Abstract
Several DNA polymerases participate in DNA synthesis during genome replication and DNA repair. PCNA, a homotrimeric ring, acts as a processivity factor for DNA polymerases. PCNA also acts as a “landing pad” for proteins that interact with chromatin and DNA at the moving fork. The interaction between PCNA and polymerase delta (Polδ) is mediated by PIPs (PCNA-interacting peptides), in particular the one on Pol32, a regulatory subunit of Polδ. Here, we demonstrate that pol3-01, an exonuclease mutant of Polδ’s catalytic subunit, exhibits a weak interaction with Pol30 compared to the WT DNA polymerase. The weak interaction activates DNA bypass pathways, leading to increased mutagenesis and sister chromatid recombination. Strengthening pol3-01′s weak interaction with PCNA suppresses most of the phenotypes. Our results are consistent with a model in which Pol3-01 tends to detach from the chromatin, allowing an easier replacement of Polδ by the trans-lesion synthesis polymerase Zeta (Polz), thus leading to the increased mutagenic phenotype.
1. Introduction
DNA synthesis is an essential and universal process. Accurate DNA synthesis has an important role in preventing spontaneous mutations and carcinogenesis [1]. In the yeast Saccharomyces cerevisiae, DNA replication is carried out by three DNA polymerases that belong to the B family [2]. DNA synthesis is initiated by the activity of Polα. This polymerase is then replaced by either Polε or Polδ. Polε is currently believed to synthesize most of the leading strand [3], whereas Polδ synthesizes the lagging strand and creates Okazaki fragments [4].
The synthesis ability of Polδ depends on PCNA, a homotrimeric clamp (composed of three copies of the Pol30 protein) that encircles the DNA, enabling better DNA processivity during DNA synthesis [5]. The interaction between PCNA and Polδ is mediated by a conserved motif called the PCNA-interacting peptide (PIP). Its consensus sequence is Q-x-x(M/L/I)-x-x-F-F, although many variations of the PIP motif have been found [6,7].
Polδ is a complex composed of three subunits; Pol3 (the catalytic subunit), Pol31, and Pol32 [8,9] (Figure 1A). POL3 and POL31 are essential genes [10], while POL32 is not [9], although its absence confers cold sensitivity and affects the repair of double-stranded breaks (DSBs) [11]. It has been shown that POL32 has a PIP motif at its C terminus, through which it physically interacts with PCNA. Mutating the PIP motif of POL32 or deleting the POL32 gene altogether does not lead to lethality or to a significant decrease in the polymerase’s processivity [12]. This suggests that additional points of interaction between the polymerase subunits are likely to exist. A crystal structure of the holoenzyme revealed that Pol3’s interaction with Pol32 is mediated by Pol31 [13]. In addition, it was shown in vitro that mutation of potential PIP motifs in POL3 or POL31 causes a decrease in DNA synthesis processivity, as well as lethality in pol32Δ cells [14]. Together with experiments showing that Pol3 physically interacts with PCNA [15], this suggests that the interaction of Polδ with PCNA does not only rely on the binding of Pol32 to PCNA but also on the binding of Pol31 and/or Pol3 to PCNA, although the interaction of Pol32 with PCNA is the strongest.
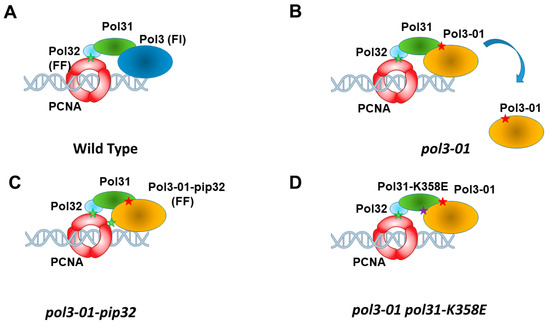
Figure 1.
Schematic representation of Polδ in contact with PCNA. (A). In wt cells, Pol32 binds PCNA via its strong PIP (PCNA-interacting peptide), which contains the sequence FF (green star). Pol3 binds Pol31, which in turn binds Pol32. Pol3 sequence contains a weak PIP (FI). (B). In pol3-01 cells, the Pol3 subunit carrying the mutation (red star) shows lower affinity and tends to disengage. (C). Changing the weak PIP of Pol3 by that of Pol32 suppresses most phenotypes of pol3-01 cells. (D). The K358E mutation in POL31 (purple star) allows higher affinity of Pol3 (and of Rev3, see text).
During cell growth, DNA synthesis may be blocked by DNA lesions, covalently linked proteins, R-loops, secondary structures of the DNA, etc.; this situation can be lethal to the cell. However, the cell can bypass these impediments by using the DNA damage tolerance (DDT, also known as post-replication repair) pathways (reviewed in [16,17]). Two main modes of lesion tolerance have been described: (1) Mono-ubiquitination of PCNA at lysine 164 by the Rad6/Rad18 E2/E3 complex [18,19] activates an “error-prone pathway” that uses alternative, trans-lesion synthesis (TLS) polymerases that are able to extend the DNA synthesis by incorporating more or less random nucleotides facing any DNA lesion. This, of course, results in the creation of mutations but prevents cell arrest and death. In yeast cells, the main TLS polymerase is Polζ, encoded by the REV3 and REV7 genes [20]. The TLS pathway is responsible for 50–70% of all spontaneous mutations and for the increased frequency of mutations seen following DNA damage [21,22,23,24]. (2) In addition, the cells may opt to bypass the lesion through an ‘error-free pathway’(also called “template switch”) mechanism that uses the undamaged sister chromatid as a template in order to copy the correct sequence and allow the cell to bypass those lesions [25,26]. This bypass pathway involves Rad5, a ubiquitin ligase/helicase that, together with Ubc13 and Mms2, poly-ubiquitinates PCNA at lysine 164 [27,28].
