HSP70 Expression Signature in Renal Cell Carcinoma: A Clinical and Bioinformatic Analysis Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
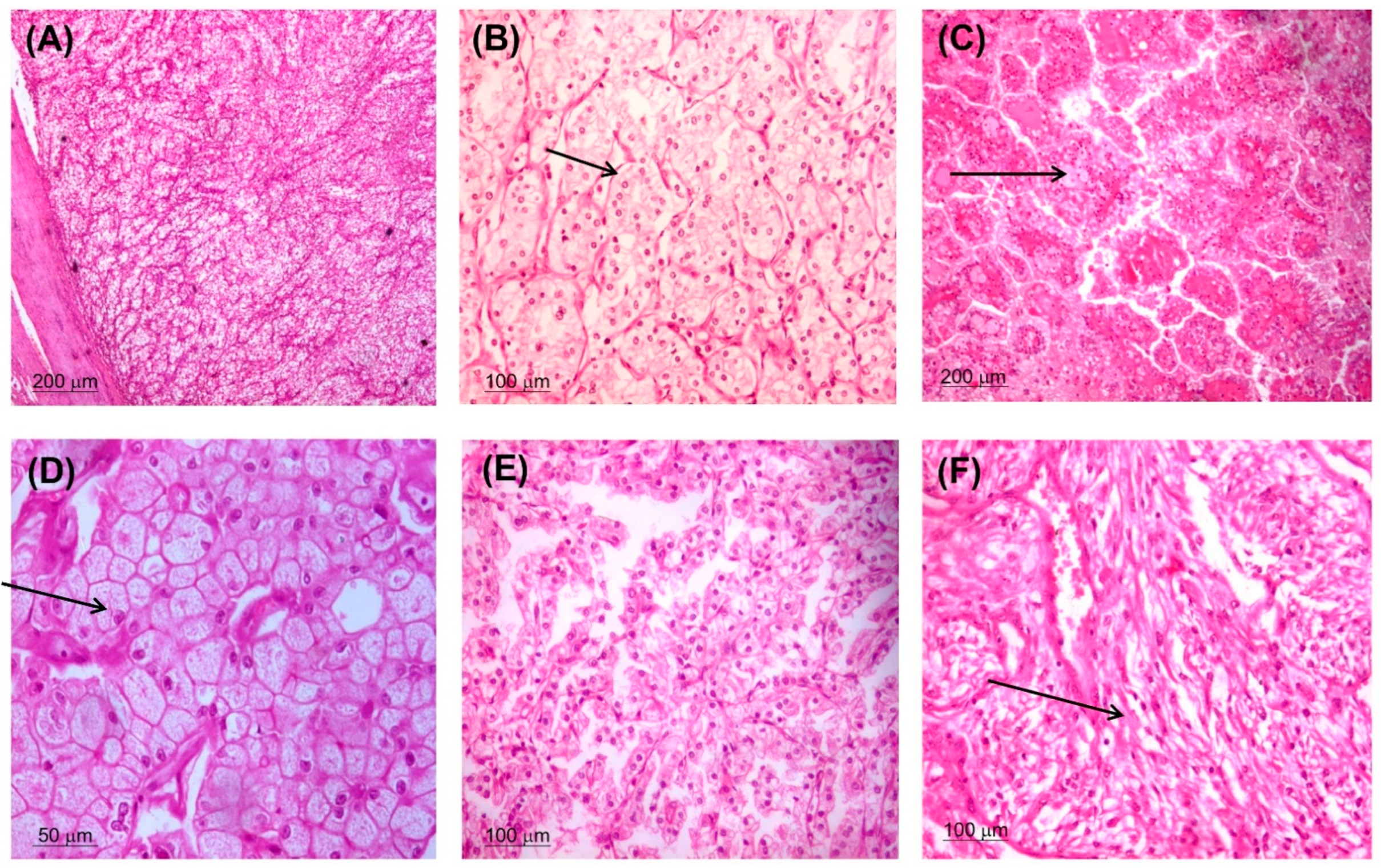
2.2. Histopathology Assessment
2.3. Gene Expression Analysis
2.4. Gene Expression Data Analysis
2.5. Bioinformatic Analysis
2.5.1. HSP70 Structural and Functional Analysis
2.5.2. In Silico Analysis Using Genotype-Tissue Expression (GTEx) and the Cancer Genome Atlas (TCGA) Tissues for Assessing the Expression of HSP70 in RCC
Gene Expression Profiling Interactive Analysis (GEPIA) Expression and Correlation Analysis
MEXPRESS Gene and Clinical Data Analysis
cBioPortal Database Analysis
2.6. Statistical Analysis
3. Results
3.1. Baseline Characteristics of the Study Group
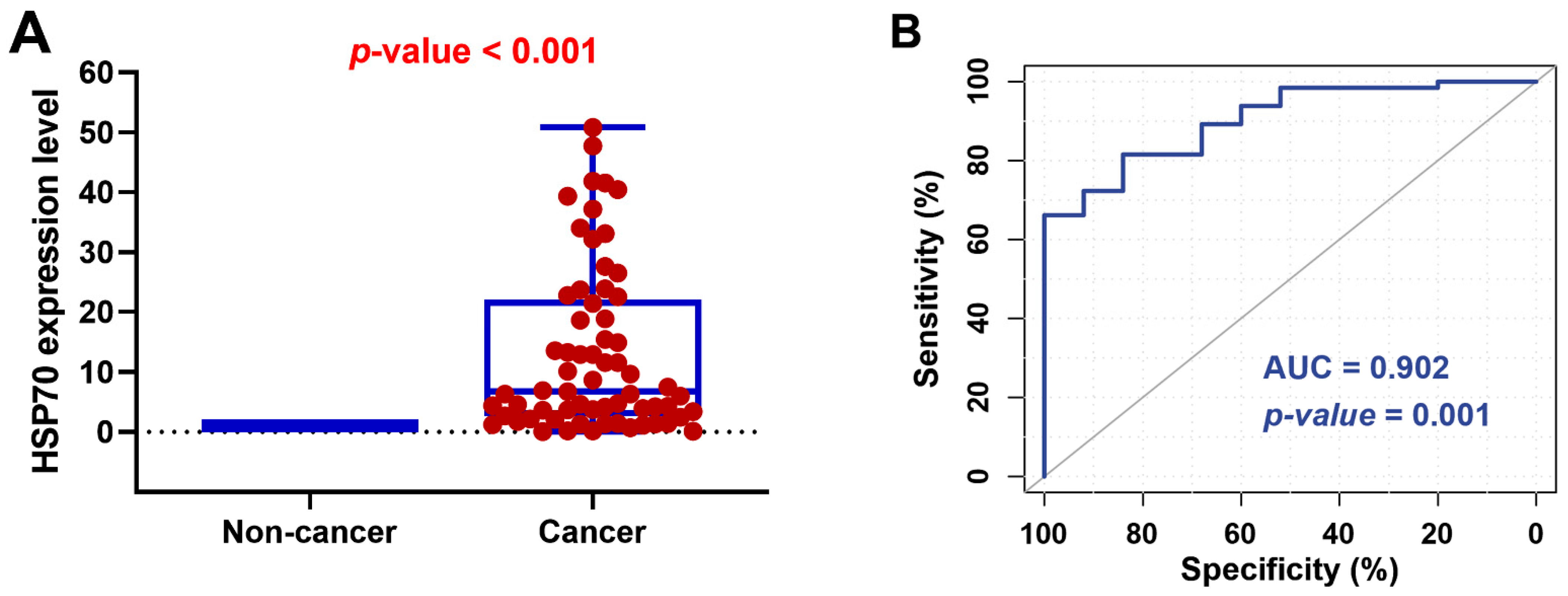
3.2. HSP70 Gene Expression Analysis in Studied Patients with RCC
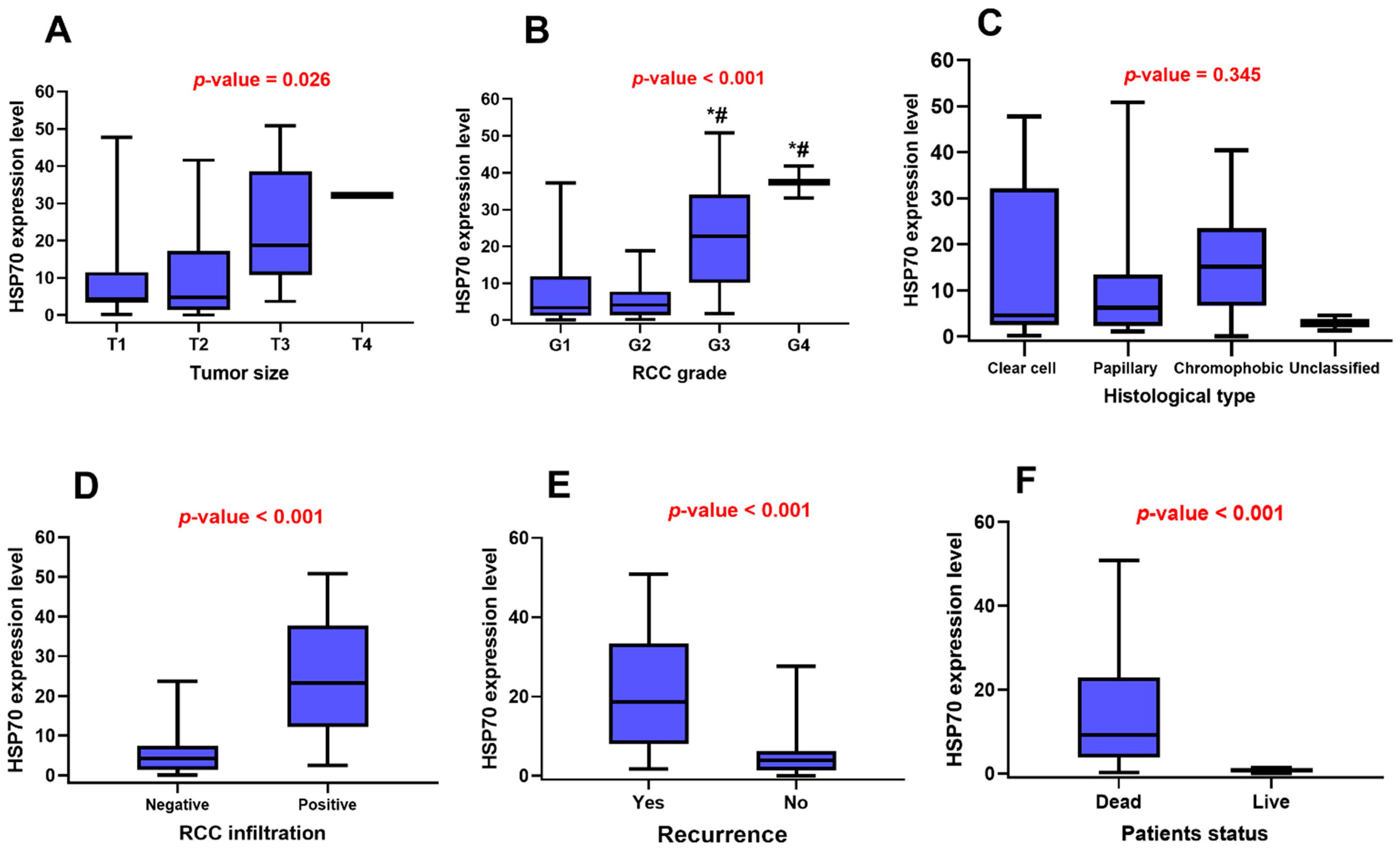
3.3. HSP70 Gene Expression Signature and the Clinicopathological Features of RCC
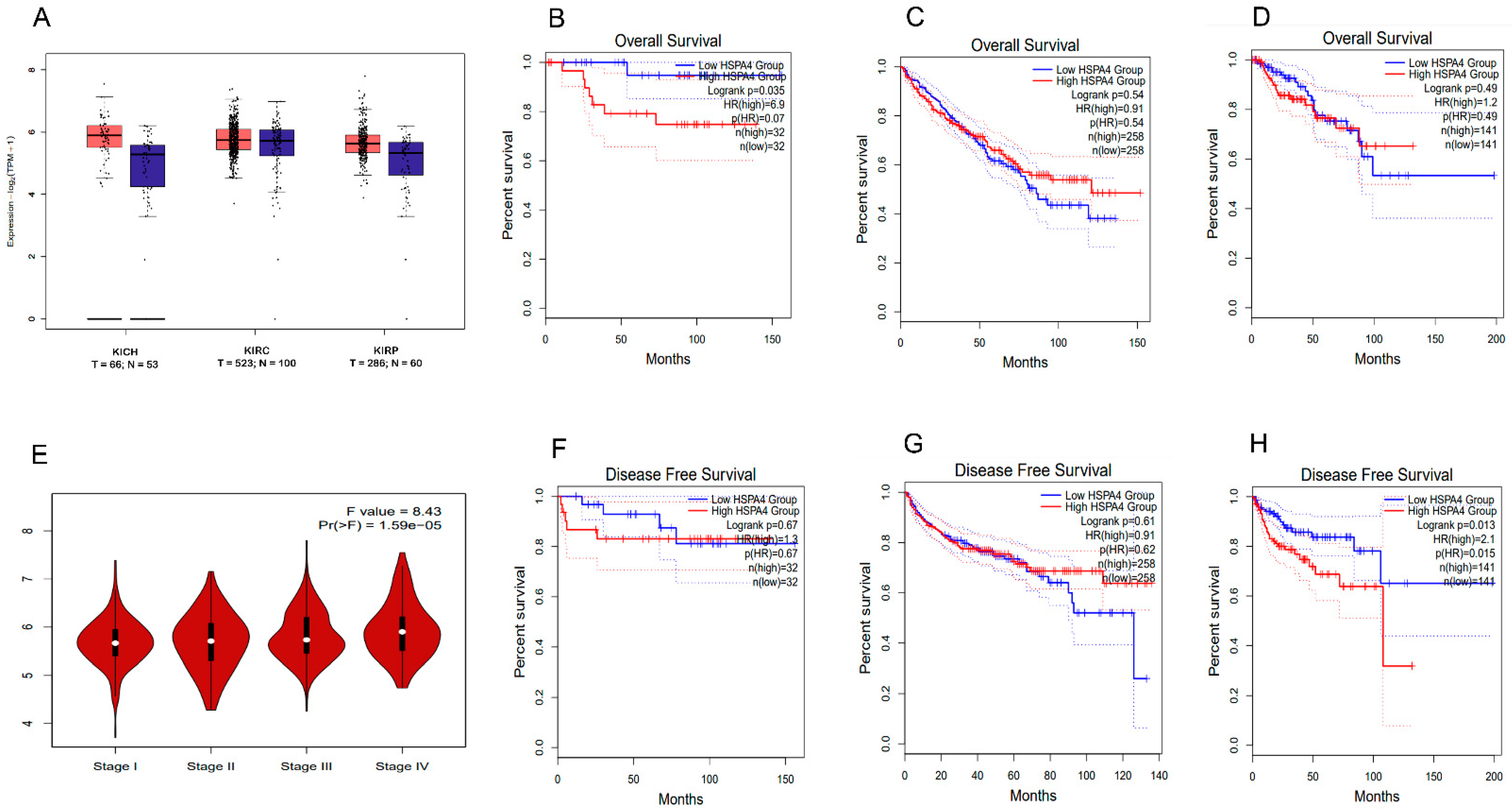
3.4. Survival Analysis
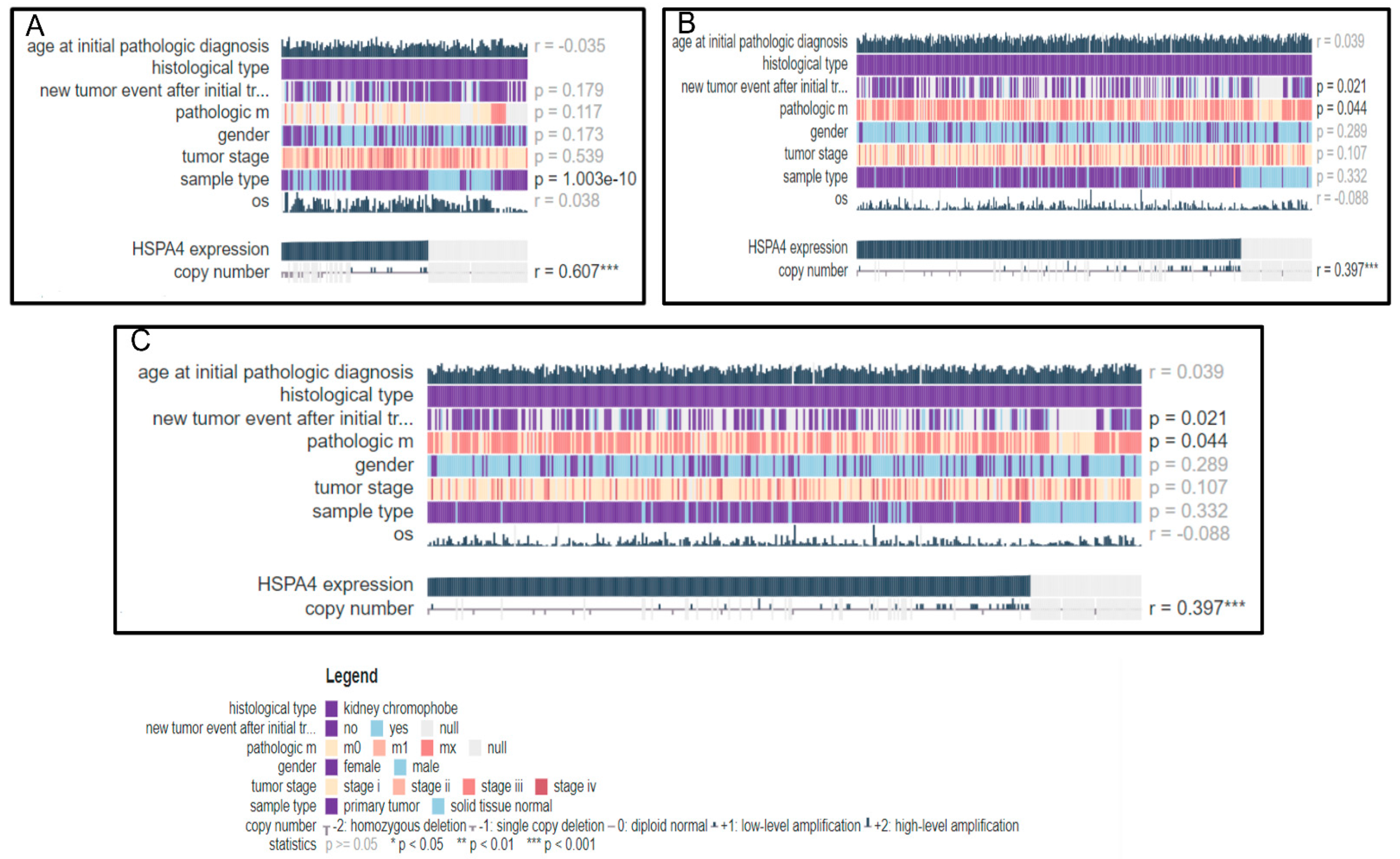
3.5. In-Silico Analysis
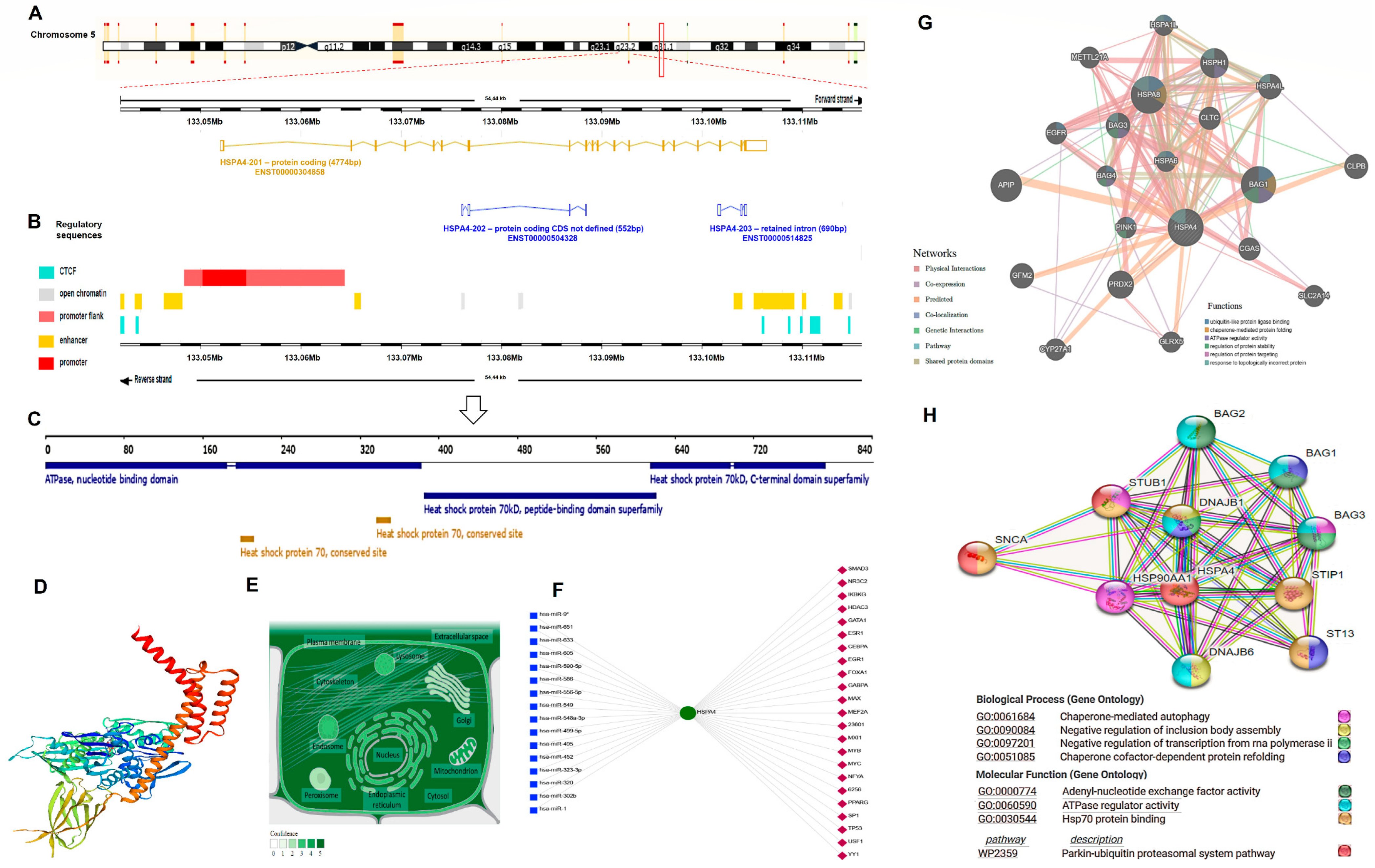
3.5.1. HSP70 Structural and Functional Analysis
3.5.2. Analysis of the GTEx Project and TCGA Tissues for the HSP70 Expression in RCC Subtypes
3.5.3. HSP70 as a Potential Prospective DNA Variation Biomarker in RCC
