Genome-Wide Identification, Evolutionary and Functional Analyses of WRKY Family Members in Ginkgo biloba
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. Identification of WRKY Family Members in G. biloba
2.3. Chromosome Localization of GbWRKYs
2.4. Prediction of Physicochemical Properties and Subcellular Localization
2.5. Gene Duplication
2.6. Nonsynonymous and Synonymous Substitution Rate Ratio (Ka/Ks)
2.7. Protein Conserved Domains and Motif Analysis
2.8. Closely Related Species Analysis
2.9. Prediction of Promoter Cis-Acting Elements
2.10. RNA Extraction and Quantitative RT-PCR Analysis
3. Results
3.1. Identification of GbWRKY Transcription Factor Members in G. biloba
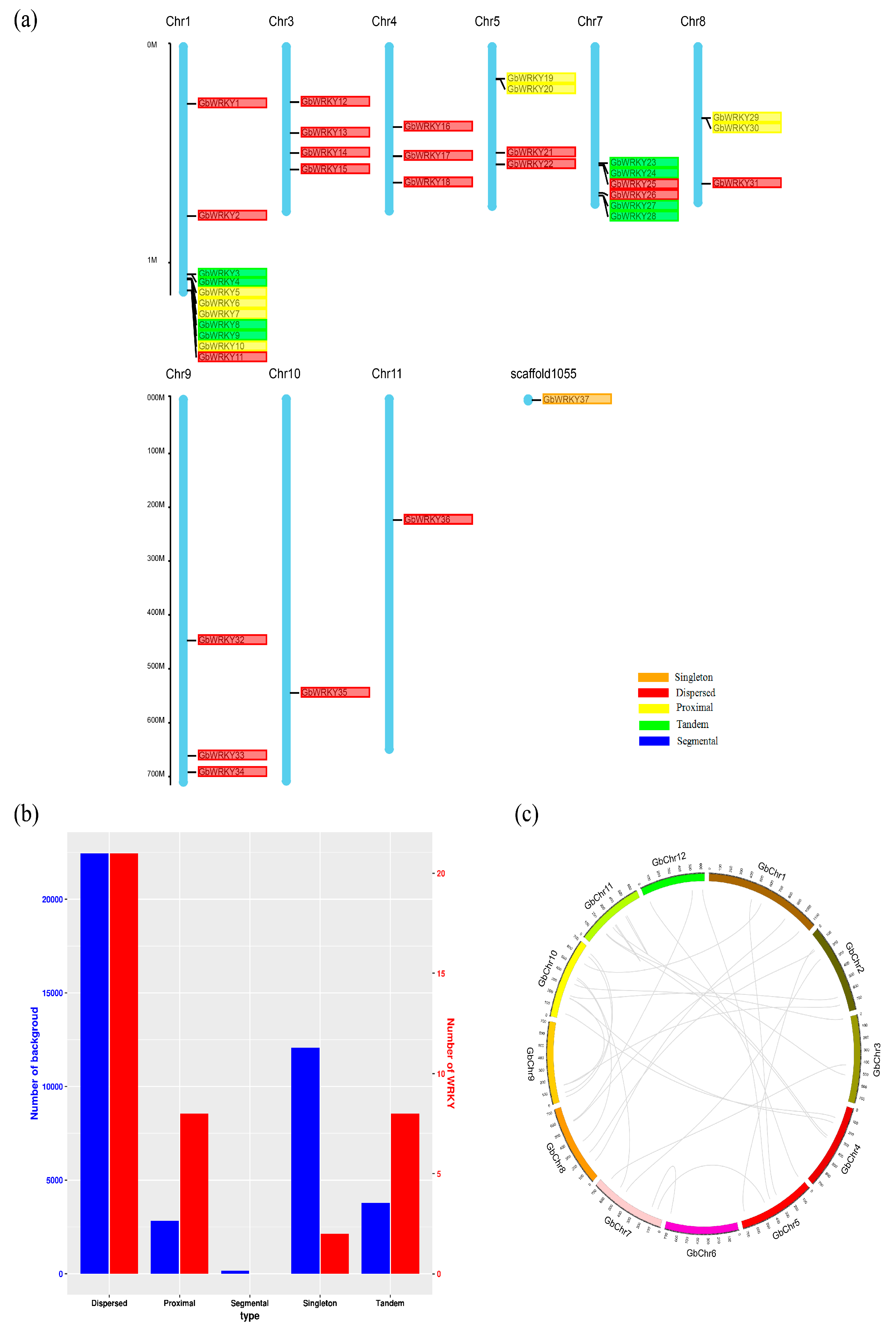
3.2. Chromosome Distribution and Gene Duplication Analysis of GbWRKY Genes
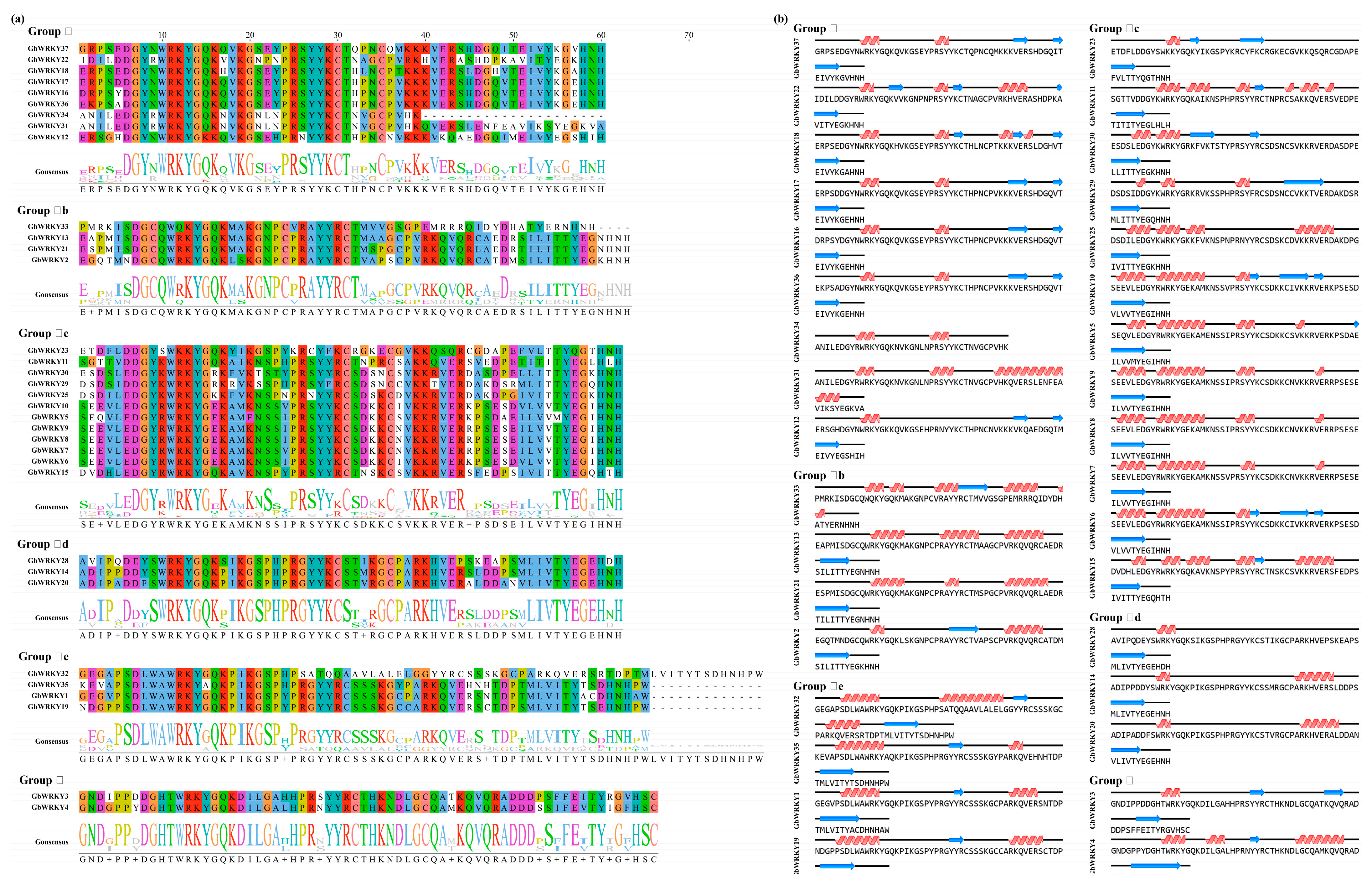
3.3. Structure and Sequence Alignment and Classification of GbWRKY Protein
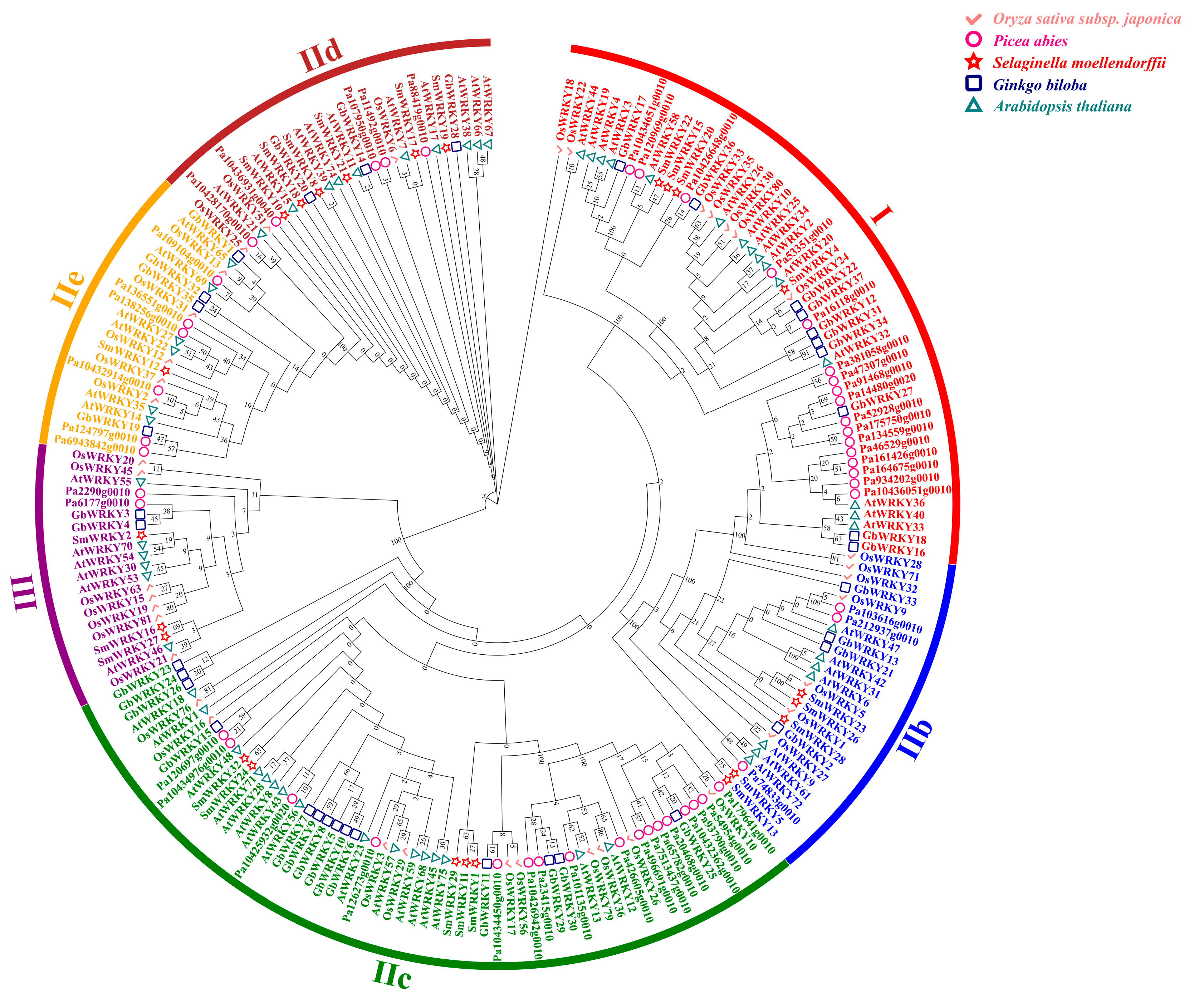
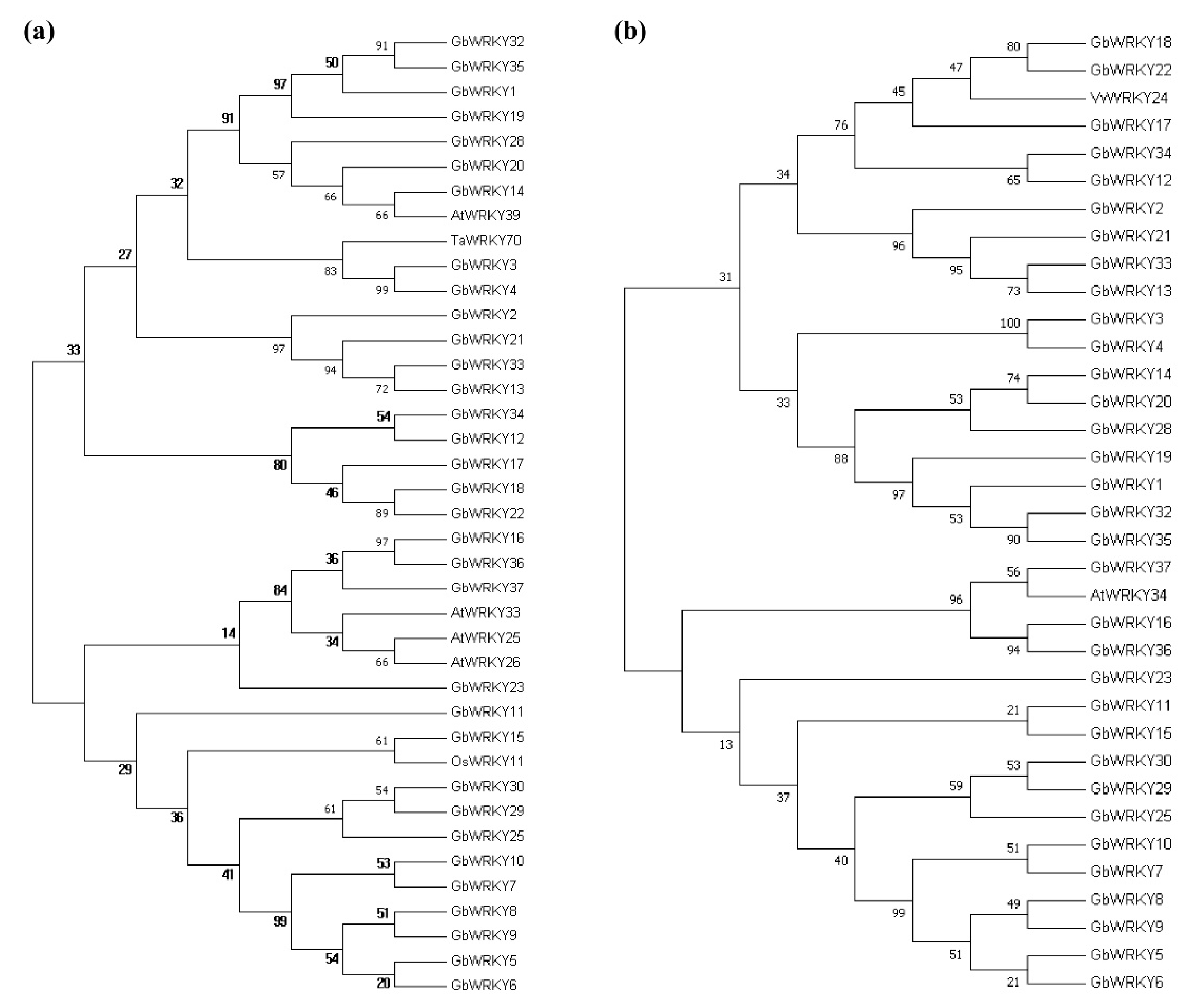
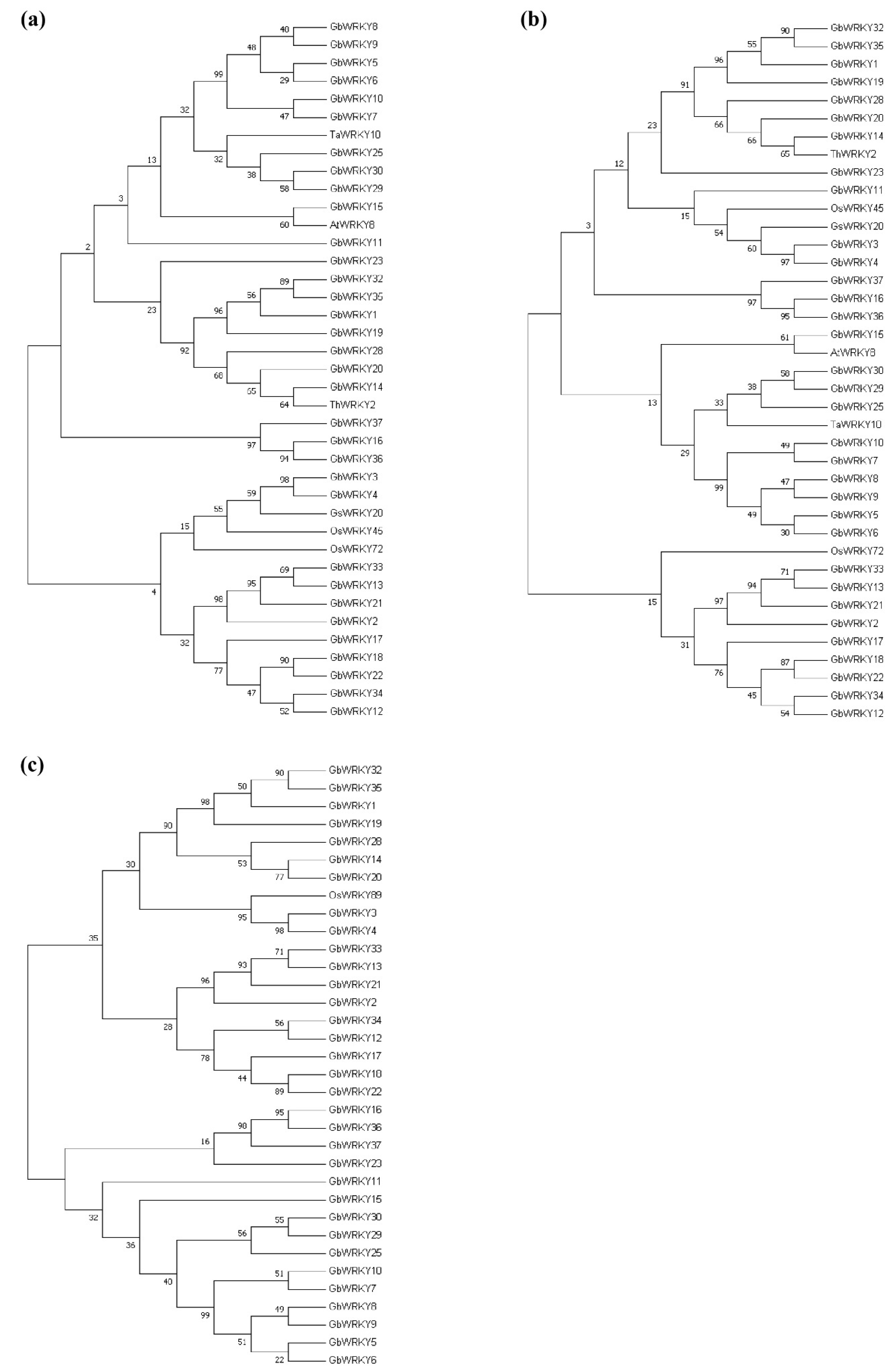
3.4. Phylogenetic Analysis, Conserved Motifs and Exon−Intron Organization of GbWRKY Family Members
3.5. Conserved Domain Evolution of G. biloba and Multispecies WRKY Proteins
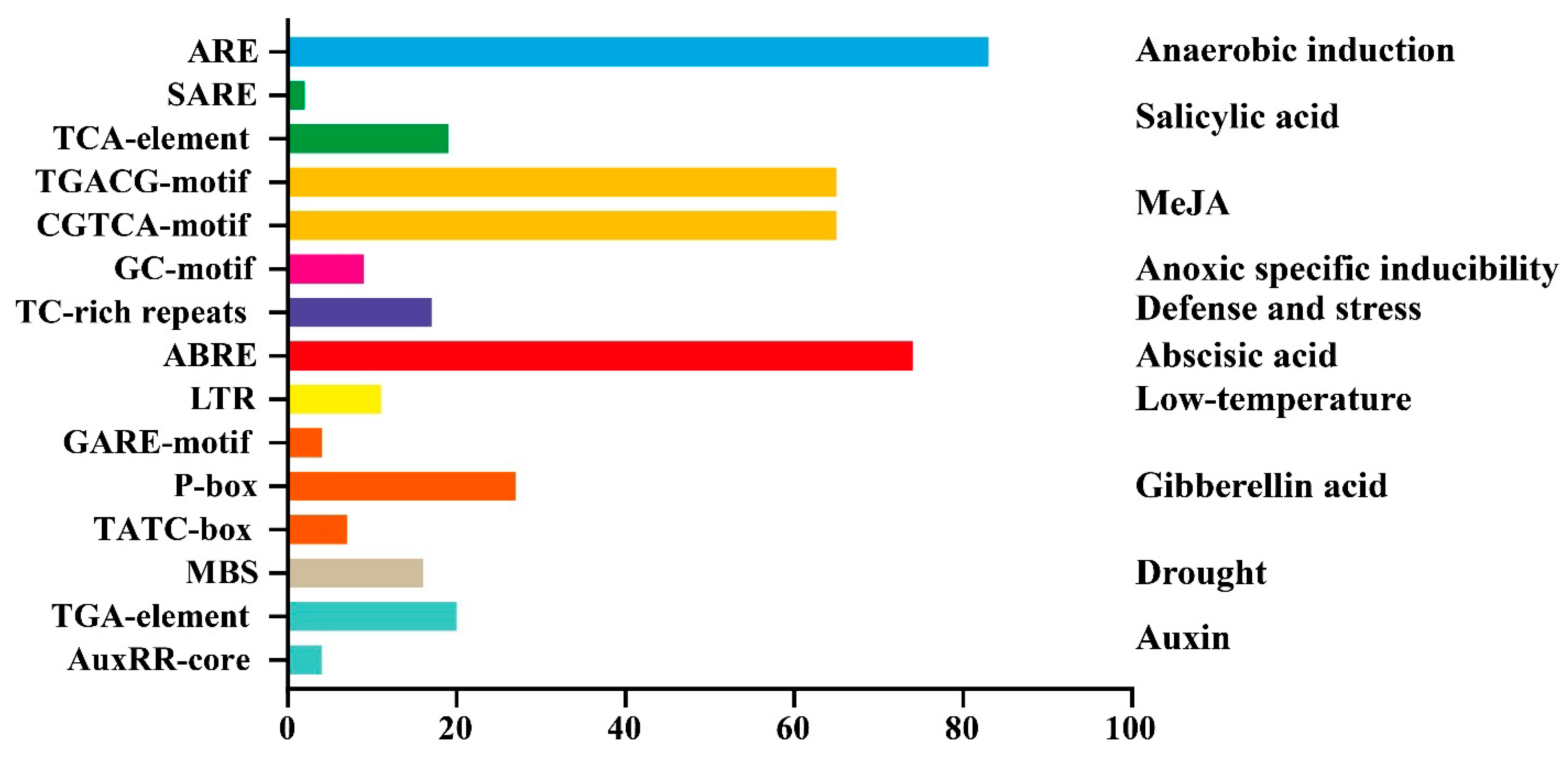
3.6. Analysis of Cis−Acting Elements in the Promoter Region of GbWRKYs
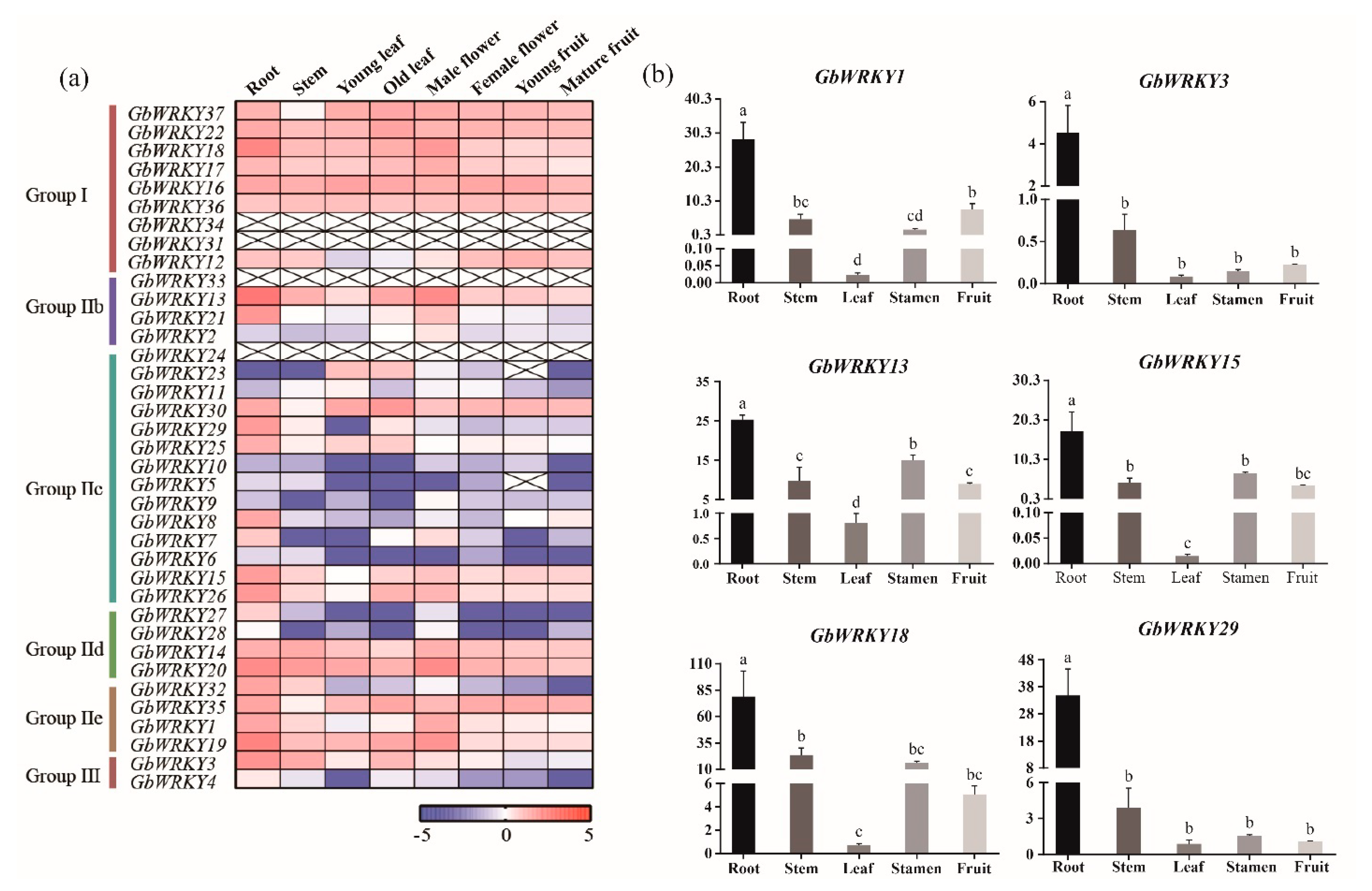
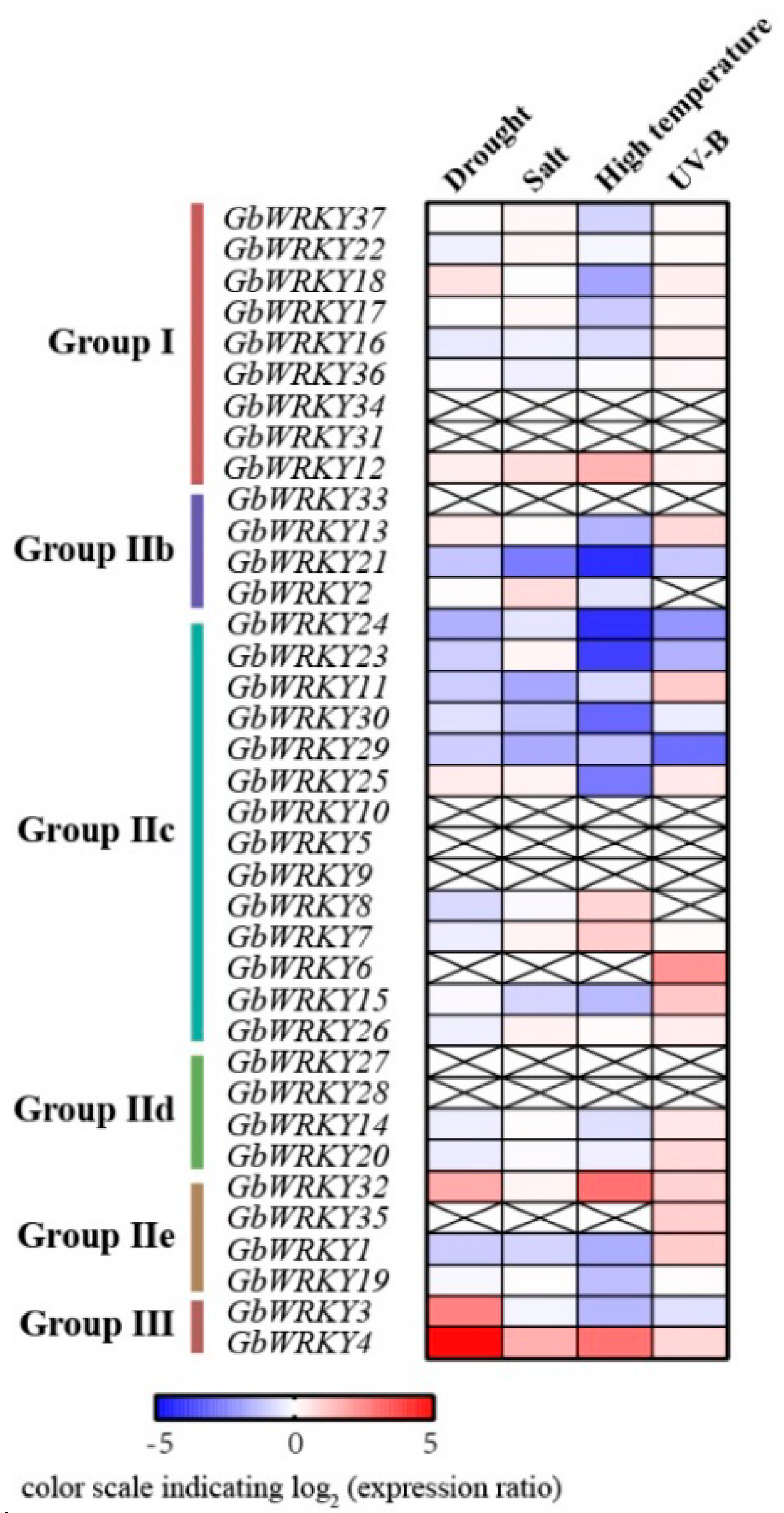
3.7. RNA−Seq Expression Analysis of GbWRKY Genes under Different Stress
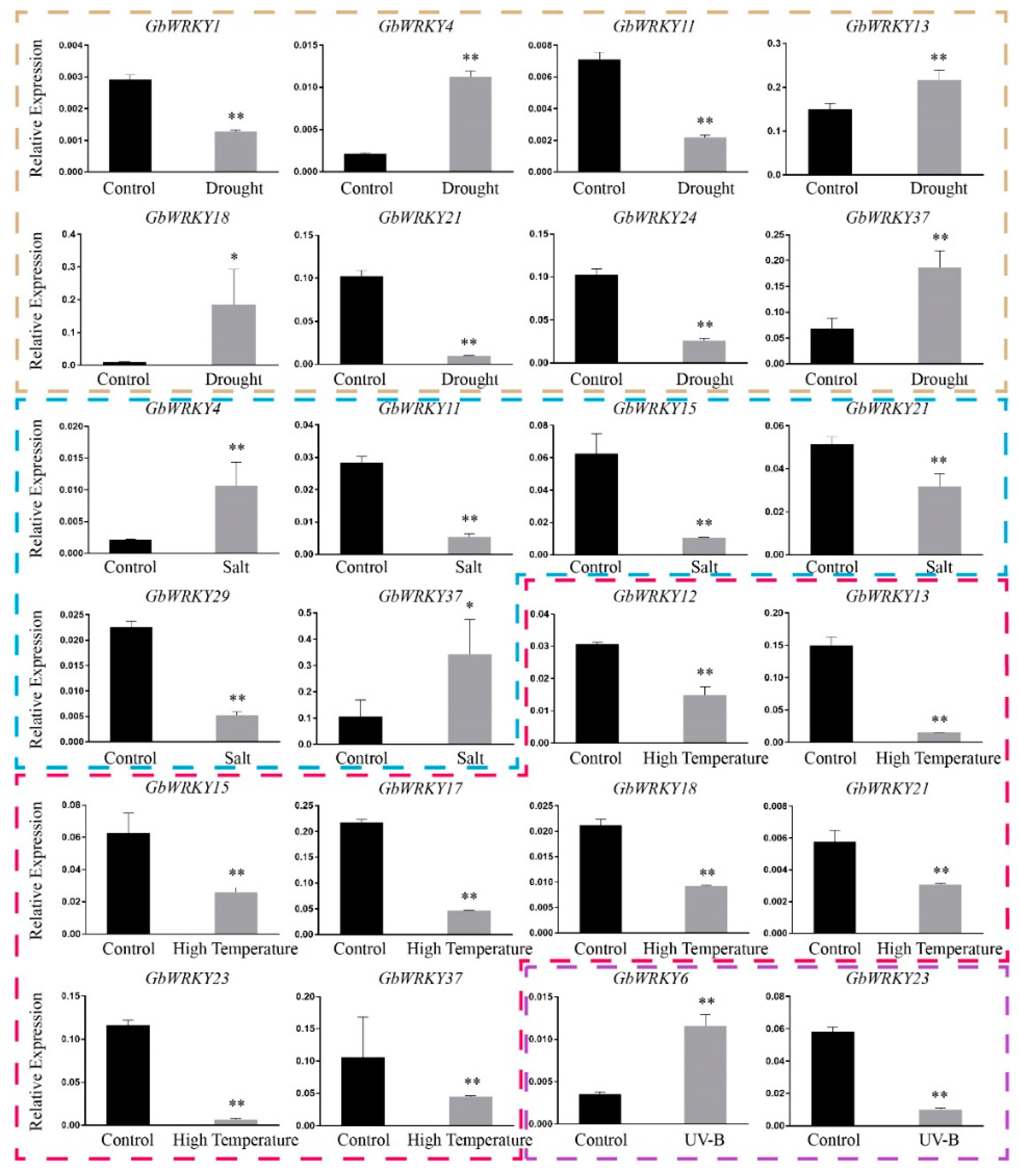
3.8. Expression Patterns of GbWRKY Genes Verified by qRT−PCR under Different Stresses
3.9. Analysis of WRKY Gene Evolution Tree of G. biloba WRKY Family and Different Species with Known Abiotic Stress
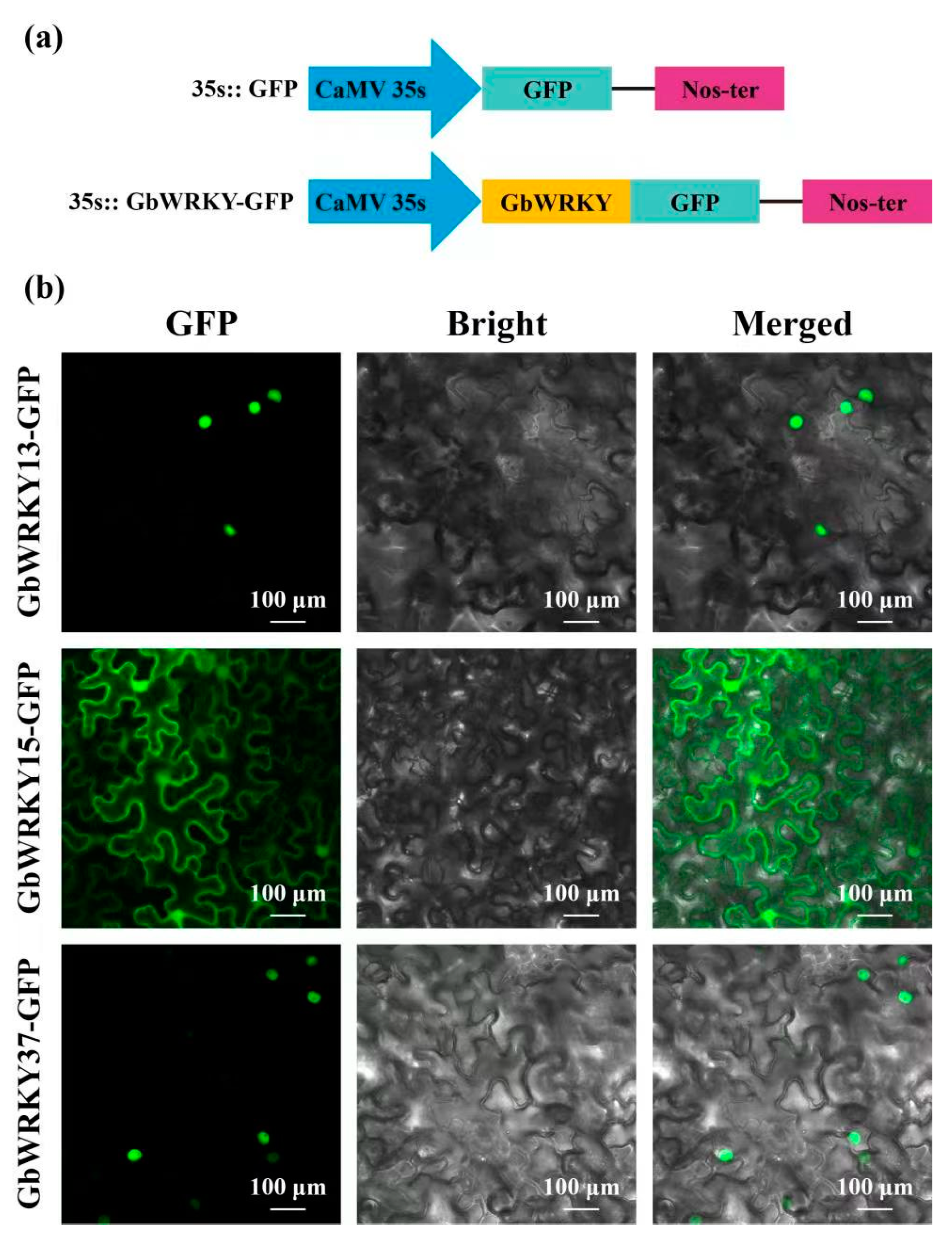
3.10. Subcellular Localization of GbWRKY13/15/37
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ülker, B.; Somssich, I.E. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004, 7, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.P.; Somssich, I.E. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef] [PubMed]
- Phukan, U.J.; Jeena, G.S.; Shukla, R.K. WRKY transcription factors: Molecular regulation and stress responses in plants. Front. Plant Sci. 2016, 7, 760. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Huang, S.; Gao, Y.; Liu, J.; Peng, X.; Niu, X.; Fei, Z.; Cao, S.; Liu, Y. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genom. 2012, 287, 495–513. [Google Scholar] [CrossRef] [PubMed]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Rinerson, C.I.; Rabara, R.C.; Tripathi, P.; Shen, Q.J.; Rushton, P.J. The evolution of WRKY transcription factors. BMC Plant Biol. 2015, 15, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Rushton, P.J.; Macdonald, H.; Huttly, A.K.; Lazarus, C.M.; Hooley, R. Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of α-Amy2 genes. Plant Mol. Biol. 1995, 29, 691–702. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Hu, Y.; Vannozzi, A.; Wu, K.; Cai, H.; Qin, Y.; Mullis, A.; Lin, Z.; Zhang, L. The WRKY transcription factor family in model plants and crops. Crit. Rev. Plant Sci. 2007, 36, 311–335. [Google Scholar] [CrossRef]