Here, we test the importance of Polδ-PCNA interactions for the different DNA bypass pathways. The exonuclease domain in DNA polymerases carries out an important proofreading function, which allows the polymerase to detect and replace errors that may have occurred during polymerization. In humans, mutations in the exonuclease domain of DNA polymerases are common in various types of cancer [29,30]. We show that yeast strains with the pol3-01 allele, which are mutated in the exonuclease domain of POL3, show elevated mutation and sister chromatid recombination phenotypes. pol3-01′s high mutation rate is usually attributed to its lack of exonuclease. In this paper, we test this assumption and show evidence that pol3-01′s high mutation rates are caused mainly by its weak interaction with PCNA.
2. Materials and Methods
2.1. Yeast Strains
The S. cerevisiae strains used in the present study (Table 1) are isogenic derivatives of BY4741 (MATa his3∆ leu2∆ met15∆ ura3∆), E134 (MAT@ ade5-1 lys2::InsEa14 trp1-289 his7-2 leu2-3,112 ura3-52) and PJ69-4a (MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4∆ gal80∆ GAL2-ADE2 LYS2:: GAL1-HIS3 met2::GAL7-lacZ). Gene deletions were performed by using a PCR-mediated one-step replacement technique. All deletions were confirmed by PCR amplification of genomic DNA and phenotypic expression. Site-directed mutagenesis was introduced by using a modified oligonucleotide in a PCR reaction, followed by transformation to yeast. All changes were confirmed by DNA sequencing. Cloning was performed by standard methods and was confirmed by restriction fragment analysis, PCR, and sequencing. The transformation was carried out by standard methods using lithium acetate.

Table 1.
Strains used in this study.
2.2. Plasmids
Yeast two-hybrid plasmids were built by cloning the relevant fragments using SalI and EcoRI for pGBKT7 and BamHI and XhoI for pACT2. The YIpAM26 pol3-01 plasmid was received from R. Kolodner [31]. The pACT2 POL32 plasmid was received from P. Burgers. All plasmids are listed in Table 2.

Table 2.
Plasmids used in this study.
2.3. Media and Growth Conditions
Saccharomyces cerevisiae strains were grown at 30 °C unless specified otherwise. The growth medium for all batch cultures was either standard minimal medium made of 6.7 g/liter yeast nitrogen base without amino acids and with ammonium sulfate, 1.5 g/liter amino acid dropout powder, and 2% of glucose: or standard YPD-rich medium containing 1% yeast extract, 2% bacto peptone, and 2% dextrose. Agar plates were made with the same growth medium plus 2% agar.
2.4. Yeast Two-Hybrid (Y2H) Assay
To detect two-hybrid interactions, yeast strain PJ69-4a was co-transformed with one LEU2-marked plasmid containing POL30/POL32 genes fused to the GAL4-activating domain (pACT2) and one TRP1-marked plasmid containing POL3/pol3-01/pol3-pip32 genes fused to the GAL4 DNA binding domain (pGBKT7). Yeast cultures were grown in SD-Trp-Leu medium and spotted on SD-Trp-Leu plates and SD-Trp-Leu-His plates. Cells were incubated for 3–4 days at 30 °C.
2.5. Fluctuation Test
Derivatives of E134 were plated on media that allowed us to detect new mutations: CAN medium is a standard minimal medium that lacks arginine and contains canavanine, a toxic agent that is an analog to arginine. If loss of function mutations occur in the CAN1 gene, the transporter loses its function, preventing the toxic canavanine from entering the cell and allowing the mutated cells to grow on this medium. The LYS2 locus of E134-derived strains contains an insertion of 14 adenine residues, which causes auxotrophy to lysine. Frameshift mutations (+1 or −2) can restore the Lys+ phenotype. SD-Lys is a standard-defined medium that lacks lysine. The trp1-289 mutation of E134-derived strains contains a single nucleotide change, which causes auxotrophy to tryptophan. Reversion to the original nucleotide allows the cells to grow on an SD-Trp plate, which is a standard-defined medium that lacks tryptophan.
Derivatives of BLS2 were plated on media that allowed us to detect new sister chromatid recombination: Strain BLS2 contains 5′ ade3 and 3′ ade3 fragments that are separated by URA3 marker. This construct allows the detection of new unequal sister chromatid recombination (USCR) by plating the cells on the appropriate medium (SD-HIS) [32]. Mutation and USCR rates were calculated as described in [33].
2.6. Fractionation Analysis
Different E134 strains were grown to O.D. 1. After reaching the desired O.D., cells were treated with 100T zymolyase 20 mg/mL and 0.01 M/mL of DTT for 1 h. Cells were then washed with SB buffer (0.02M Tris and 1M Sorbitol) and lysed using EB buffer (0.02 M Tris and 0.1 M NaCl), and fractions were separated using NIB buffer (0.02 M Tris, 0.1 M NaCl, and 1.2 M sucrose), using a sucrose gradient. A total of 2 mg of protein were extracted for each fraction: whole cell extract (WCE) and chromatin.
2.7. Protein Extraction and Immunoprecipitation Assays
Cells were grown to mid-logarithmic phase, washed once with water, and resuspended in lysis buffer (PBS, pH 7.0, 200 mM NaCl, 0.5 mM EGTA, 0.5 mM EDTA, 0.1% Triton X-100, protease inhibitor mixture, and 1 mM phenylmethylsulfonyl fluoride). Cells were broken by bead beating (45 min at 4 °C) with glass beads and centrifuged for 5 min at 1000 g, and the supernatant was collected. A total of 20 μg of total protein extract was resolved by SDS-PAGE using 10% acrylamide gels. For immunoprecipitations, 500 μg of proteins were prepared and precleared with a 20 μg protein A-Sepharose and protein G-Sepharose bead mixture (GE Healthcare). A total of 2 μg of anti-PCNA antibody were added to the cleared extract and incubated overnight at 4 °C. The beads were washed five times with lysis buffer at a NaCl concentration of 220 mM. The resulting immunoprecipitates were loaded for SDS-PAGE using acrylamide gels.