3.5.4. cBioPortal Database Genomic and Target Pathway Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Capitanio, U.; Bensalah, K.; Bex, A.; Boorjian, S.A.; Bray, F.; Coleman, J.; Gore, J.L.; Sun, M.; Wood, C.; Russo, P. Epidemiology of Renal Cell Carcinoma. Eur. Urol. 2019, 75, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Specht, H.M.; Ahrens, N.; Blankenstein, C.; Duell, T.; Fietkau, R.; Gaipl, U.S.; Günther, C.; Gunther, S.; Habl, G.; Hautmann, H.; et al. Heat Shock Protein 70 (Hsp70) Peptide Activated Natural Killer (NK) Cells for the Treatment of Patients with Non-Small Cell Lung Cancer (NSCLC) after Radiochemotherapy (RCTx)—From Preclinical Studies to a Clinical Phase II Trial. Front. Immunol. 2015, 6, 162. [Google Scholar] [CrossRef] [PubMed]
- Padala, S.A.; Barsouk, A.; Thandra, K.C.; Saginala, K.; Mohammed, A.; Vakiti, A.; Rawla, P.; Barsouk, A. Epidemiology of Renal Cell Carcinoma. World J. Oncol. 2020, 11, 79–87. [Google Scholar] [CrossRef]
- Yang, D.C.; Chen, C.-H. Potential New Therapeutic Approaches for Renal Cell Carcinoma. Semin. Nephrol. 2020, 40, 86–97. [Google Scholar] [CrossRef]
- Hsieh, J.J.; Purdue, M.P.; Signoretti, S.; Swanton, C.; Albiges, L.; Schmidinger, M.; Heng, D.Y.; Larkin, J.; Ficarra, V. Renal cell carcinoma. Nat. Rev. Dis. Prim. 2017, 3, 17009. [Google Scholar] [CrossRef]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef]
- Amato, R.J. Heat-shock protein–peptide complex-96 for the treatment of cancer. Expert Opin. Biol. Ther. 2007, 7, 1267–1273. [Google Scholar] [CrossRef]
- Koller, M.F.; Mohajeri, M.H.; Huber, M.; Wollmer, M.A.; Z’Graggen, B.V.R.; Sandmeier, E.; Moritz, E.; Tracy, J.; Nitsch, R.M.; Christen, P. Active Immunization of Mice with an Aβ-Hsp70 Vaccine. Neurodegener. Dis. 2004, 1, 20–28. [Google Scholar] [CrossRef]