- Ross, C.A.; Liu, Y.; Shen, Q.J. The WRKY gene family in rice (Oryza sativa). J. Integr. Plant Biol. 2007, 49, 827–842. [Google Scholar] [CrossRef]
- Mangelsen, E.; Kilian, J.; Berendzen, K.W.; Kolukisaoglu, Ü.H.; Harter, K.; Jansson, C.; Wanke, D. Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. BMC Genom. 2008, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Rensing, S.A.; Lang, D.; Zimmer, A.D.; Terry, A.; Salamov, A.; Shapiro, H.; Nishiyama, T.; Perroud, P.F.; Lindquist, E.A.; Kamisugi, Y.; et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 2008, 319, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.J.; Ekramoddoullah, A.K. Identification and characterization of the WRKY transcription factor family in Pinus monticola. Genome 2009, 52, 77–88. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Ling, J.; Jiang, W.; Zhang, Y.; Yu, H.; Mao, Z.; Gu, X.; Huang, S.; Xie, B. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genom. 2011, 12, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.F.; Chen, J.; Chen, Y.F.; Wu, L.J.; Xie, D.X. Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res. 2012, 19, 153–164. [Google Scholar] [CrossRef]
- Li, K.; Liu, X.; He, F.; Chen, S.; Zhou, G.; Wang, Y.; Li, L.; Zhang, S.L.; Ren, M.; Yuan, Y. Genome-wide analysis of the Tritipyrum WRKY gene family and the response of TtWRKY256 in salt-tolerance. Front Plant Sci. 2022, 13, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Li, K.; Xu, X.; Yao, Z.; Jin, C.; Zhang, S. Genome-wide analysis of WRKY transcription factors in white pear (Pyrus bretschneideri) reveals evolution and patterns under drought stress. BMC Genom. 2015, 16, 1–14. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef]
- Jiang, Y.; Deyholos, M.K. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 2009, 69, 91–105. [Google Scholar] [CrossRef]
- Li, S.; Fu, Q.; Chen, L.; Huang, W.; Yu, D. Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 2011, 233, 1237–1252. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.Y.; Tian, A.G.; Zou, H.F.; Xie, Z.M.; Lei, G.; Huang, J.; Wang, C.M.; Wang, H.W.; Zhang, J.S.; Chen, S.Y. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 2008, 6, 486–503. [Google Scholar] [CrossRef]
- Yokotani, N.; Sato, Y.; Tanabe, S.; Chujo, T.; Shimizu, T.; Okada, K.; Yamane, H.; Nishizawa, Y.; Shimono, M.; Sugano, S.; et al. WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J. Exp. Bot. 2013, 64, 5085–5097. [Google Scholar] [CrossRef]
