ABCB1 Amplicon Contains Cyclic AMP Response Element-Driven TRIP6 Gene in Taxane-Resistant MCF-7 Breast Cancer Sublines
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Culture
2.3. Collection and Processing of Breast Cancer Tissue Samples
2.4. Isolation of Nucleic Acids and Proteins
2.5. Quantitative Reverse Transcription PCR (qRT-PCR)
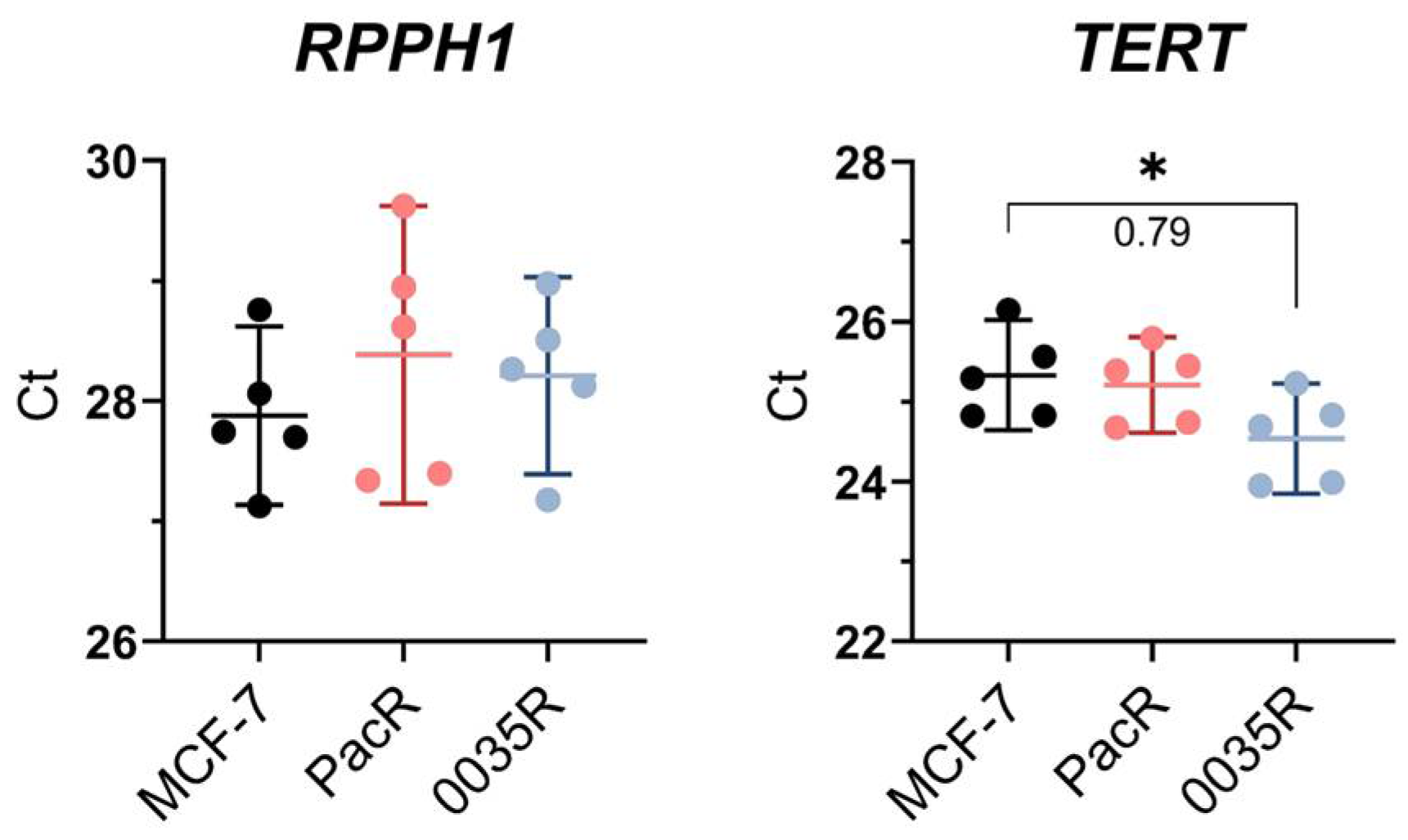
2.6. Assessment of Gene Copy Number
