Gene Regulatory Interactions at Lamina-Associated Domains
Abstract
1. Introduction
2. Materials and Methods
2.1. Adipose Stem Cell Culture and Adipogenic Induction
2.2. Immunoblotting
2.3. Fluorescence In Situ Hybridization (FISH)
2.4. Knock-Down of LMNA/C and LMNB1
2.5. Chromatin Immunoprecipitation (ChIP)
2.6. ChIP-Seq Analysis and Chromatin State Modeling
2.7. ATAC-Seq Data Reprocessing
2.8. Intersections between LADs, Genes, and Enhancer Positions
2.9. Gene Expression Analysis
3. Results
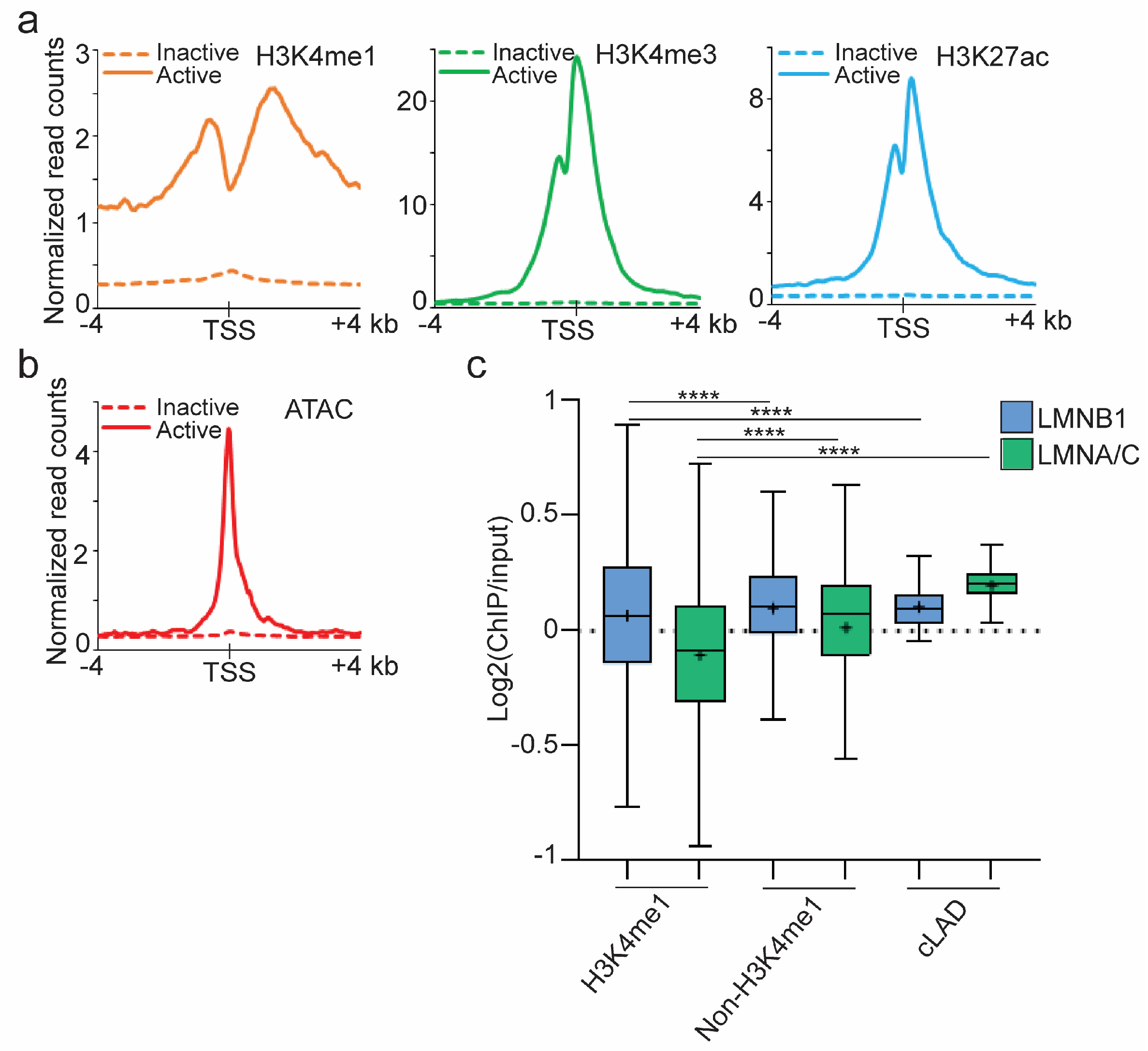
3.1. cLADs Contain Active Genes in Euchromatic Regions of Low Enrichment in A- and B-Type Lamins
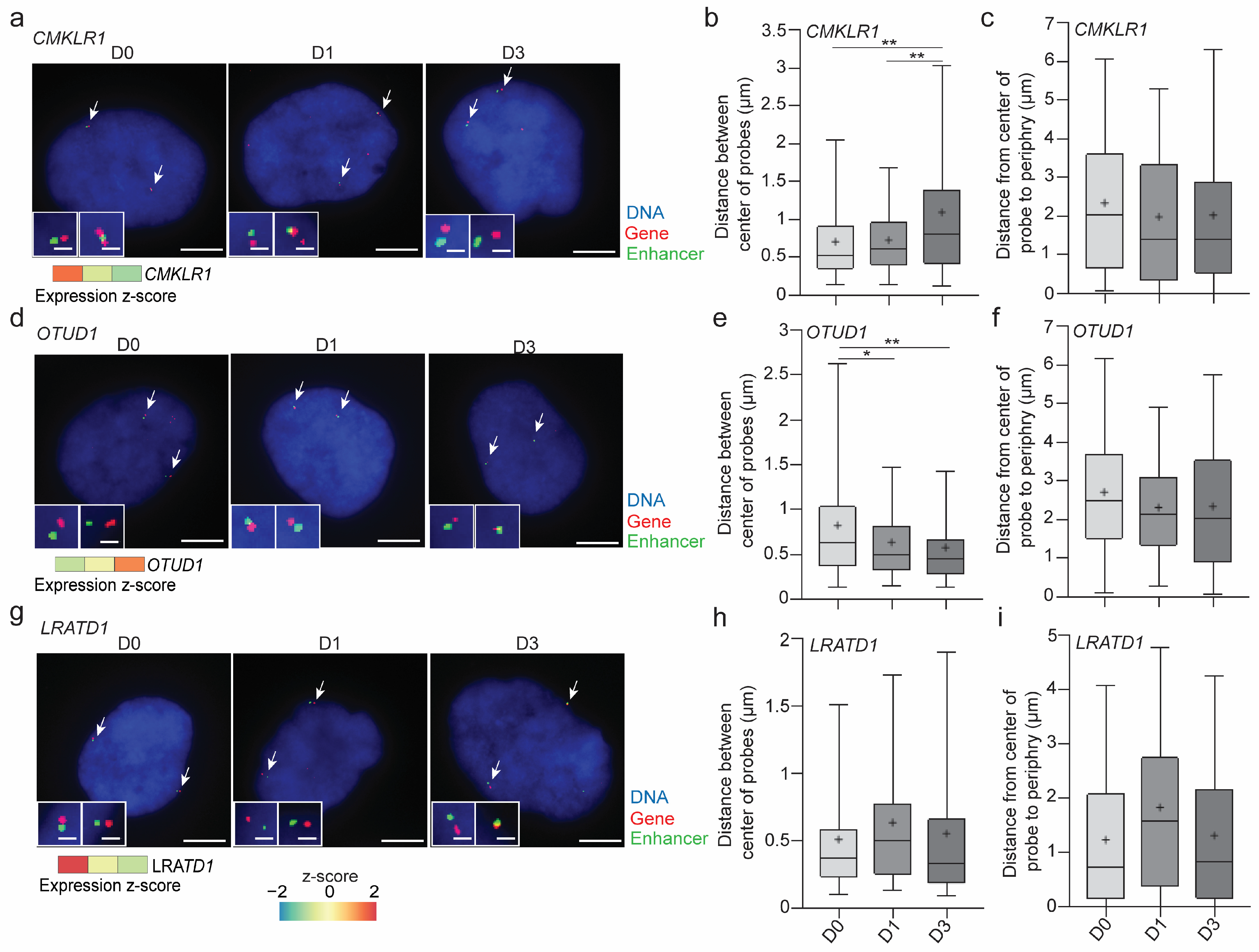
3.2. ECHi-C Data Provide Evidence of Enhancer Connectivity at the Nuclear Periphery
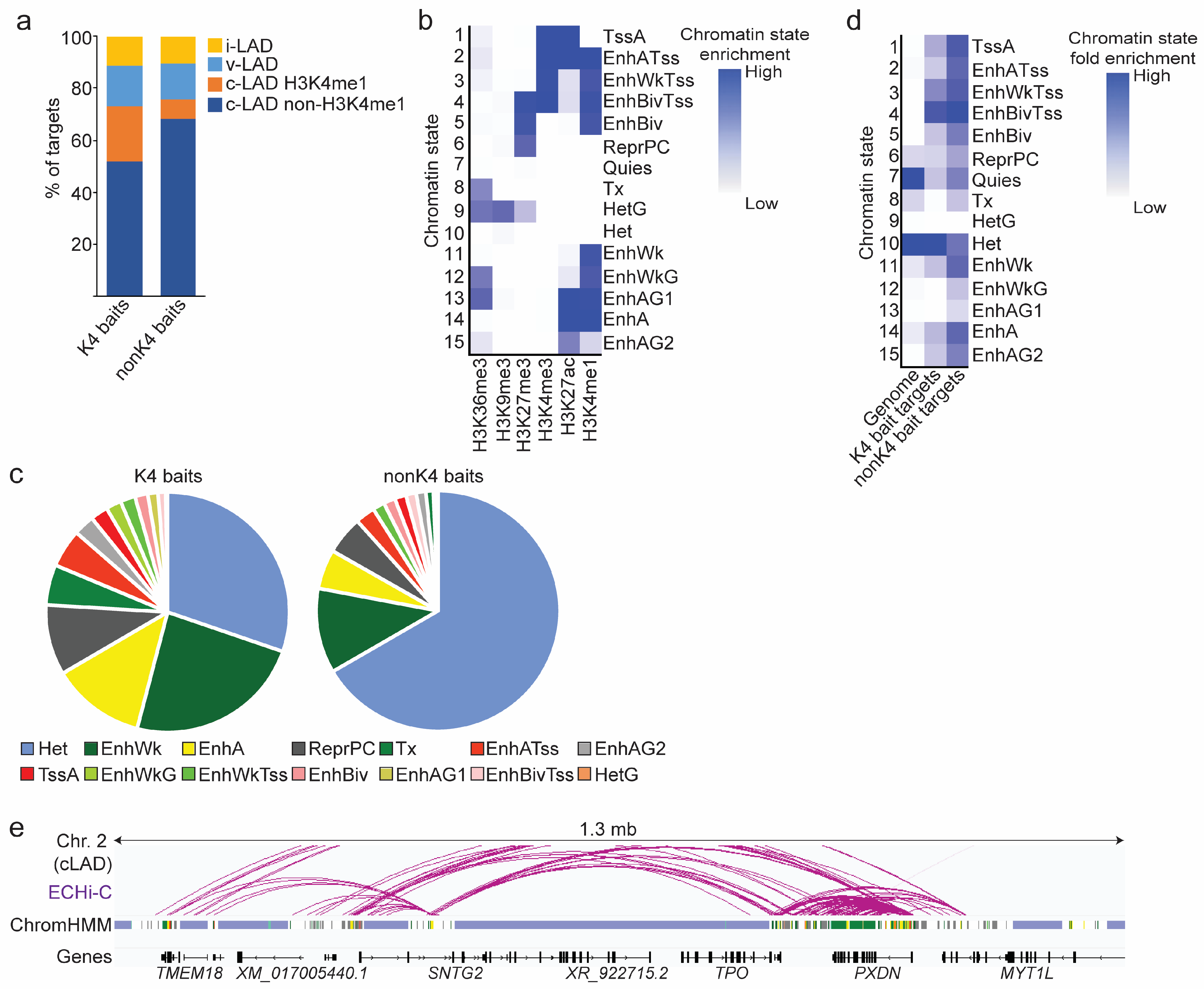
3.3. Predominance of Enhancer Connections within cLADs
3.4. Chromatin Landscape of ECHi-C Connections
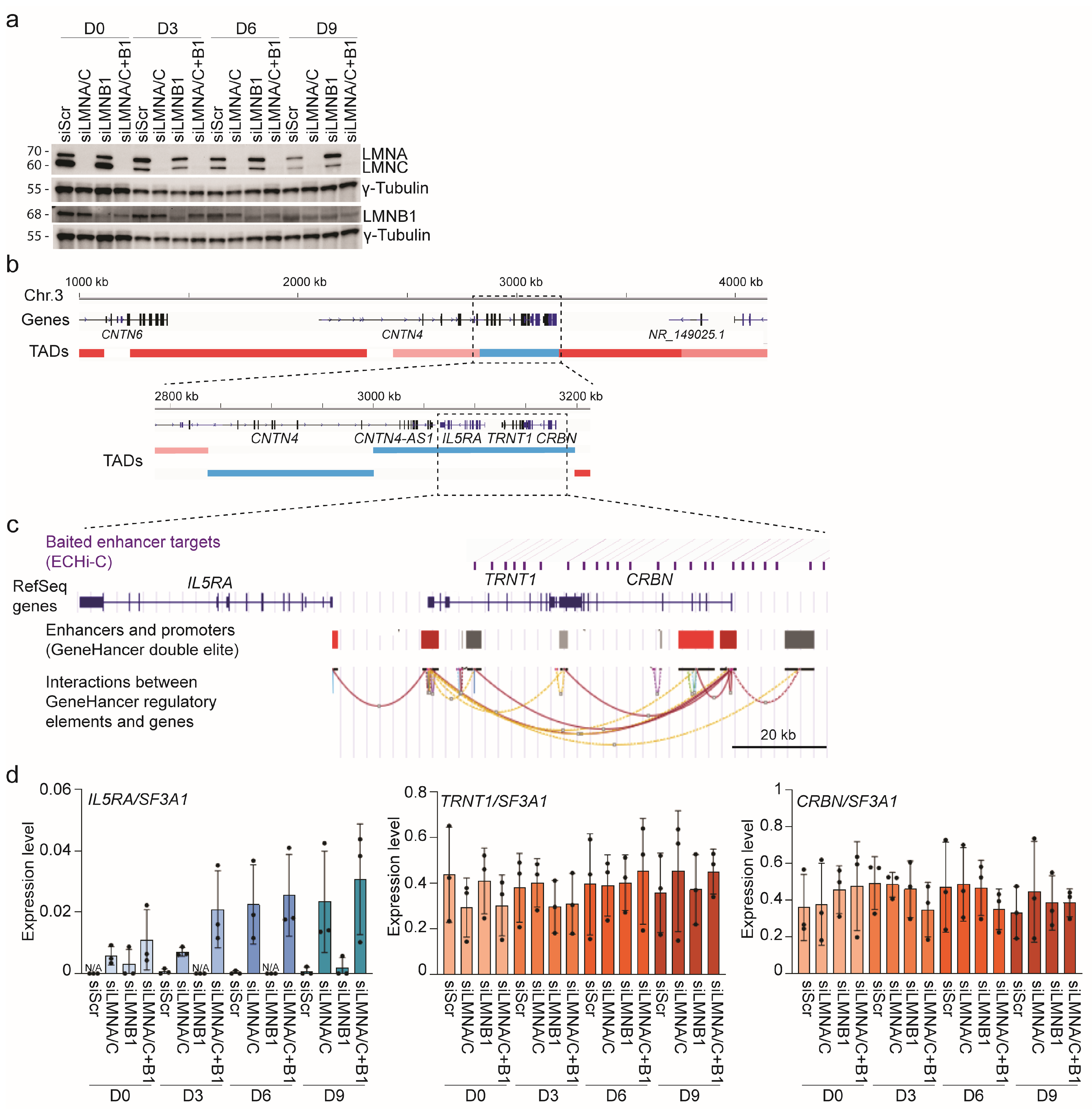
3.5. Lamin A/C Restricts Gene Expression in LADs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Buchwalter, A.; Kaneshiro, J.M.; Hetzer, M.W. Coaching from the sidelines: The nuclear periphery in genome regulation. Nat. Rev. Genet. 2019, 20, 39–50. [Google Scholar] [CrossRef]
- van Steensel, B.; Belmont, A.S. Lamina-Associated Domains: Links with Chromosome Architecture, Heterochromatin, and Gene Repression. Cell 2017, 169, 780–791. [Google Scholar] [CrossRef] [PubMed]
- Briand, N.; Collas, P. Lamina-associated domains: Peripheral matters and internal affairs. Genome Biol. 2020, 21, 85. [Google Scholar] [CrossRef] [PubMed]