- Puccio, G.; Crucitti, A.; Tiberini, A.; Mauceri, A.; Taglienti, A.; Palumbo, P.A.; Carimi, F.; Kaauwen, M.; Scholten, O.; Sunseri, F.; et al. WRKY Gene Family Drives Dormancy Release in Onion Bulbs. Cells 2022, 11, 1100. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Chen, Z.; Liu, Y.; Zhang, H.; Zhang, M.; Liu, Q.; Hong, X.; Zhu, J.; Gong, Z. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 2010, 63, 417–429. [Google Scholar] [CrossRef]
- Mahady, G.B. Ginkgo biloba for the prevention and treatment of cardiovascular disease: A review of the literature. J. Cardiovasc. Nurs. 2002, 16, 21–32. [Google Scholar] [CrossRef]
- Yao, P.; Song, F.; Li, K.; Zhou, S.; Liu, S.; Sun, X.; Nussler, A.; Liu, L. Ginkgo biloba extract prevents ethanol induced dyslipidemia. Am. J. Chin. Med. 2007, 35, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Cui, J.; Jin, B.; Zhao, J.; Xu, H.; Lu, Z.; Li, W.; Li, X.; Lin, L.; Liang, E.; et al. Multifeature analyses of vascular cambial cells reveal longevity mechanisms in old Ginkgo biloba trees. Proc. Natl. Acad. Sci. USA 2020, 117, 2201–2210. [Google Scholar] [CrossRef] [PubMed]
- Kosaki, Y.; Naito, H.; Nojima, T.; Nakao, A. Epileptic seizure from ginkgo nut intoxication in an adult. Case Rep. Emerg. Med. 2020, 2020, 5072954. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.L.; Shen, Y.B.; Chang, J.; Zhang, W.W.; Cheng, S.Y.; Xu, F. Isolation, expression, and promoter analysis of GbWRKY2: A novel transcription factor gene from Ginkgo biloba. Int. J. Genom. 2015, 2015, 607185. [Google Scholar]
- Liao, Y.L.; Xu, F.; Zang, W.W.; Cheng, S.Y.; Shen, Y.B.; Chang, J. Cloning, characterization and expression analysis of GbWRKY11, a novel transcription factor gene in Ginkgo biloba. Int. J. Agric. Biol. 2016, 18, 117–124. [Google Scholar] [CrossRef]
- Zhou, T.; Yang, X.; Wang, G.; Cao, F. Molecular cloning and expression analysis of a WRKY transcription factor gene, GbWRKY20, from Ginkgo biloba. Plant Signal. Behav. 2021, 16, 1930442. [Google Scholar] [CrossRef] [PubMed]
- Guan, R.; Zhao, Y.; Zhang, H.E.; Fan, G.; Liu, X.; Zhou, W.; Shi, C.; Wang, J.; Liu, W.; Liang, X.; et al. Draft genome of the living fossil Ginkgo biloba. Gigascience 2016, 5, 49. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Liu, X.; Liao, Y.; Zhang, W.; Ye, J.; Rao, S.; Xu, F. Genome-wide identification of WRKY family genes and analysis of their expression in response to abiotic stress in Ginkgo biloba L. Not. Bot. Horti. Agrobo. 2019, 47, 1100–1115. [Google Scholar] [CrossRef]
- Kim, J.H.; Lee, K.I.; Chang, Y.J.; Kim, S.U. Developmental pattern of Ginkgo biloba levopimaradiene synthase (GbLPS) as probed by promoter analysis in Arabidopsis thaliana. Plant Cell Rep. 2012, 31, 1119–1127. [Google Scholar] [CrossRef]
