Genetic and Structural Diversity of Prokaryotic Ice-Binding Proteins from the Central Arctic Ocean
Abstract
:1. Introduction
2. Materials and Methods
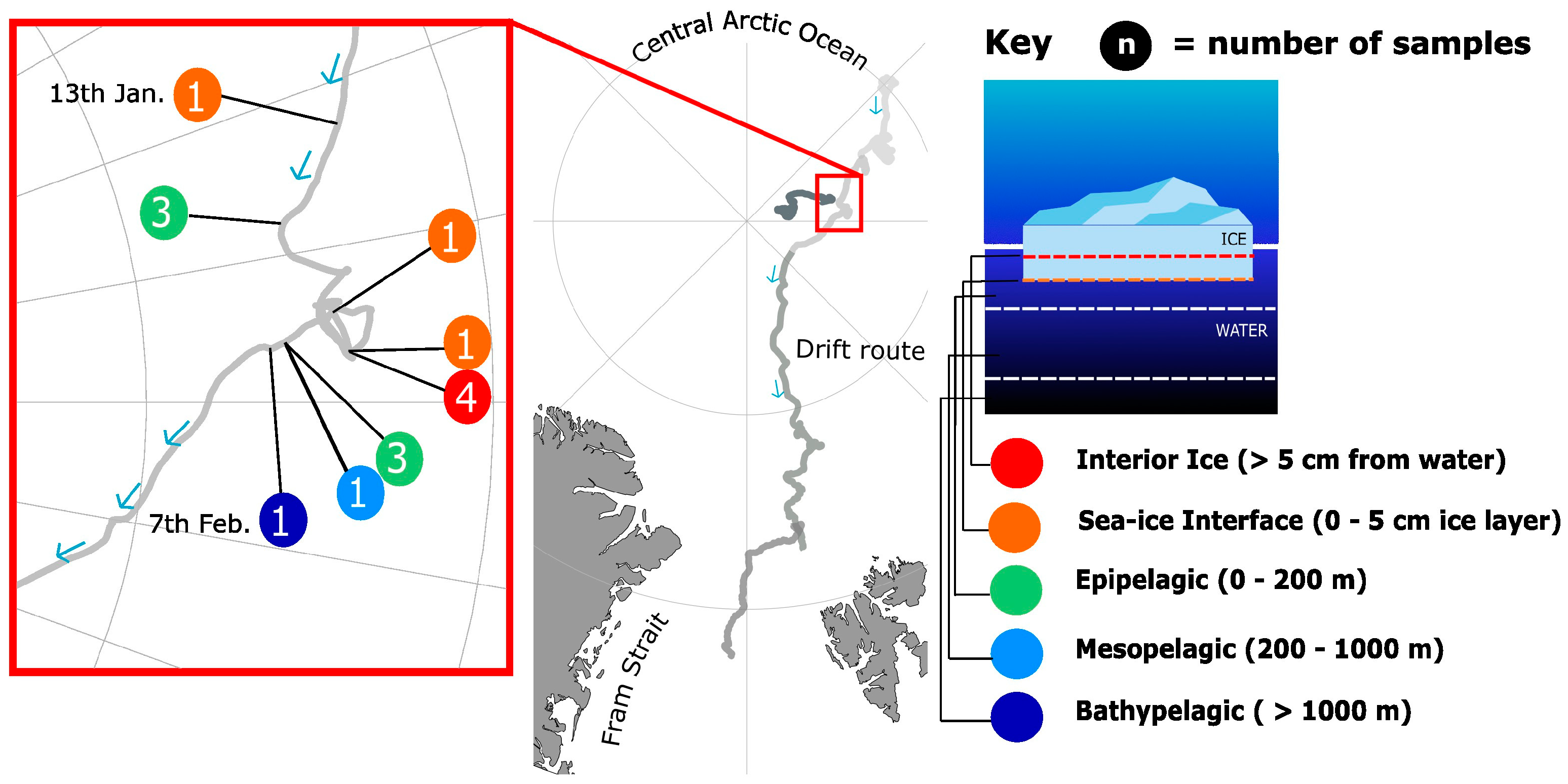
2.1. Sample Collection
2.2. DNA Extraction and Sequencing
Bioinformatics Processing of Samples
2.3. Community Analysis
2.4. Protein Structure Prediction
2.5. Upstream and Downstream Gene Analysis
2.6. Phylogenetic Analysis of IBPs
3. Results
3.1. Diverse Prokaryotic Communities and MAGs Encode IBP Genes
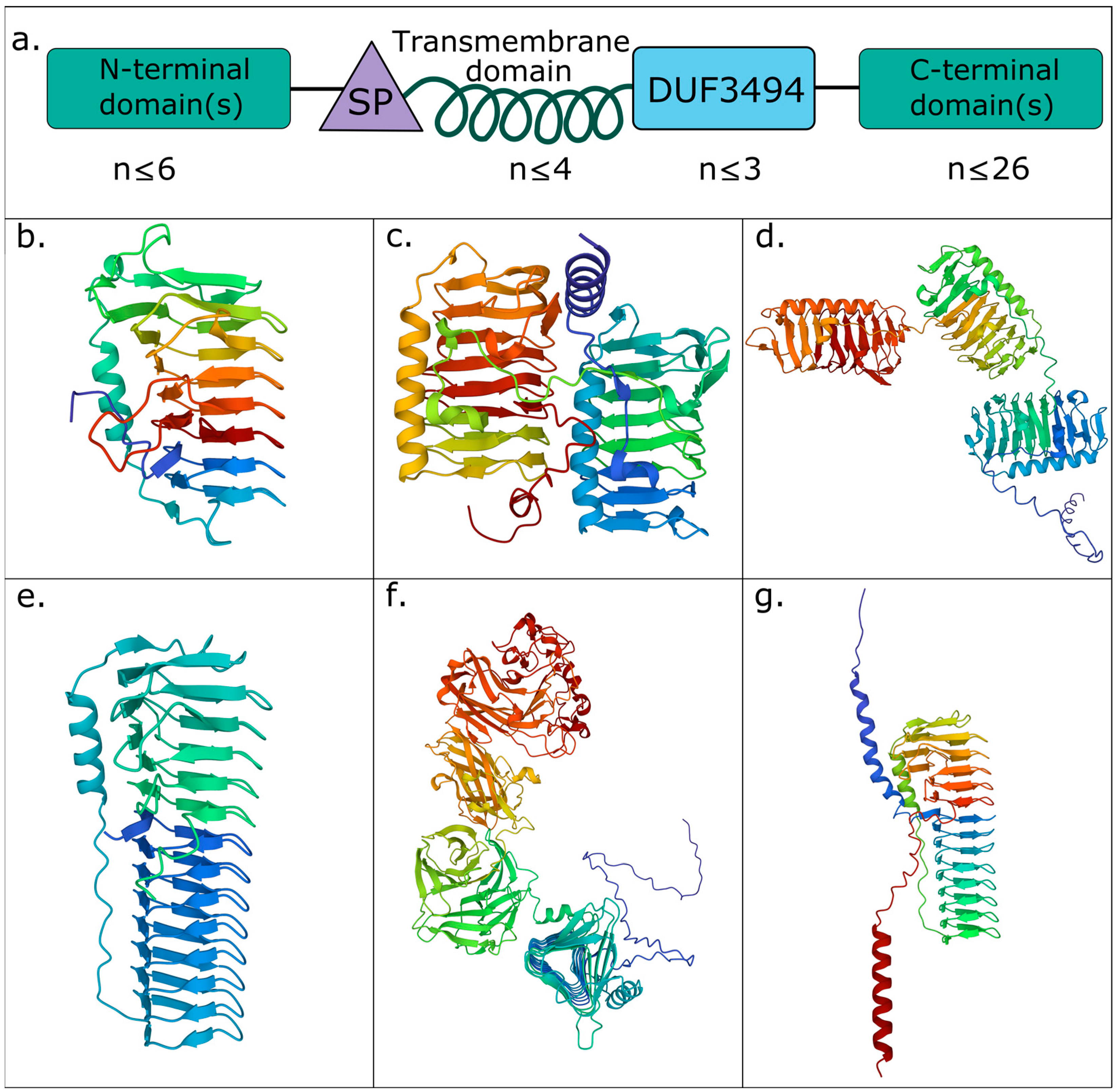
3.2. Diverse IBP Structures Are Abundant in the Natural Environment
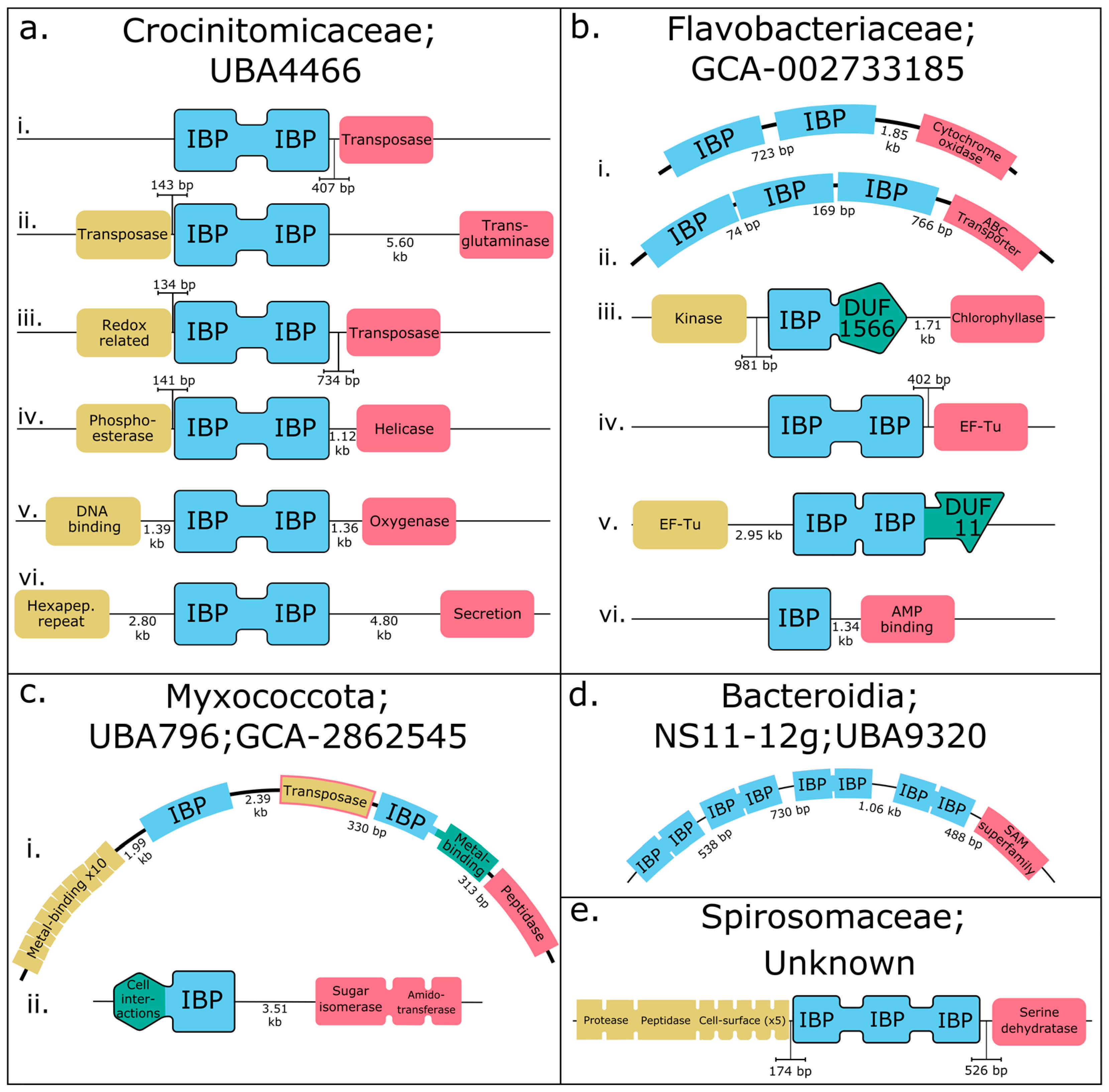
3.3. The Genomic Context of IBPs Suggests Mechanisms for Generating Diversity
3.4. Phylogenetic Distribution of Abundant Domain Architectures Implicates Domain Shuffling
3.5. IBPs from the Total Assembly Cluster Taxonomically
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gruneberg, A.K.; Graham, L.A.; Eves, R.; Agrawal, P.; Oleschuk, R.D.; Davies, P.L. Ice Recrystallization Inhibition Activity Varies with Ice-Binding Protein Type and Does Not Correlate with Thermal Hysteresis. Cryobiology 2021, 99, 28–39. [Google Scholar] [CrossRef]
- Yeh, Y.; Feeney, R.E. Antifreeze Proteins: Structures and Mechanisms of Function. Chem. Rev. 1996, 96, 601–618. [Google Scholar] [CrossRef]
- Yu, S.O.; Brown, A.; Middleton, A.J.; Tomczak, M.M.; Walker, V.K.; Davies, P.L. Ice Restructuring Inhibition Activities in Antifreeze Proteins with Distinct Differences in Thermal Hysteresis. Cryobiology 2010, 61, 327–334. [Google Scholar] [CrossRef]
- Vance, T.D.R.; Bayer-Giraldi, M.; Davies, P.L.; Mangiagalli, M. Ice-Binding Proteins and the ‘Domain of Unknown Function’ 3494 Family. FEBS J. 2019, 286, 855–873. [Google Scholar] [CrossRef] [Green Version]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro Protein Families and Domains Database: 20 Years On. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef]
- Raymond, J.A.; Janech, M.G.; Mangiagalli, M. Ice-Binding Proteins Associated with an Antarctic Cyanobacterium, Nostoc Sp. HG1. Appl. Environ. Microbiol. 2021, 87, e02499-20. [Google Scholar] [CrossRef]
- Hanada, Y.; Nishimiya, Y.; Miura, A.; Tsuda, S.; Kondo, H. Hyperactive Antifreeze Protein from an Antarctic Sea Ice Bacterium Colwellia Sp. Has a Compound Ice-Binding Site without Repetitive Sequences. FEBS J. 2014, 281, 3576–3590. [Google Scholar] [CrossRef]