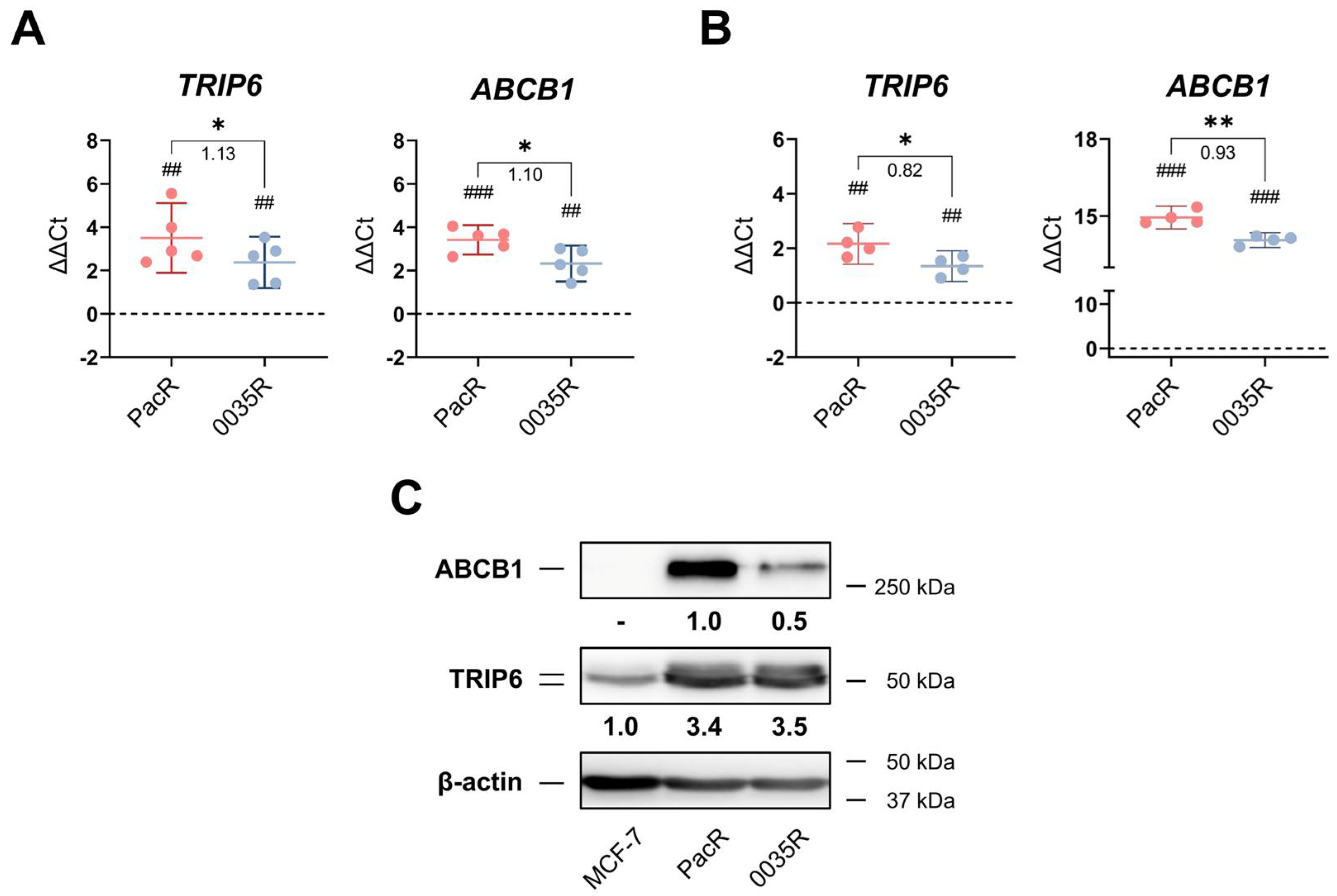
2.7. Western Blot Analysis
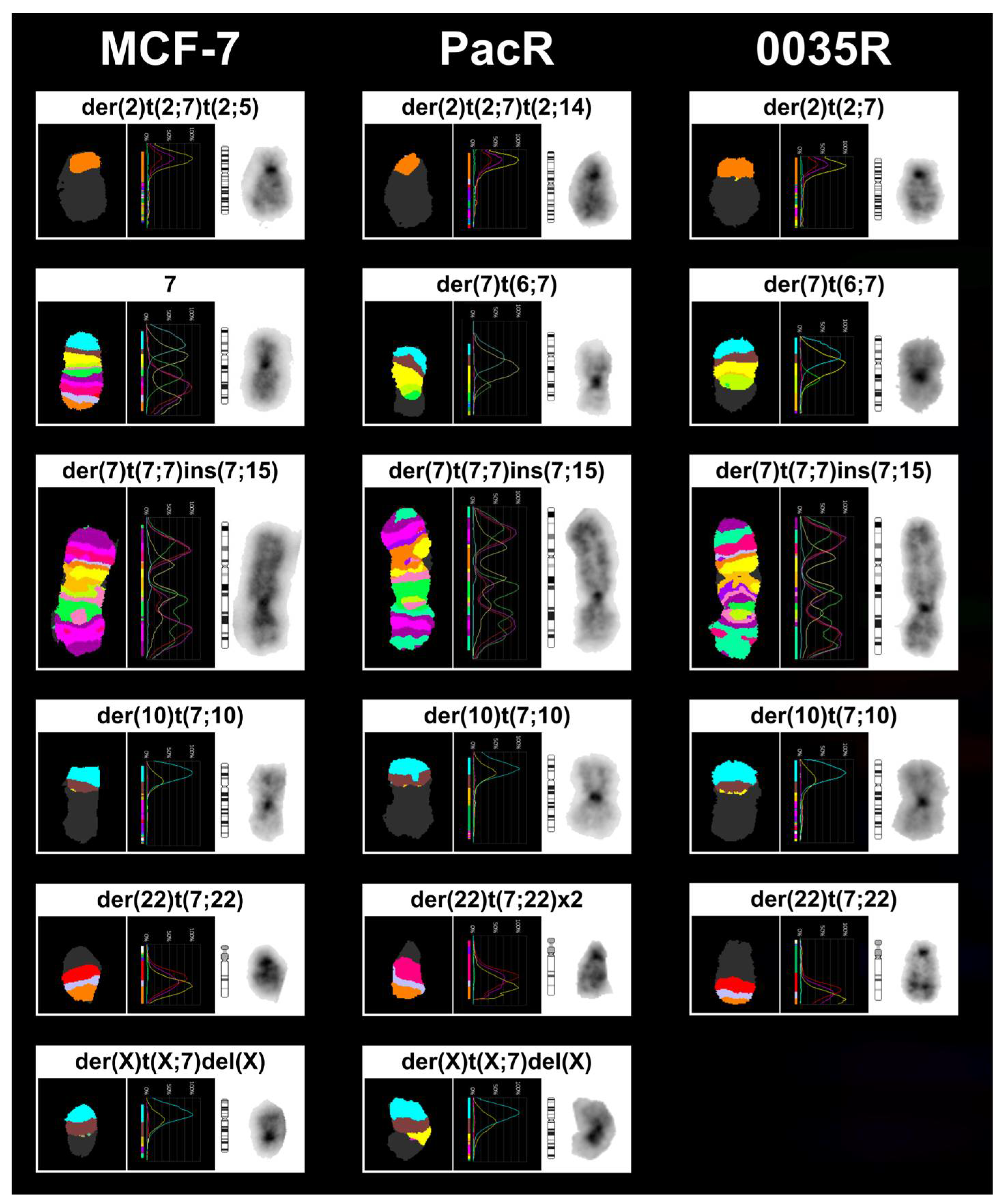
2.8. Fluorescence In Situ Hybridization (FISH) Analysis
2.9. Cloning
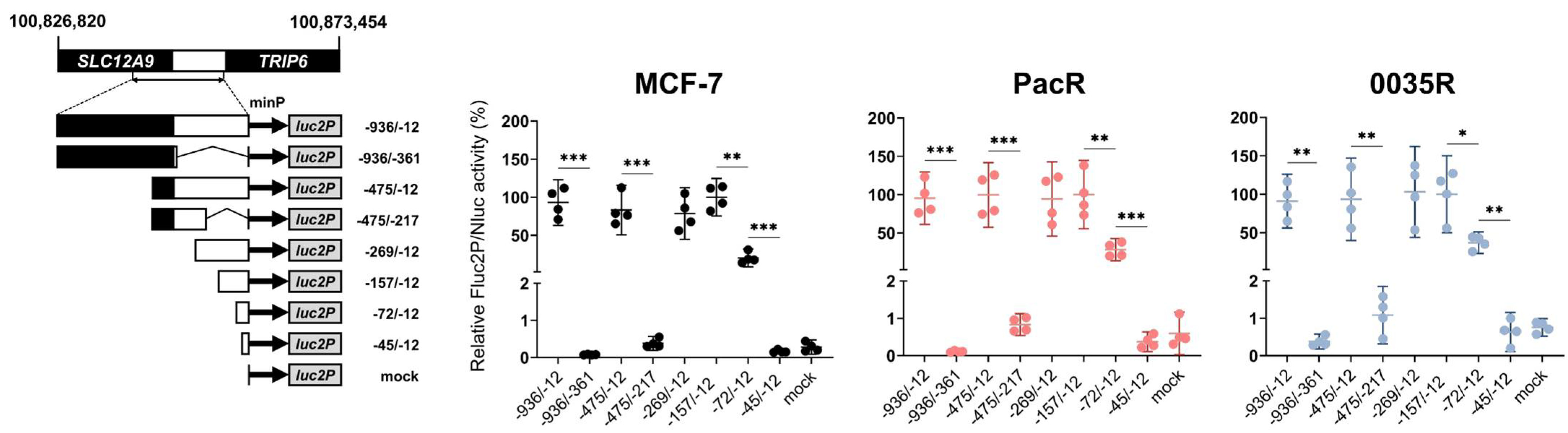