- Xu, F.; Huang, X.H.; Li, L.L.; Deng, G.; Cheng, H.; Rong, X.F.; Li, J.B.; Cheng, S.Y. Molecular cloning and characterization of GbDXS and GbGGPPS gene promoters from Ginkgo biloba. Genet. Mol. Res. 2013, 12, 293–301. [Google Scholar] [CrossRef]
- Liao, Y.; Xu, F.; Huang, X.; Zhang, W.; Cheng, H.; Linling, L.I.; Cheng, S.; Shen, Y. Promoter analysis and transcriptional profiling of Ginkgo biloba 3-hydroxy-3-methylglutaryl coenzyme a reductase (GbHMGR) gene in abiotic stress responses. Not. Bot. Horti. Agrobo. 2015, 43, 25–34. [Google Scholar] [CrossRef]
- Kang, M.K.; Nargis, S.; Kim, S.M.; Kim, S.U. Distinct expression patterns of two Ginkgo biloba 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate reductase/isopentenyl diphospahte synthase (HDR/IDS) promoters in Arabidopsis model. Plant Physiol. Biochem. 2013, 62, 47–53. [Google Scholar] [CrossRef]
- Cheng, S.Y.; Li, L.L.; Yuan, H.H.; Xu, F.; Cheng, H. Molecular cloning and characterization of GbMECT and GbMECP gene promoters from Ginkgo biloba. Genet. Mol. Res. 2015, 14, 15112–15122. [Google Scholar] [CrossRef]
- Chang, B.; Ma, K.; Lu, Z.; Lu, J.; Cui, J.; Wang, L.; Jin, B. Physiological, transcriptomic, and metabolic responses of Ginkgo biloba L. to drought, salt, and heat stresses. Biomolecules 2020, 10, 1635. [Google Scholar] [CrossRef]
- Punta, M.; Coggill, P.C.; Eberhardt, R.Y.; Mistry, J.; Tate, J.; Boursnell, C.; Pang, N.; Forslund, K.; Ceric, G.; Clements, J.; et al. The Pfam protein families database. Nucleic Acids Res. 2012, 40, D290–D301. [Google Scholar] [CrossRef]
- Schultz, J.; Milpetz, F.; Bork, P.; Ponting, C.P. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. USA 1998, 95, 5857–5864. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PloS ONE 2015, 5, e11335. [Google Scholar] [CrossRef] [PubMed]
- Jing, L.; Guo, D.; Hu, W.; Niu, X. The prediction of a pathogenesis-related secretome of Puccinia helianthi through high-throughput transcriptome analysis. BMC Bioinform. 2017, 18, 1–13. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Shen, C.; Yuan, J. Genome-wide investigation and expression analysis of K+-transport-related gene families in Chinese cabbage (Brassica rapa ssp. pekinensis). Biochem. Genet. 2021, 59, 256–282. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 1–19. [Google Scholar] [CrossRef]