- Meuleman, W.; Peric-Hupkes, D.; Kind, J.; Beaudry, J.B.; Pagie, L.; Kellis, M.; Reinders, M.; Wessels, L.; van Steensel, B. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 2013, 23, 270–280. [Google Scholar] [CrossRef]
- Keough, K.C.; Shah, P.P.; Wickramasinghe, N.M.; Dundes, C.E.; Chen, A.; Salomon, R.E.A.; Whalen, S.; Loh, K.M.; Dubois, N.; Pollard, K.S.; et al. An atlas of lamina-associated chromatin across thirteen human cell types reveals cell-type-specific and multiple subtypes of peripheral heterochromatin. Genome Biol. 2023, 24, 16. [Google Scholar]
- Peric-Hupkes, D.; Meuleman, W.; Pagie, L.; Bruggeman, S.W.; Solovei, I.; Brugman, W.; Graf, S.; Flicek, P.; Kerkhoven, R.M.; van Lohuizen, M.; et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell 2010, 38, 603–613. [Google Scholar] [CrossRef]
- Rønningen, T.; Shah, A.; Oldenburg, A.R.; Vekterud, K.; Delbarre, E.; Moskaug, J.O.; Collas, P. Prepatterning of differentiation-driven nuclear lamin A/C-associated chromatin domains by GlcNAcylated histone H2B. Genome Res. 2015, 25, 1825–1835. [Google Scholar] [CrossRef]
- Robson, M.I.; de Las Heras, J.I.; Czapiewski, R.; Sivakumar, A.; Kerr, A.R.W.; Schirmer, E.C. Constrained release of lamina-associated enhancers and genes from the nuclear envelope during T-cell activation facilitates their association in chromosome compartments. Genome Res. 2017, 27, 1126–1138. [Google Scholar] [CrossRef] [PubMed]
- Czapiewski, R.; Batrakou, D.G.; de las Heras, J.; Carter, R.N.; Sivakumar, A.; Sliwinska, M.; Dixon, C.R.; Webb, S.; Lattanzi, G.; Morton, N.M.; et al. Genomic loci mispositioning in Tmem120a knockout mice yields latent lipodystrophy. Nat. Commun. 2022, 13, 321. [Google Scholar] [CrossRef]