- Wells, A.D.; Malkovsky, M. Heat shock proteins, tumor immunogenicity and antigen presentation: An integrated view. Immunol. Today 2000, 21, 129–132. [Google Scholar] [CrossRef]
- Mosser, D.D.; Morimoto, R.I. Molecular chaperones and the stress of oncogenesis. Oncogene 2004, 23, 2907–2918. [Google Scholar] [CrossRef] [PubMed]
- Multhoff, G.; Botzler, C.; Wiesnet, M.; Müller, E.; Meier, T.; Wilmanns, W.; Issels, R.D. A stress-inducible 72-kDa heat-shock protein (HSP72) is expressed on the surface of human tumor cells, but not on normal cells. Int. J. Cancer 1995, 61, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Gastpar, R.; Gehrmann, M.; Bausero, M.A.; Asea, A.; Gross, C.; Schroeder, J.A.; Multhoff, G. Heat Shock Protein 70 Surface-Positive Tumor Exosomes Stimulate Migratory and Cytolytic Activity of Natural Killer Cells. Cancer Res 2005, 65, 5238–5247. [Google Scholar] [CrossRef] [PubMed]
- Rückert, M.; Deloch, L.; Fietkau, R.; Frey, B.; Hecht, M.; Gaipl, U.S. Immune modulatory effects of radiotherapy as basis for well-reasoned radioimmunotherapies. Strahlenther. Onkol. 2018, 194, 509–519. [Google Scholar] [CrossRef]
- Ostheimer, C.; Gunther, S.; Bache, M.; Vordermark, D.; Multhoff, G. Dynamics of Heat Shock Protein 70 Serum Levels As a Predictor of Clinical Response in Non-Small-Cell Lung Cancer and Correlation with the Hypoxia-Related Marker Osteopontin. Front. Immunol. 2017, 8, 1305. [Google Scholar] [CrossRef]