3. Results
3.1. Increased Mutagenesis and Unequal Sister Chromatid Recombination in Pol3-01 Mutants
The defective polymerase δ of pol3-01 mutants shows no detectable exonucleolytic activity in vitro [34,35]. pol3-01 strains were found to have high mutagenesis levels, a fact that was attributed to its lack of exonuclease function [31,34,36]. We decided to test this hypothesis in several ways.
First, we measured the level of mutagenesis in wild-type and pol3-01 strains using three different assays: a base substitution at the TRP1 gene, forward mutation at the CAN1 gene, and reversion of a 14A stretch insertion at the LYS2 gene. The only way to revert the trp1-289 mutation is by a very specific base substitution of the type promoted by Polζ activity. In contrast, the CAN1 assay can detect all types of mutations [37]. Most mutations in the LYS2 assay are either the deletion of two nucleotides or the insertion of a single adenine caused by polymerase slippage and independent of TLS polymerase activity [38]. Figure 2A shows that mutation levels were increased in the pol3-01 mutant cells in the three assays used (19-fold, 25-fold, and 50-fold, respectively).
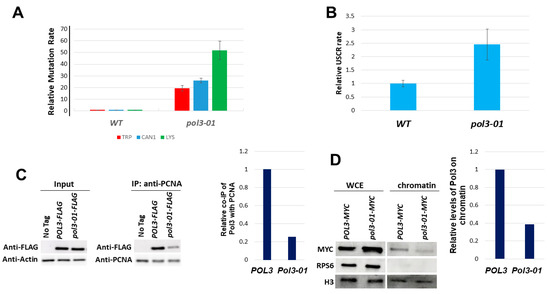
Figure 2.
Phenotypes of pol3-01 cells. (A). Relative rate of mutation of pol3-01 cells in three different mutagenesis assays. The rate of the wild type is set to 1 in each assay. The wild-type rates for Trp+, CanR, and Lys+ mutations were 1, 2, and 1 × 10−7. (B). Relative rate of unequal sister chromatid recombination (as described in Figure S1). The rate of the wild type in this assay is 5 × 10−5. (C). co-Immunoprecipitation levels of Pol3-FLAG or Pol3-01-Flag with PCNA. (D). Effect of the pol3-01 mutation on the level of Myc-tagged Pol3 protein on chromatin. Mid-log cells were fractionated into chromatin and non-chromatin fractions and probed with anti-Myc antibodies. Histone H3 served as a loading marker. WCE: whole cell extract before fractionation.
In addition to the mutagenic bypass of lesions by trans-lesion synthesis polymerases, an alternative DNA repair pathway uses the sister chromatid as a template to carry out an error-free bypass ([16,17]). This sister chromatid recombination cannot be directly detected, as the information in the two sister chromatids is identical. We, therefore, used an assay [32] that allows following unequal sister chromatid recombination (USCR, Figure S1). In this assay, pol3-01 strains show a rate 2.5-fold higher than that of the wild-type control (Figure 2B).
3.2. Reduced Interaction between Pol3-01 and PCNA
PCNA is a central regulator of lesion bypass mechanisms. To investigate the physical interaction between Pol3-01 to PCNA (Pol30), we used the Yeast Two-Hybrid assay. Figure S2 shows that Pol3-01 has a weaker interaction with PCNA than the WT Pol3 protein. These results were confirmed by co-immunoprecipitation (co-IP) experiments (Figure 2C): PCNA was immunoprecipitated, and the level of FLAG-tagged versions of Pol3 or Pol3-01 co-IPed was measured. The pol3-01 strain exhibited a 4-fold reduction in co-IP. If, indeed, Pol3-01 interacts in a weaker manner than Pol3 with PCNA, we expect it to have less accumulation of Pol3-01 on chromatin. In Figure 2D, we show that, indeed, pol3-01 accumulates less on the chromatin compared to WT (about a quarter of the wild-type level).
Polδ consists of three subunits; Pol3, Pol31, and Pol32 [8,9]. Pol3 interacts with Pol31, which interacts with Pol32 [13] (Figure 1A). The pol3-01 allele carries two mutations at residues 321 and 323 (D321A, E323A). According to the recently published crystal structure of Polδ [13], these changes may disturb the interaction of the Pol3-01 protein with Pol31, and through it, with Pol32 and PCNA. It is thus possible that the weaker interaction with PCNA seen (Figure 2C) reflects the indirect interaction with Pol32 (Figure 1B).
To test whether the elevated mutation rate and the elevated sister chromatid recombination rate of pol3-01 are due to lower attachment of the Polδ subunit to PCNA, we changed the weak PCNA-interacting peptide (PIP) motif of pol3-01 to that of POL32 (pol3-01-pip32), thus strengthening the interaction with PCNA (Figure 1C). We carried out a co-IP experiment: PCNA was immunoprecipitated, and the amount of pol3-01 or pol3-01-pip32 co-IPed was measured. As can be seen in Figure 3A, pol3-01-pip32 exhibited a 4-fold increase in co-IP compared to pol3-01. In addition, adding the PIP motif of POL32 to the wild type or the Pol3-01 protein led to a ~3-fold increase in their levels in the chromatin fraction (Figure 3B). Thus, indeed, Pol3 proteins bearing the PIP32 motif show increased affinity for PCNA and increased chromatin localization.
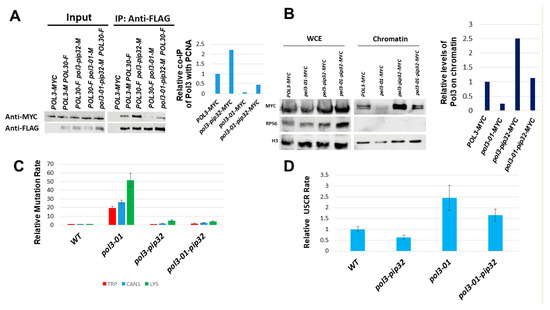
Figure 3.