- Raymond, J.A.; Fritsen, C.; Shen, K. An Ice-Binding Protein from an Antarctic Sea Ice Bacterium. FEMS Microbiol. Ecol. 2007, 61, 214–221. [Google Scholar] [CrossRef] [Green Version]
- Raymond, J.A.; Christner, B.C.; Schuster, S.C. A Bacterial Ice-Binding Protein from the Vostok Ice Core. Extremophiles 2008, 12, 713–717. [Google Scholar] [CrossRef]
- Wang, C.; Pakhomova, S.; Newcomer, M.E.; Christner, B.C.; Luo, B.-H. Structural Basis of Antifreeze Activity of a Bacterial Multi-Domain Antifreeze Protein. PLoS ONE 2017, 12, e0187169. [Google Scholar] [CrossRef] [Green Version]
- Guo, S.; Stevens, C.A.; Vance, T.D.R.; Olijve, L.L.C.; Graham, L.A.; Campbell, R.L.; Yazdi, S.R.; Escobedo, C.; Bar-Dolev, M.; Yashunsky, V.; et al. Structure of a 1.5-MDa Adhesin That Binds Its Antarctic Bacterium to Diatoms and Ice. Sci. Adv. 2017, 3, e1701440. [Google Scholar] [CrossRef] [Green Version]
- Vance, T.D.R.; Graham, L.A.; Davies, P.L. An Ice-Binding and Tandem Beta-Sandwich Domain-Containing Protein in Shewanella Frigidimarina Is a Potential New Type of Ice Adhesin. FEBS J. 2018, 285, 1511–1527. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, S.; Basak, A.J.; Nair, A.V.; Duraivelan, K.; Samanta, D. Immunoglobulin-Fold Containing Bacterial Adhesins: Molecular and Structural Perspectives in Host Tissue Colonization and Infection. FEMS Microbiol. Lett. 2021, 368, fnaa220. [Google Scholar] [CrossRef]
- Bayer-Giraldi, M.; Weikusat, I.; Besir, H.; Dieckmann, G. Characterization of an Antifreeze Protein from the Polar Diatom Fragilariopsis Cylindrus and Its Relevance in Sea Ice. Cryobiology 2011, 63, 210–219. [Google Scholar] [CrossRef]
- Gwak, Y.; Jung, W.; Lee, Y.; Kim, J.S.; Kim, C.G.; Ju, J.-H.; Song, C.; Hyun, J.-K.; Jin, E. An Intracellular Antifreeze Protein from an Antarctic Microalga That Responds to Various Environmental Stresses. FASEB J. 2014, 28, 4924–4935. [Google Scholar] [CrossRef]
- Singh, P.; Hanada, Y.; Singh, S.M.; Tsuda, S. Antifreeze Protein Activity in Arctic Cryoconite Bacteria. FEMS Microbiol. Lett. 2014, 351, 14–22. [Google Scholar] [CrossRef] [Green Version]