- Terwilliger, T.C. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 2000, 56, 965–972. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Jansson, S.; Meyer-Gauen, G.; Cerff, R.; Martin, W. Nucleotide distribution in gymnosperm nuclear sequences suggests a model for GC-content change in land-plant nuclear genomes. J. Mol. Evol. 1994, 39, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Xu, F.; Wang, X.; Liao, Y.; Chen, Q.; Meng, X. Characterization and functional analysis of a MADS-box transcription factor gene (GbMADS9) from Ginkgo biloba. Sci. Horti. 2016, 212, 104–114. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, L. The WRKY transcription factor superfamily: Its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 2005, 5, 1–12. [Google Scholar] [CrossRef]
- Sun, Y.; Zhou, X.; Ma, H. Genome-wide Analysis of Kelch Repeat-containing F-box Family. J. Integr. Plant Biol. 2007, 49, 940–952. [Google Scholar] [CrossRef]
- Vatansever, R.; Koc, I.; Ozyigit, I.I.; Sen, U.; Uras, M.E.; Anjum, N.A.; Pereira, E.; Filiz, E. Genome-wide identification and expression analysis of sulfate transporter (SULTR) genes in potato (Solanum tuberosum L.). Planta 2016, 244, 1167–1183. [Google Scholar] [CrossRef]
- Mao, D.Y.; Tang, H.; Xiao, N.; Wang, L. Uncovering the secrets of secretory fluids during the reproductive process in Ginkgo biloba. Crit. Rev. Plant Sci. 2022, 41, 161–175. [Google Scholar] [CrossRef]
- Liu, S.; Meng, Z.; Zhang, H.; Chu, Y.; Qiu, Y.; Jin, B.; Wang, L. Identification and characterization of thirteen gene families involved in flavonoid biosynthesis in Ginkgo biloba. Ind. Crop. Prod. 2022, 188, 115576. [Google Scholar] [CrossRef]
- Zhao, B.; Wang, L.; Pang, S.; Jia, Z.; Wang, L.; Li, W.; Jin, B. UV-B promotes flavonoid synthesis in Ginkgo biloba leaves. Ind. Crop. Prod. 2020, 151, 112483. [Google Scholar] [CrossRef]
- Guo, C.; Guo, R.; Xu, X.; Gao, M.; Li, X.; Song, J.; Zheng, Y.; Wang, X. Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family. J. Exp. Bot. 2014, 65, 1513–1528. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Duan, Y.; Yin, J.; Ye, S.; Zhu, J.; Zhang, F.; Lu, W.; Fan, D.; Luo, K. Genome-wide identification and characterization of the Populus WRKY transcription factor family and analysis of their expression in response to biotic and abiotic stresses. J. Exp. Bot. 2014, 65, 6629–6644. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Berri, S.; Abbruscato, P.; Faivre-Rampant, O.; Brasileiro, A.; Fumasoni, I.; Satoh, K.; Kikuchi, S.; Mizzi, L.; Morandini, P.; Pè, M.E.; et al. Characterization of WRKYco-regulatory networks in rice and Arabidopsis. BMC Plant Biol. 2009, 9, 1–22. [Google Scholar] [CrossRef]
- He, H.; Dong, Q.; Shao, Y.; Jiang, H.; Zhu, S.; Cheng, B.; Xiang, Y. Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Rep. 2012, 31, 1199–1217. [Google Scholar] [CrossRef]
- Li, J.; Yu, H.; Liu, M.; Chen, B.; Dong, N.; Chang, X.; Wang, J.; Xing, S.; Peng, H.; Zha, L.; et al. Transcriptome-wide identification of WRKY transcription factors and their expression profiles in response to methyl jasmonate in Platycodon grandiflorus. Plant Signal. Behav. 2022, 17, 2089473. [Google Scholar] [CrossRef]