Effects of changing Pol3’s PIP by that of Pol32. (A). Co-IP assay between Myc-tagged Pol3/Pol3-01/Pol3-01-pip32 and FLAG-tagged PCNA (Pol30). Proteins were immunoprecipitated with anti-FLAG antibodies. Western blotting was performed using anti-Myc to detect the presence of tagged proteins within the complexes. (B). Effect of the pol3-01 mutation on the level of Myc-tagged Pol3 protein on chromatin. Mid-log cells were fractionated into chromatin and non-chromatin fractions and probed with anti-Myc antibodies. Histone H3 served as a loading marker. WCE: whole cell extract before fractionation. (C). Relative rate of the mutation using three different mutagenesis assays. (D). Relative rate of unequal sister chromatid recombination.
Next, we checked whether strengthening pol3-01′s interaction with PCNA affects the rates of mutation and sister chromatid recombination. Figure 3C shows that adding to pol3-01 the PIP motif of Pol32 (pol3-01-pip32) reduces the mutation rate to about a tenth of its value. It also slightly reduces sister chromatid recombination (Figure 3C). Taken together, these results suggest that increasing the weak interaction between Polδ and PCNA, using a stronger PIP motif suppresses the high mutation and recombination rates of pol3-01 cells.
The CysB region of Pol3 plays an important role in the interaction between Pol3 and Pol31 [13,39]. Indeed, mutations in this region, such as pol3-11, confer a temperature-sensitive phenotype. The pol31-K358E allele of Pol31 was found to be a suppressor of pol3-11 [40,41]. The lysine 358 residue physically interacts with the Cys domains of Pol3 [39]. We, therefore, measured the level of Pol3 and Pol3-01 on chromatin in the pol31-K358E background. Figure 4A,B show that, indeed, the level of both versions of Pol3 is increased.
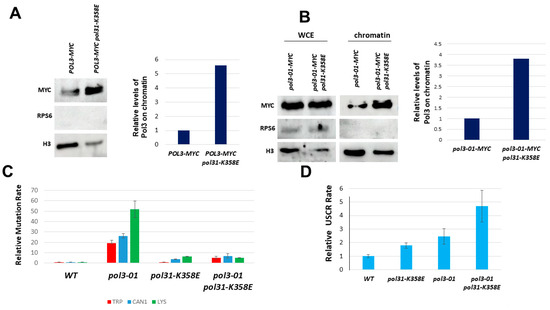
Figure 4.
Effects of the pol31-K358E mutation on Pol3-01. (A). Fractionation analysis shows increased levels of Pol3 on the chromatin fraction. (B). Fractionation analysis shows increased levels of Pol3-01 on the chromatin fraction. (C). pol31-K358E suppresses the high mutagenesis rate of pol3-01 cells. (D). pol31-K358E increases sister chromatid recombination in cells with wild-type Pol3 or with Pol3-01.
We also checked the mutation rate of pol3-01 in a pol31-K358E background. The pol31-K358E mutation completely suppresses the increased mutagenesis rates of pol3-01 (Figure 4C). In contrast, sister chromatid recombination was strongly elevated (Figure 4D)
To conclude so far, our results show that pol3-01 increases both mutagenesis and sister chromatid recombination and that this increase depends on pol3-01′s chromatin level; if we strengthen the interaction between pol3-01 and PCNA, we eliminate the increase in mutation rate. In contrast, the effects on USCR differed between pol3-01-pip32 and pol3-01 pol31-K358E. Below, we dissect their potential mechanism of action.
The trans-lesion synthesis (TLS) pathway uses an alternative polymerase that replaces the replicative polymerases and allows the bypass of the lesion. The main TLS polymerase in yeast is Polζ, composed of Rev3 (the catalytic subunit), Rev7, Pol31, and Pol32 [39]. Since we showed that pol3-01 increases the error-prone pathway, we asked whether this increase is REV3-dependent. We used the trp1-289 mutation rate assay, which measures the rate of base substitution mutations, the main type of mutations induced by Rev3 [42].
According to Figure 5A, 75% of the increased mutagenesis of pol3-01 is REV3-dependent. Thus, the lack of exonucleolytic activity in the pol3-01 mutant only accounts for a quarter of the increased mutagenesis, and the main creator of mutations is the Polζ TLS polymerase. Since other TLS polymerases (Rev1, Rad30) may also contribute to the phenotypes, the contribution of the lack of exonucleolytic activity may be even minor.
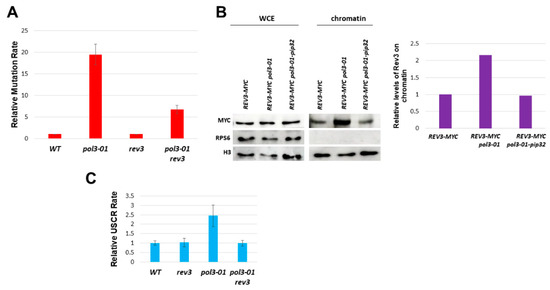
Figure 5.
Role of Rev3 in mutagenesis and sister chromatid recombination. (A). The high rate of mutagenesis of pol3-01 in the TRP assay is dependent on REV3. (B). Fractionation experiment shows increased Rev3 on the chromatin fraction in pol3-01. The pip32 mutation reduces its levels to those of the wild-type strain. (C). The increased USCR of pol3-01 also depends on REV3.
Considering these results, our hypothesis is that pol3-01′s low interaction with PCNA facilitates a more efficient replacement of the replicative Polδ by Polζ, thus increasing the mutation rate in a REV3-dependent manner. A corollary of this model is that we expect to observe a higher level of Rev3 at chromatin in pol3-01 strains in comparison to the WT.
