–716. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Patel, V.M.; Coon, M.; Nguyen, T.; Land, S.J.; Ruden, D.M.; Lu, X. Using Drosophila melanogaster as a Model for Genotoxic Chemical Mutational Studies with a New Program, SnpSift. Front. Genet. 2012, 3, 35. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, F.; Allen, J.E.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.F.; Bennet, R.; et al. Ensembl 2022. Nucleic Acids Res. 2022, 50, D988–D995. [Google Scholar] [CrossRef] [PubMed]
- UniProt C. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar]
- Somyong, S.; Phetchawang, P.; Bihi, A.K.; Sonthirod, C.; Kongkachana, W.; Sangsrakru, D.; Jomchai, N.; Pootakham, W.; Tangphatsornruang, S. A SNP variation in an expansin (EgExp4) gene affects height in oil palm. PeerJ 2022, 10, e13046. [Google Scholar] [CrossRef] [PubMed]
- Ward, N.; Moreno-Hagelsieb, G. Quickly finding orthologs as reciprocal best hits with BLAT, LAST, and UBLAST: How much do we miss? PLoS ONE 2014, 9, e101850. [Google Scholar] [CrossRef] [PubMed]
- States, D.J.; Gish, W. Combined use of sequence similarity and codon bias for coding region identification. J. Comput. Biol. 1994, 1, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Lee, J.; Hong, W.Y.; Cho, M.; Sim, M.; Lee, D.; Ko, Y.; Kim, J. Synteny Portal: A web-based application portal for synteny block analysis. Nucleic Acids Res. 2016, 44, W35–W40. [Google Scholar] [CrossRef]
- Papadopoulos, J.S.; Agarwala, R. COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics 2007, 23, 1073–1079. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Bernhofer, M.; Dallago, C.; Karl, T.; Satagopam, V.; Heinzinger, M.; Littmann, M.; Olenyi, T.; Qiu, J.; Schütze, K.; Yachdav, G.; et al. PredictProtein-Predicting Protein Structure and Function for 29 Years. Nucleic Acids Res. 2021, 49, W535–W540. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Raudsepp, T.; Chowdhary, B.P. The Eutherian Pseudoautosomal Region. Cytogenet. Genome Res. 2015, 147, 81–94. [Google Scholar] [CrossRef]
- Parmentier, H.K.; Lammers, A.; Hoekman, J.J.; De Vries Reilingh, G.; Zaanen, I.T.; Savelkoul, H.F. Different levels of natural antibodies in chickens divergently selected for specific antibody responses. Dev. Comp. Immunol. 2004, 28, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Gross, W.B.; Siegel, P.B. Effects of early environmental stresses on chicken body weight, antibody response to RBC antigens, feed efficiency, and response to fasting. Avian Dis. 1980, 24, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Boa-Amponsem, K.; Dunnington, E.A.; Baker, K.S.; Siegel, P.B. Diet and immunological memory of lines of White Leghorn chickens divergently selected for antibody response to sheep red blood cells. Poult. Sci. 1999, 78, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Watson, R.L.; Buck, J.; Levin, L.R.; Winger, R.C.; Wang, J.; Arase, H.; Muller, W.A. Endothelial CD99 signals through soluble adenylyl cyclase and PKA to regulate leukocyte transendothelial migration. J. Exp. Med. 2015, 212, 1021–1041. [Google Scholar] [CrossRef]
- Bixel, G.; Kloep, S.; Butz, S.; Petri, B.; Engelhardt, B.; Vestweber, D. Mouse CD99 participates in T-cell recruitment into inflamed skin. Blood 2004, 104, 3205–3213. [Google Scholar] [CrossRef]
- Manjula, P.; Fulton, J.E.; Seo, D.; Lee, J.H. Major histocompatibility complex B variability in Korean native chicken breeds. Poult. Sci. 2020, 99, 4704–4713. [Google Scholar] [CrossRef]
- Tarrant, K.J.; Lopez, R.; Loper, M.; Fulton, J.E. Assessing MHC-B diversity in Silkie chickens. Poult. Sci. 2020, 99, 2337–2341. [Google Scholar] [CrossRef]
- Nguyen-Phuc, H.; Fulton, J.E.; Berres, M.E. Genetic variation of major histocompatibility complex (MHC) in wild Red Junglefowl (Gallus gallus). Poult. Sci. 2016, 95, 400–411. [Google Scholar] [CrossRef] [PubMed]
- Fulton, J.E.; Lund, A.R.; McCarron, A.M.; Pinegar, K.N.; Korver, D.R.; Classen, H.L.; Aggrey, S.; Utterbach, C.; Anthony, N.B.; Berres, M.E. MHC variability in heritage breeds of chickens. Poult. Sci. 2016, 95, 393–399. [Google Scholar] [CrossRef]