- Breloer, M.; Fleischer, B.; Von Bonin, A. In vivo and in vitro activation of T cells after administration of Ag-negative heat shock proteins. J. Immunol. 1999, 162, 3141–3147. [Google Scholar] [CrossRef]
- Daniels, G.A.; Sanchez-Perez, L.; Diaz, R.M.; Kottke, T.; Thompson, J.; Lai, M.; Gough, M.; Karim, M.; Bushell, A.; Chong, H.; et al. A simple method to cure established tumors by inflammatory killing of normal cells. Nat. Biotechnol. 2004, 22, 1125–1132. [Google Scholar] [CrossRef]
- Rothammer, A.; Sage, E.K.; Werner, C.; Combs, S.E.; Multhoff, G. Increased heat shock protein 70 (Hsp70) serum levels and low NK cell counts after radiotherapy—Potential markers for predicting breast cancer recurrence? Radiat. Oncol. 2019, 14, 78. [Google Scholar] [CrossRef]
- Blachere, N.E.; Li, Z.; Chandawarkar, R.Y.; Suto, R.; Jaikaria, N.S.; Basu, S.; Udono, H.; Srivastava, P.K. Heat Shock Protein–Peptide Complexes, Reconstituted In Vitro, Elicit Peptide-specific Cytotoxic T Lymphocyte Response and Tumor Immunity. J. Exp. Med. 1997, 186, 1315–1322. [Google Scholar] [CrossRef]
- Albakova, Z.; Armeev, G.A.; Kanevskiy, L.M.; Kovalenko, E.I.; Sapozhnikov, A.M. HSP70 Multi-Functionality in Cancer. Cells 2020, 9, 587. [Google Scholar] [CrossRef]
- Santarosa, M.; Favaro, D.; Quaia, M.; Galligioni, E. Expression of heat shock protein 72 in renal cell carcinoma: Possible role and prognostic implications in cancer patients. Eur. J. Cancer 1997, 33, 873–877. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Suri, A. Targeting the testis-specific heat-shock protein 70-2 (HSP70-2) reduces cellular growth, migration, and invasion in renal cell carcinoma cells. Tumor Biol. 2014, 35, 12695–12706. [Google Scholar] [CrossRef] [PubMed]