- Madsen-Østerbye, J.; Abdelhalim, M.; Baudement, M.O.; Collas, P. Local euchromatin enrichment in lamina-associated domains anticipates their re-positioning in the adipogenic lineage. Genome Biol. 2022, 23, 91. [Google Scholar] [CrossRef]
- Shumaker, D.K.; Dechat, T.; Kohlmaier, A.; Adam, S.A.; Bozovsky, M.R.; Erdos, M.R.; Eriksson, M.; Goldman, A.E.; Khuon, S.; Collins, F.S.; et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl. Acad. Sci. USA 2006, 103, 8703–8708. [Google Scholar] [CrossRef]
- Perovanovic, J.; Dell’Orso, S.; Gnochi, V.F.; Jaiswal, J.K.; Sartorelli, V.; Vigouroux, C.; Mamchaoui, K.; Mouly, V.; Bonne, G.; Hoffman, E.P. Laminopathies disrupt epigenomic developmental programs and cell fate. Sci. Transl. Med. 2016, 8, 335ra358. [Google Scholar] [CrossRef]
- Paulsen, J.; Sekelja, M.; Oldenburg, A.R.; Barateau, A.; Briand, N.; Delbarre, E.; Shah, A.; Sørensen, A.L.; Vigouroux, C.; Buendia, B.; et al. Chrom3D: Three-dimensional genome modeling from Hi-C and lamin-genome contacts. Genome Biol. 2017, 18, 21. [Google Scholar] [CrossRef]
- Briand, N.; Guenantin, A.C.; Jeziorowska, D.; Shah, A.; Mantecon, M.; Capel, E.; Garcia, M.; Oldenburg, A.; Paulsen, J.; Hulot, J.S.; et al. The lipodystrophic hotspot lamin A p.R482W mutation deregulates the mesodermal inducer T/Brachyury and early vascular differentiation gene networks. Hum. Mol. Genet. 2018, 27, 1447–1459. [Google Scholar] [CrossRef] [PubMed]
- Bellanger, A.; Madsen-Osterbye, J.; Galigniana, N.M.; Collas, P. Restructuring of Lamina-Associated Domains in Senescence and Cancer. Cells 2022, 11, 1846. [Google Scholar] [CrossRef]
- Wu, F.; Yao, J. Identifying Novel Transcriptional and Epigenetic Features of Nuclear Lamina-associated Genes. Sci. Rep. 2017, 7, 100. [Google Scholar] [CrossRef]