- Huang, T.; Yang, J.; Yu, D.; Han, X.; Wang, X. Bioinformatics analysis of WRKY transcription factors in grape and their potential roles prediction in sugar and abscisic acid signaling pathway. J. Plant Biochem. Biot. 2021, 30, 67–80. [Google Scholar] [CrossRef]
- Di, P.; Wang, P.; Yan, M.; Han, P.; Huang, X.; Yin, L.; Yan, Y.; Xu, Y.; Wang, Y. Genome-wide characterization and analysis of WRKY transcription factors in Panax ginseng. BMC Genom. 2021, 22, 1–15. [Google Scholar] [CrossRef]
- Chen, X.; Li, C.; Wang, H.; Guo, Z. WRKY transcription factors: Evolution, binding, and action. Phytopathol. Res. 2019, 1, 1–15. [Google Scholar] [CrossRef]
- Zhang, T.; Tan, D.; Zhang, L.; Zhang, X.; Han, Z. Phylogenetic analysis and drought-responsive expression profiles of the WRKY transcription factor family in maize. Agri Gene 2017, 3, 99–108. [Google Scholar] [CrossRef]
- Jimmy, J.L.; Babu, S. Variations in the structure and evolution of rice WRKY genes in indica and japonica genotypes and their co-expression network in mediating disease resistance. Evol. Bioinform. 2019, 15, 1–12. [Google Scholar] [CrossRef]
- Tang, R.; Dong, H.; He, L.; Li, P.; Shi, Y.; Yang, Q.; Jia, X.; Li, X.Q. Genome-wide identification, evolutionary and functional analyses of KFB family members in potato. BMC Plant Biol. 2022, 22, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Wang, P.; Hou, L.; Zhao, S.; Zhao, C.; Xia, H.; Li, P.; Zhang, Y.; Bian, X.; Wang, X. Global analysis of WRKY genes and their response to dehydration and salt stress in soybean. Front. Plant Sci. 2016, 7, 9. [Google Scholar] [CrossRef]
- van Verk, M.C.; Pappaioannou, D.; Neeleman, L.; Bol, J.F.; Linthorst, H.J. A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol. 2008, 146, 1983–1995. [Google Scholar] [CrossRef]
- Wei, Y.; Shi, H.; Xia, Z.; Tie, W.; Ding, Z.; Yan, Y.; Wang, W.; Hu, W.; Li, K. Genome-wide identification and expression analysis of the WRKY gene family in cassava. Front. Plant Sci. 2016, 7, 25. [Google Scholar] [CrossRef]
- Xu, Y.H.; Sun, P.W.; Tang, X.L.; Gao, Z.H.; Zhang, Z.; Wei, J.H. Genome-wide analysis of WRKY transcription factors in Aquilaria sinensis (Lour.) Gilg. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Eulgem, T.; Somssich, I.E. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 2007, 10, 366–371. [Google Scholar] [CrossRef]
- Banks, J.A.; Nishiyama, T.; Hasebe, M.; Bowman, J.L.; Gribskov, M.; DePamphilis, C.; Albert, V.A.; Aono, N.; Aoyama, T.; Ambrose, B.A.; et al. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 2011, 332, 960–963. [Google Scholar] [CrossRef]