- Pfister, K.; Radons, J.; Busch, R.; Tidball, J.G.; Pfeifer, M.; Freitag, L.; Feldmann, H.-J.; Milani, V.; Issels, R.; Multhoff, G. Patient survival by Hsp70 membrane phenotype: Association with different routes of metastasis. Cancer 2007, 110, 926–935. [Google Scholar] [CrossRef] [PubMed]
- Ramp, U.; Mahotka, C.; Heikaus, S.; Shibata, T.; Grimm, M.O.; Willers, R.; Gabbert, H.E. Expression of heat shock protein 70 in renal cell carcinoma and its relation to tumor progression and prognosis. Histol. Histopathol. 2007, 22, 1099–1107. [Google Scholar] [CrossRef]
- Toraih, E.A.; Ibrahiem, A.T.; Fawzy, M.S.; Hussein, M.H.; Al-Qahtani, S.A.M.; Shaalan, A.A.M. MicroRNA-34a: A Key Regulator in the Hallmarks of Renal Cell Carcinoma. Oxid. Med. Cell. Longev. 2017, 2017, 3269379. [Google Scholar] [CrossRef]
- Fawzy, M.S.; Toraih, E.A.; El-Wazir, A.; Hosny, M.M.; Badran, D.I.; El Kelish, A. Long intergenic non-coding RNA, regulator of reprogramming (LINC-ROR) over-expression predicts poor prognosis in renal cell carcinoma. Arch. Med. Sci. 2021, 17, 1016–1027. [Google Scholar] [CrossRef]
- Srigley, J.R.; Delahunt, B.; Eble, J.N.; Egevad, L.; Epstein, J.I.; Grignon, D.; Hes, O.; Moch, H.; Montironi, R.; Tickoo, S.K.; et al. The International Society of Urological Pathology (ISUP) Vancouver Classification of Renal Neoplasia. Am. J. Surg. Pathol. 2013, 37, 1469–1489. [Google Scholar] [CrossRef] [PubMed]
- Fuhrman, S.A.; Lasky, L.C.; Limas, C. Prognostic significance of morphologic parameters in renal cell carcinoma. Am. J. Surg. Pathol. 1982, 6, 655–664. [Google Scholar] [CrossRef]