- Leemans, C.; van der Zwalm, M.C.H.; Brueckner, L.; Comoglio, F.; van Schaik, T.; Pagie, L.; van Arensbergen, J.; van Steensel, B. Promoter-Intrinsic and Local Chromatin Features Determine Gene Repression in LADs. Cell 2019, 177, 852–864.e814. [Google Scholar] [CrossRef] [PubMed]
- Brueckner, L.; Zhao, P.A.; van Schaik, T.; Leemans, C.; Sima, J.; Peric-Hupkes, D.; Gilbert, D.M.; van Steensel, B. Local rewiring of genome-nuclear lamina interactions by transcription. EMBO J. 2020, 39, e103159. [Google Scholar] [CrossRef] [PubMed]
- van Schaik, T.; Liu, N.Q.; Manzo, S.G.; Peric-Hupkes, D.; de Wit, E.; van Steensel, B. CTCF and cohesin promote focal detachment of DNA from the nuclear lamina. Genome Biol. 2022, 23, 185. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Liu, Y.; Liu, S.; Zhu, X.; Wu, L.; Zhu, Y.; Zhao, D.; Xu, X.; Chemparathy, A.; Wang, H.; et al. Nested epistasis enhancer networks for robust genome regulation. Science 2022, 377, 1077–1085. [Google Scholar] [CrossRef]
- Madsen, J.G.S.; Madsen, M.S.; Rauch, A.; Traynor, S.; Van Hauwaert, E.L.; Haakonsson, A.K.; Javierre, B.M.; Hyldahl, M.; Fraser, P.; Mandrup, S. Highly interconnected enhancer communities control lineage-determining genes in human mesenchymal stem cells. Nat. Genet. 2020, 52, 1227–1238. [Google Scholar] [CrossRef] [PubMed]
- Robson, M.I.; de Las Heras, J.I.; Czapiewski, R.; Le Thanh, P.; Booth, D.G.; Kelly, D.A.; Webb, S.; Kerr, A.R.W.; Schirmer, E.C. Tissue-Specific Gene Repositioning by Muscle Nuclear Membrane Proteins Enhances Repression of Critical Developmental Genes during Myogenesis. Mol. Cell 2016, 62, 834–847. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Luperchio, T.R.; Wong, X.; Doan, E.B.; Byrd, A.T.; Roy Choudhury, K.; Reddy, K.L.; Krangel, M.S. A Lamina-Associated Domain Border Governs Nuclear Lamina Interactions, Transcription, and Recombination of the Tcrb Locus. Cell Rep. 2018, 25, 1729–1740.e1726. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.L.; Poleshko, A.; Epstein, J.A. The nuclear periphery is a scaffold for tissue-specific enhancers. Nucleic Acids Res. 2021, 49, 6181–6195. [Google Scholar] [CrossRef] [PubMed]
- Potolitsyna, E.; Hazell Pickering, S.; Germier, T.; Collas, P.; Briand, N. Long non-coding RNA HOTAIR regulates cytoskeleton remodeling and lipid storage capacity during adipogenesis. Sci. Rep. 2022, 12, 10157. [Google Scholar] [CrossRef] [PubMed]
- Oldenburg, A.; Briand, N.; Sorensen, A.L.; Cahyani, I.; Shah, A.; Moskaug, J.O.; Collas, P. A lipodystrophy-causing lamin A mutant alters conformation and epigenetic regulation of the anti-adipogenic MIR335 locus. J. Cell Biol. 2017, 216, 2731–2743. [Google Scholar] [CrossRef]
- Iannuccelli, E.; Mompart, F.; Gellin, J.; Lahbib-Mansais, Y.; Yerle, M.; Boudier, T. NEMO: A tool for analyzing gene and chromosome territory distributions from 3D-FISH experiments. Bioinformatics 2010, 26, 696–697. [Google Scholar] [CrossRef]
- Paulsen, J.; Liyakat Ali, T.M.; Nekrasov, M.; Delbarre, E.; Baudement, M.O.; Kurscheid, S.; Tremethick, D.; Collas, P. Long-range interactions between topologically associating domains shape the four-dimensional genome during differentiation. Nat. Genet. 2019, 51, 835–843. [Google Scholar] [CrossRef]