As can be seen in Figure 5B, in pol3-01′s background, there is indeed an increased level of Rev3 on the chromatin, compared to POL3 and pol3-01-pip32 backgrounds.
These results strengthen our theory: pol3-01′s low interaction with PCNA allows the replacement of Pol3 by Rev3. This replacement allows the usage of the TLS pathway. When we strengthen the interaction between pol3-01 and PCNA by using PIP32, we obtain a higher level of Pol3 on the chromatin, but a lower level of Rev3 on the chromatin, compared to pol3-01, in addition to a decreased mutation rate (Figure 5B).
Interestingly, the increase in recombination seen in pol3-01 is eliminated by deleting REV3 (Figure 5C). This result implies that the replacement of Polδ by Polζ in the presence of the pol3-01 mutant also causes an elevation in the USCR rate.
We used a triply-tagged strain in order to measure the interactions between Pol31, Pol3, and Rev3. A co-immunoprecipitation experiment was carried out by immunoprecipitating HA-tagged Pol31 and measuring the level of co-IP by Western blot, using anti-Myc antibodies able to detect both Pol3 and Rev3. Figure 6A confirms that in pol3-01 cells, the interaction between Pol31 and the mutant Pol3 subunit is weak and is restored to normal levels in the pol31-K358E background. The figure also shows that the interaction between Rev3 and Pol31 is almost undetectable in wild-type cells, but it increases in pol3-01 and in both the POL3 and pol3-01 genetic backgrounds when POL31 is mutated.
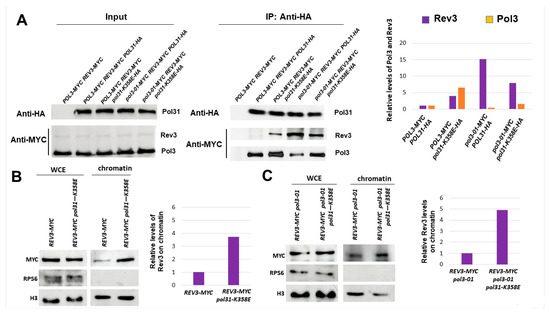
Figure 6.
pol31-K358E increases the affinity of Pol3 to Rev3. (A). Co-Immunoprecipitation between HA-tagged Pol31 and Myc-tagged Rev3 and Pol3. Mid-log cells were immunoprecipitated with anti-HA antibodies, and Western blotting was performed using anti-Myc to detect the presence of tagged proteins within the complexes. (B). Fractionation analysis shows increased levels of Rev3 on the chromatin fraction in pol31-K358E cells. (C). Fractionation analysis shows increased levels of Rev3 on the chromatin fraction in pol3-01 pol31-K358E cells.
Although we demonstrated that in the pol31-K358E background there are higher levels of Pol3 (or Pol3-01) on the chromatin (Figure 4A,B), and indeed more interaction of the catalytic subunit with Pol31 (Figure 6A), Figure 6B shows that the single pol31-K358E mutant also exhibits a 4-fold increase in accumulation of Rev3 on chromatin. The increase in Rev3 at the chromatin fraction can also be seen in the pol3-01 pol31-K358E double mutant (Figure 6C).
In summary, our results show two very different mechanisms by which the increased mutagenesis of pol3-01 can be suppressed: Strengthening the interactions with PCNA by mutating Pol3’s PIP reduces both mutation and sister chromatid recombination, and this result can be explained by the increase in Pol3-01 and a decrease in Rev3 at the chromatin. In contrast, the pol31-K358E mutation increases Pol31’s interactions with both Pol3 and Rev3. Although we see a reduction in mutagenesis, we also see an increase in the rate of sister chromatid recombination, both in the single pol31-K358E and in the pol3-01 pol31-K358E double mutant (Figure 4D).
4. Discussion
Timely and accurate replication of the genome is essential for life. DNA polymerases must balance the need for speedy activity with a requirement for accuracy. The proofreading mechanism of DNA polymerases, which detects their own errors while copying the genome, prevents the incorporation of mismatched nucleotides. The exonuclease activity removes the mistaken nucleotide, and a new, suitable nucleotide is incorporated instead [36,43,44]. This requires a change in the pace of progression and probably a change in the 3D architecture of the enzyme [45].
We have investigated pol3-01, a POL3 mutant that lacks exonuclease function and exhibits increased rates of mutation and sister chromatid recombination. Our results show that in addition to a defect in exonucleolytic activity, Pol3-01 has reduced interaction with PCNA and a lower protein level at the chromatin (Figure 2).
To test whether the interaction with the replicative clamp plays a role in the increased levels of mutation and unequal sister chromatid recombination, we looked for different ways to strengthen the interaction between Pol3-01 and PCNA. We used two different methods:
(1) A change of the weak PCNA-interacting motif to the stronger PIP of Pol32. (2) Introducing pol31-K358E, a mutation in Pol31 that strengthens the interactions between Pol3 and Pol31, and thus ensures better attachment of Polδ to PCNA. Our results show that both strategies were successful in stabilizing Pol3 (Figure 3 and Figure 4) and substantially reduced the high mutation rate of pol3-01.
Since the exonucleolytic activity of Pol3 plays an important role in providing accuracy to the enzyme, it is logical to assume that the increased mutagenesis of pol3-01 is a direct result of the misincorporation by the mutated DNA polymerase. However, we show (Figure 5A) that two-thirds of the base substitutions in pol3-01 strains depend on the activity of the Polζ trans-lesion synthesis polymerase. Our results may thus explain why mutations in the exonucleolytic domain of Polδ result in a much higher rate of mutation than similar defects in the catalytic subunit of Polε [46] if only the first and not the latter facilitate the exchange with Polζ.