- Goordial, J.; Davila, A.; Greer, C.W.; Cannam, R.; DiRuggiero, J.; McKay, C.P.; Whyte, L.G. Comparative Activity and Functional Ecology of Permafrost Soils and Lithic Niches in a Hyper-Arid Polar Desert. Environ. Microbiol. 2017, 19, 443–458. [Google Scholar] [CrossRef]
- Krembs, C.; Eicken, H.; Junge, K.; Deming, J.W. High Concentrations of Exopolymeric Substances in Arctic Winter Sea Ice: Implications for the Polar Ocean Carbon Cycle and Cryoprotection of Diatoms. Deep Sea Res. Part Oceanogr. Res. Pap. 2002, 49, 2163–2181. [Google Scholar] [CrossRef]
- Uhlig, C.; Kilpert, F.; Frickenhaus, S.; Kegel, J.U.; Krell, A.; Mock, T.; Valentin, K.; Beszteri, B. In Situ Expression of Eukaryotic Ice-Binding Proteins in Microbial Communities of Arctic and Antarctic Sea Ice. ISME J. 2015, 9, 2537–2540. [Google Scholar] [CrossRef] [Green Version]
- Raymond, J.A. Dependence on Epiphytic Bacteria for Freezing Protection in an Antarctic Moss, Bryum Argenteum. Environ. Microbiol. Rep. 2016, 8, 14–19. [Google Scholar] [CrossRef]
- Koo, H.; Hakim, J.A.; Bej, A.K. Metagenomic Analysis of Microbial Cold Stress Proteins in Polar Lacustrine Ecosystems. In Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2016; pp. 837–844. ISBN 978-1-119-00481-3. [Google Scholar]
- Overbeek, R.; Fonstein, M.; D’Souza, M.; Pusch, G.D.; Maltsev, N. The Use of Gene Clusters to Infer Functional Coupling. Proc. Natl. Acad. Sci. USA 1999, 96, 2896–2901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, P.H.; Touchon, M.; Cury, J.; Rocha, E.P.C. The Chromosomal Organization of Horizontal Gene Transfer in Bacteria. Nat. Commun. 2017, 8, 841. [Google Scholar] [CrossRef] [Green Version]
- Parks, D.H.; Rinke, C.; Chuvochina, M.; Chaumeil, P.-A.; Woodcroft, B.J.; Evans, P.N.; Hugenholtz, P.; Tyson, G.W. Recovery of Nearly 8,000 Metagenome-Assembled Genomes Substantially Expands the Tree of Life. Nat. Microbiol. 2017, 2, 1533–1542. [Google Scholar] [CrossRef] [PubMed]
- Tully, B.J.; Sachdeva, R.; Graham, E.D.; Heidelberg, J.F. 290 Metagenome-Assembled Genomes from the Mediterranean Sea: A Resource for Marine Microbiology. PeerJ 2017, 5, e3558. [Google Scholar] [CrossRef] [Green Version]