- Sobin, L.H.; Gospodarowicz, M.K.; Wittekind, C. TNM Classification of Malignant Tumours, 7th 2009 ed.; Wiley-Blackwell: Chichester, UK, 2010. [Google Scholar]
- Toraih, E.A.; Fawzy, M.S.; El-Falouji, A.I.; Hamed, E.O.; Nemr, N.A.; Hussein, M.H.; Fadeal, N.M.A.E. Stemness-Related Transcriptional Factors and Homing Gene Expression Profiles in Hepatic Differentiation and Cancer. Mol. Med. 2016, 22, 653–663. [Google Scholar] [CrossRef]
- Toraih, E.; Alrefai, H.G.; Hussein, M.H.; Helal, G.M.; Khashana, M.; Fawzy, M.S. Overexpression of heat shock protein HSP90AA1 and translocase of the outer mitochondrial membrane TOM34 in HCV-induced hepatocellular carcinoma: A pilot study. Clin. Biochem. 2018, 63, 10–17. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Koch, A.; Jeschke, J.; Van Criekinge, W.; Van Engeland, M.; De Meyer, T. MEXPRESS update 2019. Nucleic Acids Res. 2019, 47, W561–W565. [Google Scholar] [CrossRef] [PubMed]
- Kang, H. Sample size determination and power analysis using the G*Power software. J. Educ. Eval. Health Prof. 2021, 18, 17. [Google Scholar] [CrossRef] [PubMed]
- Vostakolaei, M.A.; Hatami-Baroogh, L.; Babaei, G.; Molavi, O.; Kordi, S.; Abdolalizadeh, J. Hsp70 in cancer: A double agent in the battle between survival and death. J. Cell. Physiol. 2020, 236, 3420–3444. [Google Scholar] [CrossRef]
- Tsai, C.-H.; Weng, J.-R.; Lin, H.-W.; Lu, M.-T.; Liu, Y.-C.; Chu, P.-C. Targeting Triple Negative Breast Cancer Stem Cells by Heat Shock Protein 70 Inhibitors. Cancers 2022, 14, 4898. [Google Scholar] [CrossRef]