Since the high mutagenesis observed depends on the interactions with PCNA and on Rev3, we propose that the high level of mutations observed in pol3-01 strains is due to the fact that the Pol3-01 protein tends to fall off chromatin, allowing an easier replacement of Polδ by Polζ (Figure 1B). Misincorporation of nucleotides by Polζ thus accounts for most of the increased mutagenesis.
In support of this hypothesis, we show that the level of Pol3-01 on chromatin is reduced (Figure 2), whereas a higher amount of Rev3 can be seen in the chromatin fraction in pol3-01 cells (Figure 5B). Changing the PIP motif of Pol3-01 to that of the Pol32 protein increases the stability of Polδ, leading to higher protein levels in chromatin (Figure 3) and reducing the levels of Rev3 to those of the wild type (Figure 5B).
The facilitated exchange of DNA polymerases (between Pol3-01 and Polζ) may not necessitate PCNA ubiquitination. Indeed, trans-lesion synthesis in the absence of PCNA ubiquitination has been observed both in yeast and human cells [47,48].
An alternative to the error-prone lesion bypass mechanism is the error-free template switch (TS) pathway. The molecular details of this process are still being delineated; we only know that it is catalyzed by Rad5, a protein that has both helicase and poly-ubiquitination activities [16,17]. Our current understanding is that during TS, information is copied from the recently created sister chromatid in a process that results in a sister chromatid recombination. Since the two chromatids created during DNA synthesis are identical and thus indistinguishable from each other, we used unequal sister chromatid recombination (USCR) as an assay for TS. pol3-01 strains showed increased USCR levels (Figure 2B), and these were decreased by replacing the PIP of Pol3 with a stronger PIP motif (Figure 3D). Thus, increasing Polδ affinity for PCNA via its PIP motif reduces the use of both the error-prone and the error-free branches of the DNA Damage Tolerance pathway.
We took advantage of the pol31-K358E mutation, which strengthens the interactions between Pol31 and Pol3 and, thus, indirectly, with Pol32 and PCNA [39]. Figure 4A,B and Figure 6A show that, similar to the PIP mutation, pol31-K358E leads to increased interactions with Pol3-01 and elevated levels of Polδ on the chromatin. Consistently, we also see a dramatic reduction in the level of mutations in a pol3-01 pol31-K358E double mutant (Figure 4C). However, in contrast to the pol3-01-pip32 strain, the double mutant exhibits higher, not lower, levels of USCR (Figure 4D). Thus, although both the POL3 PIP change and the POL31 mutation have similar effects on the error-prone branch, they differ in their effect on the error-free branch of the DDT.
When we measured the level of Rev3 on the chromatin, we saw that in contrast to cells carrying the pol3-01-pip32 allele, pol3-01 pol31-K358E strains exhibited higher, not lower, levels of Rev3, in addition to having higher levels of Pol3-01 (Figure 6). Thus, pol31-K358E strengthens the interaction of Pol31 with both Pol3 and Rev3. The reduced levels of mutation are thus probably due to a higher affinity of Pol3 over Rev3 in the binding to the mutant Pol31 subunit.
Deletion of REV3 in the pol3-01 strain resulted in a reduction in USCR (Figure 5C). This result uncovers an unexpected role of Polζ in the TS branch of the DDT. This role is minor in the wild type (no changes in USCR are observed in single rev3∆ mutants) but becomes visible when the stability of Pol3 is compromised by the pol3-01 mutation. It is possible that the instability of Pol3-01 facilitates the use of alternative repair pathways, such as the error-free DDT branch or the microhomology-mediated break-induced replication (MMBIR), which also depends on Rev3 [49].
In summary, we present evidence for the fact that the increased mutagenesis in pol3-01 mutants is mainly due to its lower stability on the chromatin and not only due to lack of exonucleolytic activity. Since mutations in the exonuclease domain of DNA polymerases are common in various types of cancer in humans [29,30], it is particularly interesting to determine whether they also lead to increased mutagenesis that depends on TLS polymerases.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes14020391/s1, Figure S1: Assay for measuring unequal sister chromatid recombination (USCR); Figure S2: Yeast two-hybrid experiment to test interactions between POL30 (PCNA) fused to the activating domain (AD)of Gal4, and different POL3 alleles fused to the DNA binding domain of Gal4 (DBD).