- Tytgat, B.; Verleyen, E.; Obbels, D.; Peeters, K.; Wever, A.D.; D’hondt, S.; Meyer, T.D.; Criekinge, W.V.; Vyverman, W.; Willems, A. Bacterial Diversity Assessment in Antarctic Terrestrial and Aquatic Microbial Mats: A Comparison between Bidirectional Pyrosequencing and Cultivation. PLoS ONE 2014, 9, e97564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duncan, A.; Barry, K.; Daum, C.; Eloe-Fadrosh, E.; Roux, S.; Schmidt, K.; Tringe, S.G.; Valentin, K.U.; Varghese, N.; Salamov, A.; et al. Metagenome-Assembled Genomes of Phytoplankton Microbiomes from the Arctic and Atlantic Oceans. Microbiome 2022, 10, 67. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Zhang, W.; Ding, W.; Wang, M.; Fan, S.; Yang, B.; Mcminn, A.; Wang, M.; Xie, B.; Qin, Q.-L.; et al. Structure and Function of the Arctic and Antarctic Marine Microbiota as Revealed by Metagenomics. Microbiome 2020, 8, 47. [Google Scholar] [CrossRef] [Green Version]
- Royo-Llonch, M.; Sánchez, P.; Ruiz-González, C.; Salazar, G.; Pedrós-Alió, C.; Sebastián, M.; Labadie, K.; Paoli, L.; Ibarbalz, F.M.; Zinger, L.; et al. Compendium of 530 Metagenome-Assembled Bacterial and Archaeal Genomes from the Polar Arctic Ocean. Nat. Microbiol. 2021, 6, 1561–1574. [Google Scholar] [CrossRef] [PubMed]
- Mock, T.; Boulton, W.; Balmonte, J.-P.; Barry, K.; Bertilsson, S.; Bowman, J.; Buck, M.; Bratbak, G.; Chamberlain, E.J.; Cunliffe, M.; et al. Multiomics in the Central Arctic Ocean for Benchmarking Biodiversity Change. PLoS Biol. 2022, 20, e3001835. [Google Scholar] [CrossRef]
- Ottesen, A.; Kocurek, B. QIAGEN DNeasy Power Water SOP. Available online: https://www.protocols.io/view/qiagen-dneasy-power-water-sop-bztap6ie (accessed on 6 December 2022).
- Clum, A.; Huntemann, M.; Bushnell, B.; Foster, B.; Foster, B.; Roux, S.; Hajek, P.P.; Varghese, N.; Mukherjee, S.; Reddy, T.B.K.; et al. DOE JGI Metagenome Workflow. mSystems 2021, 6, e00804-20. [Google Scholar] [CrossRef] [PubMed]
- Bushnell, B. BBMap: A Fast, Accurate, Splice-Aware Aligner. 2014. Available online: https://www.osti.gov/biblio/1241166 (accessed on 7 December 2022).
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. MetaSPAdes: A New Versatile Metagenomic Assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef] [Green Version]
- Lukashin, A.V.; Borodovsky, M. GeneMark.Hmm: New Solutions for Gene Finding. Nucleic Acids Res. 1998, 26, 1107–1115. [Google Scholar] [CrossRef] [Green Version]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-Fold Faster RNA Homology Searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef] [Green Version]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic Gene Recognition and Translation Initiation Site Identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [Green Version]