- Zerbino, D.R.; Johnson, N.; Juettemann, T.; Wilder, S.P.; Flicek, P. WiggleTools: Parallel processing of large collections of genome-wide datasets for visualization and statistical analysis. Bioinformatics 2014, 30, 1008–1009. [Google Scholar] [CrossRef]
- Ramírez, F.; Dündar, F.; Diehl, S.; Grüning, B.A.; Manke, T. deepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014, 42, W187–W191. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nussbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef] [PubMed]
- Ernst, J.; Kellis, M. ChromHMM: Automating chromatin-state discovery and characterization. Nat. Methods 2012, 9, 215–216. [Google Scholar] [CrossRef]
- Miklos, I.; Meyer, I.M. A linear memory algorithm for Baum-Welch training. BMC Bioinform. 2005, 6, 231. [Google Scholar] [CrossRef]
- Thorvaldsdottir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Divoux, A.; Sandor, K.; Bojcsuk, D.; Yi, F.; Hopf, M.E.; Smith, J.S.; Balint, B.L.; Osborne, T.F.; Smith, S.R. Fat Distribution in Women Is Associated With Depot-Specific Transcriptomic Signatures and Chromatin Structure. J. Endocr. Soc. 2020, 4, bvaa042. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Neph, S.; Kuehn, M.S.; Reynolds, A.P.; Haugen, E.; Thurman, R.E.; Johnson, A.K.; Rynes, E.; Maurano, M.T.; Vierstra, J.; Thomas, S. BEDOPS: High-performance genomic feature operations. Bioinformatics 2012, 28, 1919–1920. [Google Scholar] [CrossRef]
- Khan, A.; Mathelier, A. Intervene: A tool for intersection and visualization of multiple gene or genomic region sets. BMC Bioinform. 2017, 18, 287. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Huang, X.; Ebert, D.; Mills, C.; Guo, X.; Thomas, P.D. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 2019, 14, 703–721. [Google Scholar] [CrossRef] [PubMed]