- Storry, J.R.; Reid, M.E.; Yazer, M.H. The Cromer blood group system: A review. Immunohematology 2010, 26, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Petty, A.C.; Green, C.A.; Poole, J.; Daniels, G.L. Analysis of Knops blood group antigens on CR1 (CD35) by the MAIEA test and by immunoblotting. Transfus. Med. 1997, 7, 55–62. [Google Scholar] [CrossRef]
- Moulds, J.M. The Knops blood-group system: A review. Immunohematology 2010, 26, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Cooling, L. Blood Groups in Infection and Host Susceptibility. Clin. Microbiol. Rev. 2015, 28, 801–870. [Google Scholar] [CrossRef] [PubMed]
- Moller, M.; Lee, Y.Q.; Vidovic, K.; Kjellstrom, S.; Bjorkman, L.; Storry, J.R.; Olsson, M.L. Disruption of a GATA1-binding motif upstream of XG/PBDX abolishes Xg(a) expression and resolves the Xg blood group system. Blood 2018, 132, 334–338. [Google Scholar] [CrossRef] [PubMed]


| Source | Breed 1 | D alleles present | Samples (n) | Reference 2 |
|---|---|---|---|---|
| NIU DNA Bank; Pedigree | WL, Ancona | D1, D2 | 53 | none |
| NIU DNA Bank | WL, Ancona | D1, D2, D3 | 51 | none |
| HAS/LAS | WL | D1, D3, D4 | 44 | [15] |
| UCD001 | RJF | unk | 10 | [17] 3 |
| UCD003 | WL | D3 | 20 | [19,20] |
| WL1 | WL | D2, D3 | 58 | none |
| WL2 | WL | D3 | 90 | none |
| WL3 | WL | D1, D3 | 120 | none |
| WL4 | WL | D1, D3 | 119 | none |
| WL5 | WL | D3 | 120 | none |
| RIR1 | RIR | unk | 64 | none |
| WPR1 | WPR | unk | 320 | none |
| WPR1 | WPR | unk | 272 | none |
| Sequence only | Avg. coverage | Source | ||
| UCD001 | RJF | unk | 6.6 | [21] |
| UCD003 | WL | D3 | 18.7 | Hans Cheng, pers. comm. |
| IAH-61 | WL | D3 | 15.2 | [18] |
| IAH-72 | WL | D1 | 17.6 | [18] |
| IAH-RHC | WL | D3 | 15.9 | [18] |
| Haplotype | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP Name | Location (bp) * | Exon Gene Location | Codon Change | Nucleotide Change | aa | Ref | Alt | H01 | H02 | H03 | H04 | H05 | H06 A | H07 | H08 | H09 | H10 | H11 |
| rs74153692 | 129,875,094 | 2 | GTG > ATG | G > A | V27M | G | A | G | G | G | G | G | G | A | G | G | G | G |
| rs314496839 | 129,871,024 | 3 | GTT > ATT | G > A | V37I | G | A | A | G | G | G | A | G | G | G | A | G | A |
| rs312634736 | 129,870,489 | 4 | GGC > GAC | G > A | G48D | G | A | G | A | G | G | A | G | G | G | A | G | G |
| rs316308207 | 129,867,634 | 7 | GGT > GGC | T > C | G95G | T | C | C | C | C | C | C | T | C | T | T | T | C |
| rs10730300 | 129,867,625 | 7 | CCA > CCG | A > G | P98P | A | G | A | A | G | A | A | A | A | A | A | A | A |
| rs735363747 | 129,866,188 | 8 | GAG > GGG | A > G | E124G | A | G | A | A | A | A | A | A | A | A | A | A | A |
| rs13623448 | 129,866,175 | intron | splice region | ref | del | del | del | ref | ref | ref | ref | del | del | del | ref | del | ||
| rs735519530 | 129,864,266 | 11 | TTG > CTG | T > C | L185L | T | C | T | T | T | T | T | T | T | T | T | T | C |
| D blood system serological allele | D1 | D2 | D3 | D3 | unk | D3 | unk | unk | unk | unk | unk | |||||||
| A Allele | Haplotype | Source(s) |
|---|---|---|
| D1 | CD99-H01 | NIU DNA Bank and NIU pedigree, IAH-72, HAS, LAS, WL4, WL3 |
| D2 | CD99-H02 | NIU DNA Bank and NIU pedigree, WL1 |
| D3 | CD99-H03 | NIU DNA Bank, IAH-61, IAH-RHC, UCD003, HAS, LAS, WL1, WL2, WL3, WL4, WL5 |
| D3 | CD99-H04 | NIU DNA Bank |
| D3 | CD99-H06 | HAS, WL3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fulton, J.E.; Drobik-Czwarno, W.; Lund, A.R.; Schmidt, C.J.; Taylor, R.L., Jr. CD99 and the Chicken Alloantigen D Blood System. Genes 2023, 14, 402. https://doi.org/10.3390/genes14020402
Fulton JE, Drobik-Czwarno W, Lund AR, Schmidt CJ, Taylor RL Jr. CD99 and the Chicken Alloantigen D Blood System. Genes. 2023; 14(2):402. https://doi.org/10.3390/genes14020402
Chicago/Turabian StyleFulton, Janet E., Wiola Drobik-Czwarno, Ashlee R. Lund, Carl J. Schmidt, and Robert L. Taylor, Jr. 2023. "CD99 and the Chicken Alloantigen D Blood System" Genes 14, no. 2: 402. https://doi.org/10.3390/genes14020402
APA StyleFulton, J. E., Drobik-Czwarno, W., Lund, A. R., Schmidt, C. J., & Taylor, R. L., Jr. (2023). CD99 and the Chicken Alloantigen D Blood System. Genes, 14(2), 402. https://doi.org/10.3390/genes14020402