- Chan, P.P.; Lowe, T.M. TRNAscan-SE: Searching for TRNA Genes in Genomic Sequences. In Gene Prediction: Methods and Protocols; Kollmar, M., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2019; pp. 1–14. ISBN 978-1-4939-9173-0. [Google Scholar]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in Homology Search: HMMER3 and Convergent Evolution of Coiled-Coil Regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef] [Green Version]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam Protein Families Database: Towards a More Sustainable Future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Kang, D.D.; Li, F.; Kirton, E.; Thomas, A.; Egan, R.; An, H.; Wang, Z. MetaBAT 2: An Adaptive Binning Algorithm for Robust and Efficient Genome Reconstruction from Metagenome Assemblies. PeerJ 2019, 7, e7359. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [Green Version]
- Chaumeil, P.-A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A Toolkit to Classify Genomes with the Genome Taxonomy Database. Bioinformatics 2020, 36, 1925–1927. [Google Scholar] [CrossRef]
- Käll, L.; Krogh, A.; Sonnhammer, E.L.L. Advantages of Combined Transmembrane Topology and Signal Peptide Prediction—The Phobius Web Server. Nucleic Acids Res. 2007, 35, W429–W432. [Google Scholar] [CrossRef] [Green Version]
- Steinegger, M.; Söding, J. Mmseqs2 Enables Sensitive Protein Sequence Searching for the Analysis of Massive Data Sets. Nat. Biotechnol. 2017, 35, 1026–1028. [Google Scholar] [CrossRef] [Green Version]
- Keeling, P.J.; Burki, F.; Wilcox, H.M.; Allam, B.; Allen, E.E.; Amaral-Zettler, L.A.; Armbrust, E.V.; Archibald, J.M.; Bharti, A.K.; Bell, C.J.; et al. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): Illuminating the Functional Diversity of Eukaryotic Life in the Oceans through Transcriptome Sequencing. PLoS Biol. 2014, 12, e1001889. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P. Vegan: Community Ecology Package. R Package Version 2.5–7 2020. 2022. Available online: https://cran.r-project.org/package=vegan (accessed on 9 December 2022).