- Turgay, Y.; Eibauer, M.; Goldman, A.E.; Shimi, T.; Khayat, M.; Ben-Harush, K.; Dubrovsky-Gaupp, A.; Sapra, K.T.; Goldman, R.D.; Medalia, O. The molecular architecture of lamins in somatic cells. Nature 2017, 543, 261–264. [Google Scholar] [CrossRef] [PubMed]
- Forsberg, F.; Brunet, A.; Ali, T.M.L.; Collas, P. Interplay of lamin A and lamin B LADs on the radial positioning of chromatin. Nucleus 2019, 10, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Therizols, P.; Illingworth, R.S.; Courilleau, C.; Boyle, S.; Wood, A.J.; Bickmore, W.A. Chromatin decondensation is sufficient to alter nuclear organization in embryonic stem cells. Science 2014, 346, 1238–1242. [Google Scholar] [CrossRef] [PubMed]
- Fishilevich, S.; Nudel, R.; Rappaport, N.; Hadar, R.; Plaschkes, I.; Iny Stein, T.; Rosen, N.; Kohn, A.; Twik, M.; Safran, M.; et al. GeneHancer: Genome-wide integration of enhancers and target genes in GeneCards. Database 2017, 2017, bax028. [Google Scholar] [CrossRef] [PubMed]
- Ernst, J.; Kellis, M. Large-scale imputation of epigenomic datasets for systematic annotation of diverse human tissues. Nat. Biotechnol. 2015, 33, 364–376. [Google Scholar] [CrossRef]
- Bronshtein, I.; Kepten, E.; Kanter, I.; Berezin, S.; Lindner, M.; Redwood, A.B.; Mai, S.; Gonzalo, S.; Foisner, R.; Shav-Tal, Y.; et al. Loss of lamin A function increases chromatin dynamics in the nuclear interior. Nat. Commun. 2015, 6, 8044. [Google Scholar] [CrossRef]
- Cesarini, E.; Mozzetta, C.; Marullo, F.; Gregoretti, F.; Gargiulo, A.; Columbaro, M.; Cortesi, A.; Antonelli, L.; Di Pelino, S.; Squarzoni, S.; et al. Lamin A/C sustains PcG protein architecture, maintaining transcriptional repression at target genes. J. Cell Biol. 2015, 211, 533–551. [Google Scholar] [CrossRef]