Author Contributions
Conceptualization, S.N.H., M.G., B.L. and M.K.; formal analysis, S.N.H., M.G., B.L. and M.K.; investigation, S.N.H., M.G., B.L. and M.K.; writing—original draft preparation, S.N.H., M.G., B.L. and M.K.; writing—review and editing, S.N.H., M.G., B.L. and M.K.; supervision, M.K.; funding acquisition, M.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Israel Science Foundation, grant number 140/18, the Israel Cancer Research Fund, grant 458, the Minerva Stiftung, grant #238, and the DFG-Middle East, grant 188/4-1.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank all members of the Kupiec lab for support and ideas.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pavlov, Y.I.; Frahm, C.; Nick McElhinny, S.A.; Niimi, A.; Suzuki, M.; Kunkel, T.A. Evidence that errors made by DNA polymerase α are corrected by DNA polymerase δ. Curr. Biol. 2006, 16, 202–207. [Google Scholar] [CrossRef] [PubMed]
- Burgers, P.M.J.; Kunkel, T.A. Eukaryotic DNA Replication Fork. Annu. Rev. Biochem. 2017, 86, 417–438. [Google Scholar] [CrossRef] [PubMed]
- Pursell, Z.F.; Isoz, I.; Lundstrom, E.B.; Johansson, E.; Kunkel, T.A. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science 2007, 317, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Nick McElhinny, S.A.; Gordenin, D.A.; Stith, C.M.; Burgers, P.M.; Kunkel, T.A. Division of labor at the eukaryotic replication fork. Mol. Cell 2008, 30, 137–144. [Google Scholar] [CrossRef]
- Bauer, G.A.; Burgers, P.M. The yeast analog of mammalian cyclin/proliferating-cell nuclear antigen interacts with mammalian DNA polymerase delta. Proc. Natl. Acad. Sci. USA 1988, 85, 7506–7510. [Google Scholar] [CrossRef]
- Jonsson, Z.O.; Hindges, R.; Hubscher, U. Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen. EMBO J. 1998, 17, 2412–2425. [Google Scholar] [CrossRef]
- Warbrick, E. The puzzle of PCNA’s many partners. BioEssays 2000, 22, 997–1006. [Google Scholar] [CrossRef]
- Boulet, A.; Simon, M.; Faye, G.; Bauer, G.A.; Burgers, P.M. Structure and function of the Saccharomyces cerevisiae CDC2 gene encoding the large subunit of DNA polymerase III. EMBO J 1989, 8, 1849–1854. [Google Scholar] [CrossRef]
- Gerik, K.J.; Li, X.; Pautz, A.; Burgers, P.M. Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase δ. J. Biol. Chem. 1998, 273, 19747–19755. [Google Scholar] [CrossRef]
- Johansson, E.; Majka, J.; Burgers, P.M. Structure of DNA polymerase δ from Saccharomyces cerevisiae. J. Biol. Chem. 2001, 276, 43824–43828. [Google Scholar] [CrossRef]
- Huang, M.E.; Cadieu, E.; Souciet, J.L.; Galibert, F. Disruption of six novel yeast genes reveals three genes essential for vegetative growth and one required for growth at low temperature. Yeast 1997, 13, 1181–1194. [Google Scholar] [CrossRef]
- Johansson, E.; Garg, P.; Burgers, P.M. The Pol32 subunit of DNA polymerase δ contains separable domains for processive replication and proliferating cell nuclear antigen (PCNA) binding. J. Biol. Chem. 2004, 279, 1907–1915. [Google Scholar] [CrossRef]
- Jain, R.; Rice, W.J.; Malik, R.; Johnson, R.E.; Prakash, L.; Prakash, S.; Ubarretxena-Belandia, I.; Aggarwal, A.K. Cryo-EM structure and dynamics of eukaryotic DNA polymerase δ holoenzyme. Nat. Struct. Mol. Biol. 2019, 26, 955–962. [Google Scholar] [CrossRef]
- Acharya, N.; Klassen, R.; Johnson, R.E.; Prakash, L.; Prakash, S. PCNA binding domains in all three subunits of yeast DNA polymerase δ modulate its function in DNA replication. Proc. Natl. Acad. Sci. USA 2011, 108, 17927–17932. [Google Scholar] [CrossRef]
- Burkovics, P.; Sebesta, M.; Sisakova, A.; Plault, N.; Szukacsov, V.; Robert, T.; Pinter, L.; Marini, V.; Kolesar, P.; Haracska, L.; et al. Srs2 mediates PCNA-SUMO-dependent inhibition of DNA repair synthesis. EMBO J. 2013, 32, 742–755. [Google Scholar] [CrossRef]
- Arbel, M.; Liefshitz, B.; Kupiec, M. How yeast cells deal with stalled replication forks. Curr. Genet. 2020, 66, 911–915. [Google Scholar] [CrossRef]
- Arbel, M.; Liefshitz, B.; Kupiec, M. DNA damage bypass pathways and their effect on mutagenesis in yeast. FEMS Microbiol. Rev. 2021, 45, fuaa038. [Google Scholar] [CrossRef]
- Bailly, V.; Lauder, S.; Prakash, S.; Prakash, L. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J. Biol. Chem. 1997, 272, 23360–23365. [Google Scholar] [CrossRef]
- Hoege, C.; Pfander, B.; Moldovan, G.L.; Pyrowolakis, G.; Jentsch, S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 2002, 419, 135–141. [Google Scholar] [CrossRef]
- Nelson, J.R.; Lawrence, C.W.; Hinkle, D.C. Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science 1996, 272, 1646–1649. [Google Scholar] [CrossRef]
- Cassier, C.; Chanet, R.; Henriques, J.A.; Moustacchi, E. The effects of three PSO genes on induced mutagenesis: A novel class of mutationally defective yeast. Genetics 1980, 96, 841–857. [Google Scholar] [CrossRef] [PubMed]
- Quah, S.K.; von Borstel, R.C.; Hastings, P.J. The origin of spontaneous mutation in Saccharomyces cerevisiae. Genetics 1980, 96, 819–839. [Google Scholar] [CrossRef] [PubMed]
- Gan, G.N.; Wittschieben, J.P.; Wittschieben, B.O.; Wood, R.D. DNA polymerase zeta (pol ζ) in higher eukaryotes. Cell Res. 2008, 18, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Goodman, M.F. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 2002, 71, 17–50. [Google Scholar] [CrossRef]
- Branzei, D.; Szakal, B. DNA damage tolerance by recombination: Molecular pathways and DNA structures. DNA Repair 2016, 44, 68–75. [Google Scholar] [CrossRef]