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A Multiple Sequence Alignment Method with Reduced Time and Space Complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. TrimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Stephens, T.G.; González-Pech, R.A.; Cheng, Y.; Mohamed, A.R.; Burt, D.W.; Bhattacharya, D.; Ragan, M.A.; Chan, C.X. Genomes of the Dinoflagellate Polarella Glacialis Encode Tandemly Repeated Single-Exon Genes with Adaptive Functions. BMC Biol. 2020, 18, 56. [Google Scholar] [CrossRef]
- Grossart, H.-P.; Massana, R.; McMahon, K.D.; Walsh, D.A. Linking Metagenomics to Aquatic Microbial Ecology and Biogeochemical Cycles. Limnol. Oceanogr. 2020, 65, S2–S20. [Google Scholar] [CrossRef] [Green Version]
- Foster, T.J.; Geoghegan, J.A.; Ganesh, V.K.; Höök, M. Adhesion, Invasion and Evasion: The Many Functions of the Surface Proteins of Staphylococcus Aureus. Nat. Rev. Microbiol. 2014, 12, 49–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawashima, T.; Kawashima, S.; Tanaka, C.; Murai, M.; Yoneda, M.; Putnam, N.H.; Rokhsar, D.S.; Kanehisa, M.; Satoh, N.; Wada, H. Domain Shuffling and the Evolution of Vertebrates. Genome Res. 2009, 19, 1393–1403. [Google Scholar] [CrossRef] [Green Version]
- Loftus, J.C.; Smith, J.W.; Ginsberg, M.H. Integrin-Mediated Cell Adhesion: The Extracellular Face. J. Biol. Chem. 1994, 269, 25235–25238. [Google Scholar] [CrossRef]
- Lukomski, S.; Bachert, B.A.; Squeglia, F.; Berisio, R. Collagen-like Proteins of Pathogenic Streptococci. Mol. Microbiol. 2017, 103, 919–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Underwood, G.J.C.; Fietz, S.; Papadimitriou, S.; Thomas, D.N.; Dieckmann, G.S. Distribution and Composition of Dissolved Extracellular Polymeric Substances (EPS) in Antarctic Sea Ice. Mar. Ecol. Prog. Ser. 2010, 404, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Roukaerts, A.; Deman, F.; Van der Linden, F.; Carnat, G.; Bratkic, A.; Moreau, S.; Lannuzel, D.; Dehairs, F.; Delille, B.; Tison, J.-L.; et al. The Biogeochemical Role of a Microbial Biofilm in Sea Ice: Antarctic Landfast Sea Ice as a Case Study. Elem. Sci. Anthr. 2021, 9, 00134. [Google Scholar] [CrossRef]
- Kuchler, K.; Thorner, J. Membrane Translocation of Proteins without Hydrophobic Signal Peptides. Curr. Opin. Cell Biol. 1990, 2, 617–624. [Google Scholar] [CrossRef]
- Krembs, C.; Deming, J.W. The Role of Exopolymers in Microbial Adaptation to Sea Ice. In Psychrophiles: From Biodiversity to Biotechnology; Margesin, R., Schinner, F., Marx, J.-C., Gerday, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 247–264. ISBN 978-3-540-74335-4. [Google Scholar]
- Lamers, J.; van der Meer, T.; Testerink, C. How Plants Sense and Respond to Stressful Environments1 [OPEN]. Plant Physiol. 2020, 182, 1624–1635. [Google Scholar] [CrossRef] [Green Version]
- Weeks, W.F.; Gow, A.J. Preferred Crystal Orientations in the Fast Ice along the Margins of the Arctic Ocean. J. Geophys. Res. Oceans 1978, 83, 5105–5121. [Google Scholar] [CrossRef] [Green Version]