- Solovei, I.; Wang, A.S.; Thanisch, K.; Schmidt, C.S.; Krebs, S.; Zwerger, M.; Cohen, T.V.; Devys, D.; Foisner, R.; Peichl, L.; et al. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 2013, 152, 584–598. [Google Scholar] [CrossRef]
- Lund, E.; Oldenburg, A.; Delbarre, E.; Freberg, C.; Duband-Goulet, I.; Eskeland, R.; Buendia, B.; Collas, P. Lamin A/C-promoter interactions specify chromatin state-dependent transcription outcomes. Genome Res. 2013, 23, 1580–1589. [Google Scholar] [CrossRef]
- van Schaik, T.; Manzo, S.G.; Vouzas, A.E.; Liu, N.Q.; Teunissen, H.; de Wit, E.; Gilbert, D.M.; van Steensel, B. Dynamic chromosomal interactions and control of heterochromatin positioning by Ki-67. EMBO Rep. 2022, 23, e55782. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Madsen-Østerbye, J.; Abdelhalim, M.; Pickering, S.H.; Collas, P. Gene Regulatory Interactions at Lamina-Associated Domains. Genes 2023, 14, 334. https://doi.org/10.3390/genes14020334
Madsen-Østerbye J, Abdelhalim M, Pickering SH, Collas P. Gene Regulatory Interactions at Lamina-Associated Domains. Genes. 2023; 14(2):334. https://doi.org/10.3390/genes14020334
Chicago/Turabian StyleMadsen-Østerbye, Julia, Mohamed Abdelhalim, Sarah Hazell Pickering, and Philippe Collas. 2023. "Gene Regulatory Interactions at Lamina-Associated Domains" Genes 14, no. 2: 334. https://doi.org/10.3390/genes14020334
APA StyleMadsen-Østerbye, J., Abdelhalim, M., Pickering, S. H., & Collas, P. (2023). Gene Regulatory Interactions at Lamina-Associated Domains. Genes, 14(2), 334. https://doi.org/10.3390/genes14020334