- Kabakov, A.E.; Gabai, V.L. HSP70s in Breast Cancer: Promoters of Tumorigenesis and Potential Targets/Tools for Therapy. Cells 2021, 10, 3446. [Google Scholar] [CrossRef]
- Tustumi, F.; Agareno, G.A.; Galletti, R.P.; da Silva, R.B.R.; Quintas, J.G.; Sesconetto, L.D.A.; Szor, D.J.; Wolosker, N. The Role of the Heat-Shock Proteins in Esophagogastric Cancer. Cells 2022, 11, 2664. [Google Scholar] [CrossRef]
- Liu, J.; Liu, J.; Guo, S.-Y.; Liu, H.-L.; Li, S.-Z. HSP70 inhibitor combined with cisplatin suppresses the cervical cancer proliferation in vitro and transplanted tumor growth: An experimental study. Asian Pac. J. Trop. Med. 2017, 10, 184–188. [Google Scholar] [CrossRef]
- Garg, M.; Kanojia, D.; Seth, A.; Kumar, R.; Gupta, A.; Surolia, A.; Suri, A. Heat-shock protein 70-2 (HSP70-2) expression in bladder urothelial carcinoma is associated with tumour progression and promotes migration and invasion. Eur. J. Cancer 2010, 46, 207–215. [Google Scholar] [CrossRef]
- Dall’Oglio, M.F.; Coelho, R.F.; Leite, K.R.M.; Sousa-Canavez, J.M.; Oliveira, P.S.L.; Srougi, M. Gene expression profile of renal cell carcinoma clear cell type. Int. Braz J. Urol. 2010, 36, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Ghosn, M.; Jaloudi, M.; Larbaoui, B.; Salim, A.; Dreosti, L. Insights into the epidemiology of renal cell carcinoma in North Africa and the Middle East. J. Fac. Med. Or. 2017, 1, 65. Available online: https://confajol3.ajol.info/index.php/jmfo/article/view/485 (accessed on 2 December 2022).