- Huynen, M.; Snel, B.; Lathe, W.; Bork, P. Predicting Protein Function by Genomic Context: Quantitative Evaluation and Qualitative Inferences. Genome Res. 2000, 10, 1204–1210. [Google Scholar] [CrossRef] [Green Version]
- Suyama, M.; Bork, P. Evolution of Prokaryotic Gene Order: Genome Rearrangements in Closely Related Species. Trends Genet. 2001, 17, 10–13. [Google Scholar] [CrossRef] [PubMed]
- Yelton, A.P.; Thomas, B.C.; Simmons, S.L.; Wilmes, P.; Zemla, A.; Thelen, M.P.; Justice, N.; Banfield, J.F. A Semi-Quantitative, Synteny-Based Method to Improve Functional Predictions for Hypothetical and Poorly Annotated Bacterial and Archaeal Genes. PLoS Comput. Biol. 2011, 7, e1002230. [Google Scholar] [CrossRef] [PubMed]
- Jung, W.; Campbell, R.L.; Gwak, Y.; Kim, J.I.; Davies, P.L.; Jin, E. New Cysteine-Rich Ice-Binding Protein Secreted from Antarctic Microalga, Chloromonas Sp. PLoS ONE 2016, 11, e0154056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, Y.; DeVries, A.L. Antifreeze Glycoprotein Levels in Antarctic Notothenioid Fishes Inhabiting Different Thermal Environments and the Effect of Warm Acclimation. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2006, 144, 290–300. [Google Scholar] [CrossRef] [PubMed]
- Scott, G.K.; Hayes, P.H.; Fletcher, G.L.; Davies, P.L. Wolffish Antifreeze Protein Genes Are Primarily Organized as Tandem Repeats That Each Contain Two Genes in Inverted Orientation. Mol. Cell. Biol. 1988, 8, 3670–3675. [Google Scholar] [CrossRef]
- Mahatabuddin, S.; Hanada, Y.; Nishimiya, Y.; Miura, A.; Kondo, H.; Davies, P.L.; Tsuda, S. Concentration-Dependent Oligomerization of an Alpha-Helical Antifreeze Polypeptide Makes It Hyperactive. Sci. Rep. 2017, 7, 42501. [Google Scholar] [CrossRef] [Green Version]
- Sun, T.; Lin, F.-H.; Campbell, R.L.; Allingham, J.S.; Davies, P.L. An Antifreeze Protein Folds with an Interior Network of More Than 400 Semi-Clathrate Waters. Science 2014, 343, 795–798. [Google Scholar] [CrossRef] [PubMed]
- Sorhannus, U. Evolution of Antifreeze Protein Genes in the Diatom Genus Fragilariopsis: Evidence for Horizontal Gene Transfer, Gene Duplication and Episodic Diversifying Selection. Evol. Bioinform. 2011, 7, EBO.S8321. [Google Scholar] [CrossRef]
- Haft, D.H.; Paulsen, I.T.; Ward, N.; Selengut, J.D. Exopolysaccharide-Associated Protein Sorting in Environmental Organisms: The PEP-CTERM/EpsH System. Application of a Novel Phylogenetic Profiling Heuristic. BMC Biol. 2006, 4, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, N.; Xia, M.; Dai, J.; Yu, D.; An, W.; Li, S.; Liu, S.; He, P.; Zhang, L.; Wu, Z.; et al. Both Widespread PEP-CTERM Proteins and Exopolysaccharides Are Required for Floc Formation of Zoogloea Resiniphila and Other Activated Sludge Bacteria. Environ. Microbiol. 2018, 20, 1677–1692. [Google Scholar] [CrossRef] [PubMed]

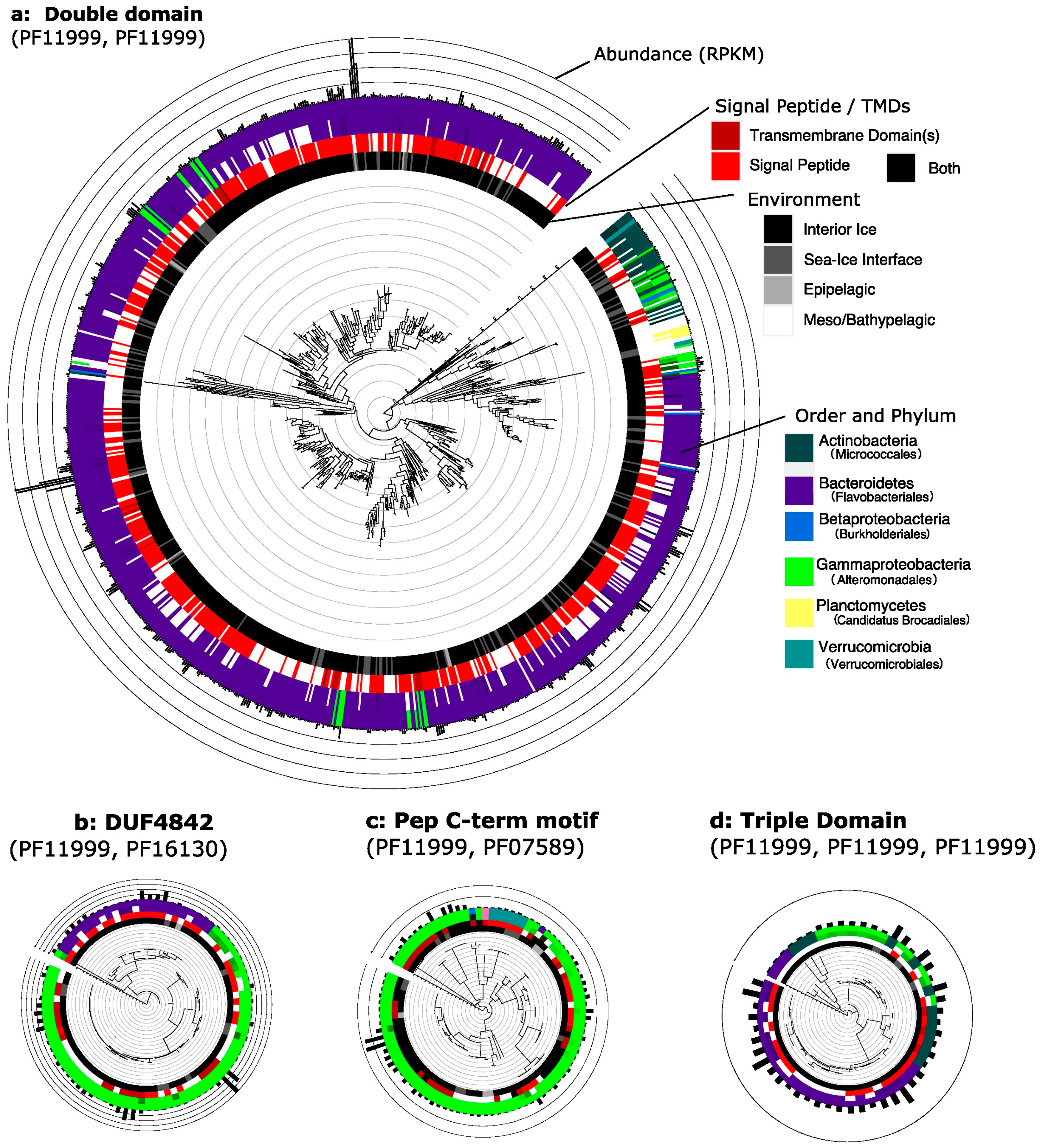