- Glöckner, G.; Eichinger, L.; Szafranski, K.; Pachebat, J.A.; Bankier, A.T.; Dear, P.H.; Lehmann, R.; Baumgart, C.; Parra, G.; Abril, J.F.; et al. Sequence and analysis of chromosome 2 of Dictyostelium discoideum. Nature 2002, 418, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.J.; Cho, C.C.; Kao, Y.Y.; Sun, C.H. A novel WRKY-like protein involved in transcriptional activation of cyst wall protein genes in Giardia lamblia. J. Biol. Chem. 2009, 284, 17975–17988. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zeng, J.; Li, Y.; Rong, X.; Sun, J.; Sun, T.; Li, M.; Wang, L.; Feng, Y.; Chai, R.; et al. Expression of TaWRKY44, a wheat WRKY gene, in transgenic tobacco confers multiple abiotic stress tolerances. Front. Plant Sci. 2015, 6, 615. [Google Scholar] [CrossRef] [PubMed]
| Name | ID | Chromosomal Location | CDS (bp) | GRAVY | Subcellular Localization | MW (KDa) | pI |
|---|---|---|---|---|---|---|---|
| GbWRKY1 | Gb_05176 | 261561178-261563292 | 1494 | −0.814 | Nucleus | 54.67 | 6.53 |
| GbWRKY2 | Gb_02351 | 783125879-783138739 | 1581 | −0.593 | Nucleus | 56.24 | 7.20 |
| GbWRKY3 | Gb_00547 | 1053172260-1053174875 | 1740 | −0.538 | Nucleus | 63.67 | 6.58 |
| GbWRKY4 | Gb_00545 | 1053573972-1053576514 | 1446 | −0.620 | Nucleus | 53.13 | 5.38 |
| GbWRKY5 | Gb_40207 | 1072194964-1072196293 | 801 | −0.666 | Nucleus | 29.95 | 7.69 |
| GbWRKY6 | Gb_26411 | 1075330508-1075331449 | 747 | −0.793 | Nucleus | 27.90 | 7.70 |
| GbWRKY7 | Gb_26412 | 1075829444-1075830197 | 573 | −0.949 | Nucleus | 21.53 | 9.03 |
| GbWRKY8 | Gb_26413 | 1075925548-1075926489 | 747 | −0.776 | Nucleus | 27.96 | 8.20 |
| GbWRKY9 | Gb_40261 | 1077671233-1077672112 | 681 | −0.711 | Nucleus | 25.77 | 8.83 |
| GbWRKY10 | Gb_40257 | 1078258461-1078259371 | 723 | −0.830 | Nucleus | 27.48 | 6.76 |
| GbWRKY11 | Gb_17074 | 1129012414-1129014214 | 1404 | −0.739 | Nucleus | 51.72 | 4.89 |
| GbWRKY12 | Gb_36273 | 253690691-253784117 | 2621 | −0.573 | Nucleus | 96.08 | 8.83 |
| GbWRKY13 | Gb_39366 | 397260590-397263274 | 2124 | −0.651 | Nucleus | 76.52 | 6.11 |
| GbWRKY14 | Gb_16513 | 490181118-490183897 | 1083 | −0.786 | Nucleus | 40.49 | 9.66 |
| GbWRKY15 | Gb_01873 | 567507034-567508969 | 1458 | −0.721 | Nucleus | 52.55 | 5.73 |
| GbWRKY16 | Gb_20926 | 369244325-369246968 | 1443 | −0.651 | Nucleus | 50.98 | 8.53 |
| GbWRKY17 | Gb_31953 | 505067926-505086750 | 1980 | −0.866 | Nucleus | 71.11 | 6.86 |
| GbWRKY18 | Gb_25118 | 628096908-628105595 | 3046 | −0.410 | Nucleus | 110.84 | 7.18 |
| GbWRKY19 | Gb_25547 | 145264191-145266777 | 1152 | −0.717 | Nucleus | 42.17 | 4.72 |
| GbWRKY20 | Gb_01527 | 147519959-147521498 | 1104 | −0.507 | Nucleus | 39.72 | 9.22 |
| GbWRKY21 | Gb_23334 | 489258288-489261466 | 2064 | −0.745 | Nucleus | 74.81 | 5.92 |
| GbWRKY22 | Gb_32055 | 543409418-543471462 | 2328 | −0.686 | Nucleus | 83.42 | 6.11 |
| GbWRKY23 | Gb_17623 | 537103484-537103996 | 396 | −1.130 | Nucleus | 14.68 | 8.87 |