- Zhang, H.; Lawrence, C.W. The error-free component of the RAD6/RAD18 DNA damage tolerance pathway of budding yeast employs sister-strand recombination. Proc. Natl. Acad. Sci. USA 2005, 102, 15954–15959. [Google Scholar] [CrossRef]
- Ulrich, H.D.; Jentsch, S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 2000, 19, 3388–3397. [Google Scholar] [CrossRef]
- Torres-Ramos, C.A.; Prakash, S.; Prakash, L. Requirement of RAD5 and MMS2 for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 2002, 22, 2419–2426. [Google Scholar] [CrossRef]
- Church, D.N.; Briggs, S.E.; Palles, C.; Domingo, E.; Kearsey, S.J.; Grimes, J.M.; Gorman, M.; Martin, L.; Howarth, K.M.; Hodgson, S.V.; et al. DNA polymerase ε and δ exonuclease domain mutations in endometrial cancer. Hum. Mol. Genet. 2013, 22, 2820–2828. [Google Scholar] [CrossRef]
- Palles, C.; Cazier, J.B.; Howarth, K.M.; Domingo, E.; Jones, A.M.; Broderick, P.; Kemp, Z.; Spain, S.L.; Guarino, E.; Salguero, I.; et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat. Genet. 2013, 45, 136–144. [Google Scholar] [CrossRef]
- Morrison, A.; Johnson, A.L.; Johnston, L.H.; Sugino, A. Pathway correcting DNA replication errors in Saccharomyces cerevisiae. EMBO J. 1993, 12, 1467–1473. [Google Scholar] [CrossRef]
- Kadyk, L.C.; Hartwell, L.H. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 1992, 132, 387–402. [Google Scholar] [CrossRef]
- Lea, D.E.; Coulson, C.A. The distribution of the numbers of mutants in bacterial populations. J. Genet. 1949, 49, 264–285. [Google Scholar] [CrossRef]
- Simon, M.; Giot, L.; Faye, G. The 3′ to 5′ exonuclease activity located in the DNA polymerase delta subunit of Saccharomyces cerevisiae is required for accurate replication. EMBO J. 1991, 10, 2165–2170. [Google Scholar] [CrossRef]
- Jin, Y.H.; Obert, R.; Burgers, P.M.; Kunkel, T.A.; Resnick, M.A.; Gordenin, D.A. The 3′-->5′ exonuclease of DNA polymerase δ can substitute for the 5′ flap endonuclease Rad27/Fen1 in processing Okazaki fragments and preventing genome instability. Proc. Natl. Acad. Sci. USA 2001, 98, 5122–5127. [Google Scholar] [CrossRef]
- Tran, H.T.; Gordenin, D.A.; Resnick, M.A. The 3′-->5′ exonucleases of DNA polymerases delta and epsilon and the 5'-->3' exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999, 19, 2000–2007. [Google Scholar] [CrossRef]
- Lang, G.I.; Murray, A.W. Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics 2008, 178, 67–82. [Google Scholar] [CrossRef]
- Tran, H.T.; Keen, J.D.; Kricker, M.; Resnick, M.A.; Gordenin, D.A. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol. Cell. Biol. 1997, 17, 2859–2865. [Google Scholar] [CrossRef]
- Johnson, R.E.; Prakash, L.; Prakash, S. Pol31 and Pol32 subunits of yeast DNA polymerase delta are also essential subunits of DNA polymerase zeta. Proc. Natl. Acad. Sci. USA 2012, 109, 12455–12460. [Google Scholar] [CrossRef]
- Giot, L.; Chanet, R.; Simon, M.; Facca, C.; Faye, G. Involvement of the yeast DNA polymerase delta in DNA repair in vivo. Genetics 1997, 146, 1239–1251. [Google Scholar] [CrossRef]
- Giot, L.; Simon, M.; Dubois, C.; Faye, G. Suppressors of thermosensitive mutations in the DNA polymerase delta gene of Saccharomyces cerevisiae. Mol. Gen. Genet. 1995, 246, 212–222. [Google Scholar] [CrossRef] [PubMed]
- Volkova, N.V.; Meier, B.; Gonzalez-Huici, V.; Bertolini, S.; Gonzalez, S.; Vohringer, H.; Abascal, F.; Martincorena, I.; Campbell, P.J.; Gartner, A.; et al. Mutational signatures are jointly shaped by DNA damage and repair. Nat. Commun. 2020, 11, 2169. [Google Scholar] [CrossRef] [PubMed]
- Brutlag, D.; Kornberg, A. Enzymatic synthesis of deoxyribonucleic acid. 36. A proofreading function for the 3′ leads to 5′ exonuclease activity in deoxyribonucleic acid polymerases. J. Biol. Chem. 1972, 247, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Kunkel, T.A. Exonucleolytic proofreading. Cell 1988, 53, 837–840. [Google Scholar] [CrossRef]
- Jin, Y.H.; Garg, P.; Stith, C.M.; Al-Refai, H.; Sterling, J.F.; Murray, L.J.; Kunkel, T.A.; Resnick, M.A.; Burgers, P.M.; Gordenin, D.A. The multiple biological roles of the 3′-->5′ exonuclease of Saccharomyces cerevisiae DNA polymerase delta require switching between the polymerase and exonuclease domains. Mol. Cell. Biol. 2005, 25, 461–471. [Google Scholar] [CrossRef]
- Morrison, A.; Sugino, A. The 3′-->5′ exonucleases of both DNA polymerases delta and epsilon participate in correcting errors of DNA replication in Saccharomyces cerevisiae. Mol. Gen. Genet. 1994, 242, 289–296. [Google Scholar] [CrossRef]
- Hendel, A.; Krijger, P.H.; Diamant, N.; Goren, Z.; Langerak, P.; Kim, J.; Reissner, T.; Lee, K.Y.; Geacintov, N.E.; Carell, T.; et al. PCNA ubiquitination is important, but not essential for translesion DNA synthesis in mammalian cells. PLoS Genet. 2011, 7, e1002262. [Google Scholar] [CrossRef]
- Tellier-Lebegue, C.; Dizet, E.; Ma, E.; Veaute, X.; Coic, E.; Charbonnier, J.B.; Maloisel, L. The translesion DNA polymerases Pol ζ and Rev1 are activated independently of PCNA ubiquitination upon UV radiation in mutants of DNA polymerase δ. PLoS Genet. 2017, 13, e1007119. [Google Scholar] [CrossRef]
- Sakofsky, C.J.; Ayyar, S.; Deem, A.K.; Chung, W.H.; Ira, G.; Malkova, A. Translesion Polymerases Drive Microhomology-Mediated Break-Induced Replication Leading to Complex Chromosomal Rearrangements. Mol. Cell 2015, 60, 860–872. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).