| Domain Architecture | Protein Family (Pfam) | Broader Function | Abundance (RPKM) | Abundance (%) |
|---|---|---|---|---|
| pfam11999 | DUF3494 | IBP | 4983.46 | 61.54 |
| pfam11999_pfam11999 | DUF3494 | IBP | 1660.68 | 20.51 |
| pfam11999_pfam16130 | DUF4842 * | β-barrel Ig fold | 413.49 | 5.11 |
| pfam11999_pfam07589 | PEP C-term motif | Sorting/Exopolysaccharides | 209.68 | 2.59 |
| pfam11999_pfam11999_pfam11999 | DUF3494 | IBP | 93.11 | 1.15 |
| pfam11999_pfam11999_pfam01345 | DUF11 | Cell wall-related | 63.74 | 0.79 |
| pfam11999_pfam02010 | REJ domain | Membrane associated | 58.44 | 0.72 |
| pfam04519_pfam11999 | Polymer-forming cytoskeletal | Cytoskeleton | 47.68 | 0.59 |
| pfam11999_pfam11999_pfam13517_pfam13517_pfam07593 | FG-GAP-like repeat | Cell adhesion | 44.35 | 0.55 |
| ASPIC and UnbV | Cell adhesion | |||
| pfam11999_pfam11999_pfam13517_pfam13517_pfam13517_pfam07593 | FG-GAP-like repeat | Cell adhesion | 25.93 | 0.32 |
| ASPIC and UnbV | Cell adhesion | 0.31 | ||
| pfam11999_pfam03797 | Autotransporter β-domain | Secretion | 24.83 | 0.30 |
| pfam13205_pfam13205_pfam11999 | BIg-like domain * | Tethering | 24.45 | 0.26 |
| pfam11999_pfam11999_pfam02412_pfam02412_pfam02412_pfam02412_pfam02412 | Thrombospondin type 3 repeat | Cell adhesion | 21.36 | 0.25 |
| pfam11999_pfam01391 | Collagen triple helix repeat | Cell adhesion | 20.53 | 0.24 |
| pfam11999_pfam07603 | DUF1566 | Unknown | 19.42 | 0.23 |
| pfam11999_pfam02494_pfam02494_pfam02494 | HYR domain | Cell adhesion | 18.99 | 0.23 |
| pfam14341_pfam11999 | PilX N-terminal | Cell adhesion | 18.87 | 0.21 |
| pfam04862_pfam11999 | DUF642 | Unknown (thought to be exclusive to plants) | 16.73 | 0.20 |
| pfam11999_pfam04862 | DUF642 | Unknown (thought to be exclusive to plants) | 13.87 | 0.17 |
| pfam11999_pfam01345 | DUF11 | Cell wall-related | 13.17 | 0.16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Winder, J.C.; Boulton, W.; Salamov, A.; Eggers, S.L.; Metfies, K.; Moulton, V.; Mock, T. Genetic and Structural Diversity of Prokaryotic Ice-Binding Proteins from the Central Arctic Ocean. Genes 2023, 14, 363. https://doi.org/10.3390/genes14020363
Winder JC, Boulton W, Salamov A, Eggers SL, Metfies K, Moulton V, Mock T. Genetic and Structural Diversity of Prokaryotic Ice-Binding Proteins from the Central Arctic Ocean. Genes. 2023; 14(2):363. https://doi.org/10.3390/genes14020363
Chicago/Turabian StyleWinder, Johanna C., William Boulton, Asaf Salamov, Sarah Lena Eggers, Katja Metfies, Vincent Moulton, and Thomas Mock. 2023. "Genetic and Structural Diversity of Prokaryotic Ice-Binding Proteins from the Central Arctic Ocean" Genes 14, no. 2: 363. https://doi.org/10.3390/genes14020363
APA StyleWinder, J. C., Boulton, W., Salamov, A., Eggers, S. L., Metfies, K., Moulton, V., & Mock, T. (2023). Genetic and Structural Diversity of Prokaryotic Ice-Binding Proteins from the Central Arctic Ocean. Genes, 14(2), 363. https://doi.org/10.3390/genes14020363