| GbWRKY24 | Gb_15790 | 537465845-537467456 | 534 | −0.490 | Extracellular | 20.01 | 9.30 |
| GbWRKY25 | Gb_16917 | 546983507-546984721 | 822 | −0.841 | Nucleus | 30.88 | 8.10 |
| GbWRKY26 | Gb_08731 | 675694495-675695882 | 1194 | −0.598 | Nucleus | 43.05 | 5.60 |
| GbWRKY27 | Gb_12538 | 687806524-687807590 | 651 | −0.875 | Nucleus | 24.26 | 8.79 |
| GbWRKY28 | Gb_12539 | 688432818-688434000 | 759 | −0.892 | Nucleus | 28.21 | 9.27 |
| GbWRKY29 | Gb_05024 | 327509310-327510274 | 702 | −0.952 | Nucleus | 27.02 | 6.75 |
| GbWRKY30 | Gb_05026 | 328511178-328512884 | 1459 | −0.553 | Nucleus | 56.47 | 8.60 |
| GbWRKY31 | Gb_28473 | 633231750-633232705 | 801 | −0.395 | Nucleus | 29.58 | 8.52 |
| GbWRKY32 | Gb_03346 | 447558574-447562360 | 1866 | −0.515 | Nucleus | 67.99 | 5.71 |
| GbWRKY33 | Gb_07810 | 661423975-661424813 | 483 | −0.430 | Nucleus | 18.14 | 9.44 |
| GbWRKY34 | Gb_41027 | 692001348-692001991 | 474 | −0.690 | Nucleus | 17.52 | 9.39 |
| GbWRKY35 | Gb_25334 | 545181619-545183218 | 825 | −0.858 | Nucleus | 30.34 | 5.82 |
| GbWRKY36 | Gb_36184 | 223808232-223819523 | 3123 | −0.260 | Nucleus | 111.54 | 8.62 |
| GbWRKY37 | Gb_01391 | 836924-893231 | 2589 | −0.691 | Nucleus | 92.73 | 6.05 |
| Gene Pairs | Type of Gene Duplication | Chr. Location | Ka | Ks | Ka/Ks | Approximate Duplication Date (Mya) |
|---|---|---|---|---|---|---|
| GbWRKY3/GbWRKY4 | Tandem duplication | Chr01 | 0.44224 | 0.58682 | 0.753622 | 0.475293 |
| GbWRKY8/GbWRKY9 | Tandem duplication | Chr01 | 0.399267 | 0.702224 | 0.568575 | 0.468139 |
| GbWRKY23/GbWRKY24 | Tandem duplication | Chr07 | 0.272219 | 0.328896 | 0.827675 | 0.286152 |
| GbWRKY27/GbWRKY28 | Tandem duplication | Chr07 | 0.185017 | 0.399258 | 0.463401 | 0.234514 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Xiao, N.; Wang, Y.; Liu, X.; Chen, Z.; Gu, X.; Chen, Y. Genome-Wide Identification, Evolutionary and Functional Analyses of WRKY Family Members in Ginkgo biloba. Genes 2023, 14, 343. https://doi.org/10.3390/genes14020343
Li W, Xiao N, Wang Y, Liu X, Chen Z, Gu X, Chen Y. Genome-Wide Identification, Evolutionary and Functional Analyses of WRKY Family Members in Ginkgo biloba. Genes. 2023; 14(2):343. https://doi.org/10.3390/genes14020343
Chicago/Turabian StyleLi, Weixing, Nan Xiao, Yawen Wang, Ximeng Liu, Zhaoyu Chen, Xiaoyin Gu, and Yadi Chen. 2023. "Genome-Wide Identification, Evolutionary and Functional Analyses of WRKY Family Members in Ginkgo biloba" Genes 14, no. 2: 343. https://doi.org/10.3390/genes14020343
APA StyleLi, W., Xiao, N., Wang, Y., Liu, X., Chen, Z., Gu, X., & Chen, Y. (2023). Genome-Wide Identification, Evolutionary and Functional Analyses of WRKY Family Members in Ginkgo biloba. Genes, 14(2), 343. https://doi.org/10.3390/genes14020343