- Garg, M.; Kanojia, D.; Saini, S.; Suri, S.; Gupta, A.; Surolia, A.; Suri, A. Germ cell-specific heat shock protein 70-2 is expressed in cervical carcinoma and is involved in the growth, migration, and invasion of cervical cells. Cancer 2010, 116, 3785–3796. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chen, W.; Duan, C.-J.; Zhang, C.-F. Overexpression of HSPA2 is correlated with poor prognosis in esophageal squamous cell carcinoma. World J. Surg. Oncol. 2013, 11, 141–148. [Google Scholar] [CrossRef]
- Yeh, C.-H.; Tseng, R.; Hannah, A.; Estrov, Z.; Estey, E.; Kantarjian, H.; Albitar, M. Clinical correlation of circulating heat shock protein 70 in acute leukemia. Leuk. Res. 2010, 34, 605–609. [Google Scholar] [CrossRef] [PubMed]
- Yun, C.W.; Kim, H.J.; Lim, J.H.; Lee, S.H. Heat Shock Proteins: Agents of Cancer Development and Therapeutic Targets in Anti-Cancer Therapy. Cells 2019, 9, 60. [Google Scholar] [CrossRef]



| Characteristics | Levels | Number | Percentage |
|---|---|---|---|
| Age, years | ≤20 | 7 | 10.8 |
| 21–40 | 38 | 58.5 | |
| 41–60 | 20 | 30.7 | |
| Sex | Male | 22 | 33.8 |
| Female | 43 | 66.2 | |
| Location | Right side | 44 | 67.7 |
| Left side | 21 | 32.3 | |
| Tumor size | T1 | 19 | 29.3 |
| T2 | 33 | 50.7 | |
| T3 | 12 | 18.5 | |
| T4 | 1 | 1.5 | |
| Pathological grade | G1 | 10 | 15.4 |
| G2 | 30 | 46.1 | |
| G3 | 23 | 35.4 | |
| G4 | 2 | 3.1 | |
| Histological type | KIRC | 32 | 49.2 |
| KIRP | 19 | 29.2 | |
| KICH | 12 | 18.5 | |
| Unclassified | 2 | 3.1 | |
| Capsular infiltration | Absent | 39 | 60 |
| Present | 26 | 40 | |
| Recurrence | Negative | 34 | 52.3 |
| Positive | 31 | 47.7 | |
| Mortality | Alive | 7 | 10.8 |
| Died | 58 | 89.2 | |
| Overall survival, months | ≤20 | 35 | 53.8 |
| >20 | 30 | 46.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abd El-Fadeal, N.M.; Ellawindy, A.; Jeraiby, M.A.; Qusti, S.Y.; Alshammari, E.M.; Alzahrani, A.K.; Ismail, E.A.; Ehab, Z.; Toraih, E.A.; Fawzy, M.S.; et al. HSP70 Expression Signature in Renal Cell Carcinoma: A Clinical and Bioinformatic Analysis Approach. Genes 2023, 14, 355. https://doi.org/10.3390/genes14020355
Abd El-Fadeal NM, Ellawindy A, Jeraiby MA, Qusti SY, Alshammari EM, Alzahrani AK, Ismail EA, Ehab Z, Toraih EA, Fawzy MS, et al. HSP70 Expression Signature in Renal Cell Carcinoma: A Clinical and Bioinformatic Analysis Approach. Genes. 2023; 14(2):355. https://doi.org/10.3390/genes14020355
Chicago/Turabian StyleAbd El-Fadeal, Noha M., Alia Ellawindy, Mohammed A. Jeraiby, Safaa Y. Qusti, Eida M. Alshammari, Ahmad Khuzaim Alzahrani, Ezzat A. Ismail, Ziad Ehab, Eman A. Toraih, Manal S. Fawzy, and et al. 2023. "HSP70 Expression Signature in Renal Cell Carcinoma: A Clinical and Bioinformatic Analysis Approach" Genes 14, no. 2: 355. https://doi.org/10.3390/genes14020355
APA StyleAbd El-Fadeal, N. M., Ellawindy, A., Jeraiby, M. A., Qusti, S. Y., Alshammari, E. M., Alzahrani, A. K., Ismail, E. A., Ehab, Z., Toraih, E. A., Fawzy, M. S., & Mohamed, M. H. (2023). HSP70 Expression Signature in Renal Cell Carcinoma: A Clinical and Bioinformatic Analysis Approach. Genes, 14(2), 355. https://doi.org/10.3390/genes14020355