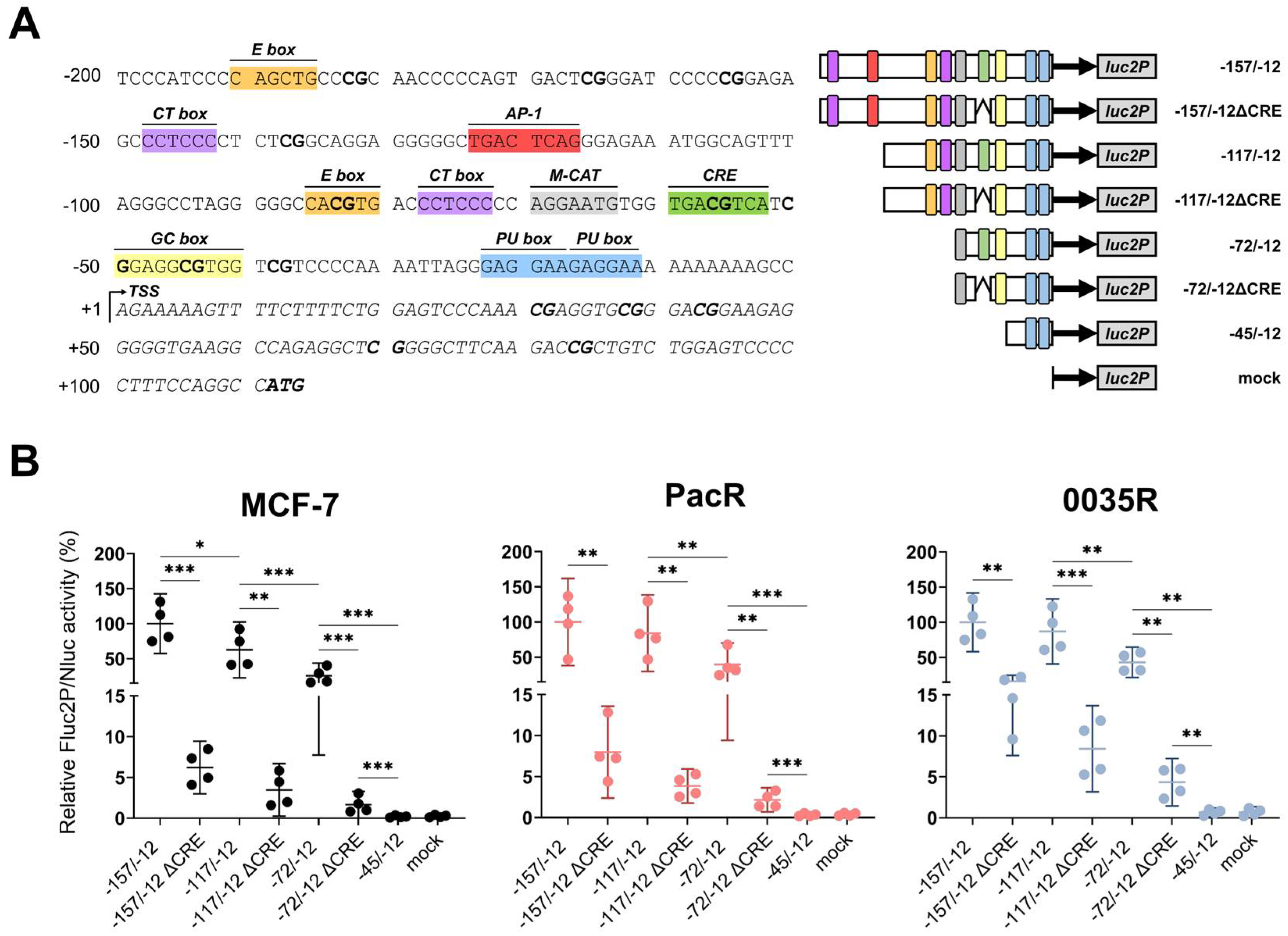
2.10. Dual-Luciferase Reporter Assay
2.11. DNA Methylation Profiling
2.12. Bisulfite Sequencing
2.13. Statistical Analysis
3. Results
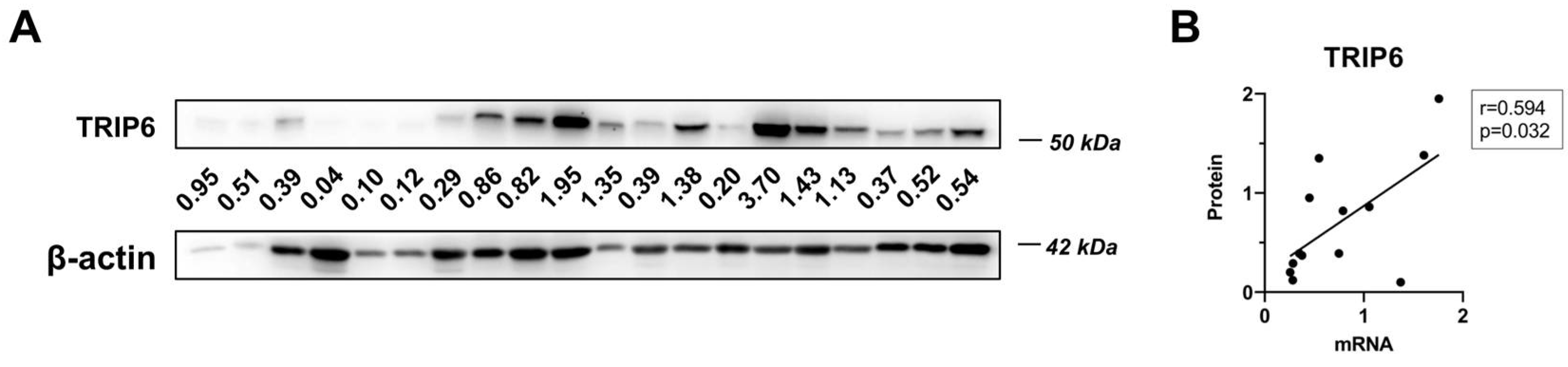
3.1. TRIP6 as Well as ABCB1 Are Overexpressed in Taxane-Resistant MCF-7 Sublines
3.2. A Few TRIP6 Loci Pre-Exists in MCF-7 Cell Line
3.3. Chromosome 7 Is Rearranged in Taxane-Resistant MCF-7 Sublines
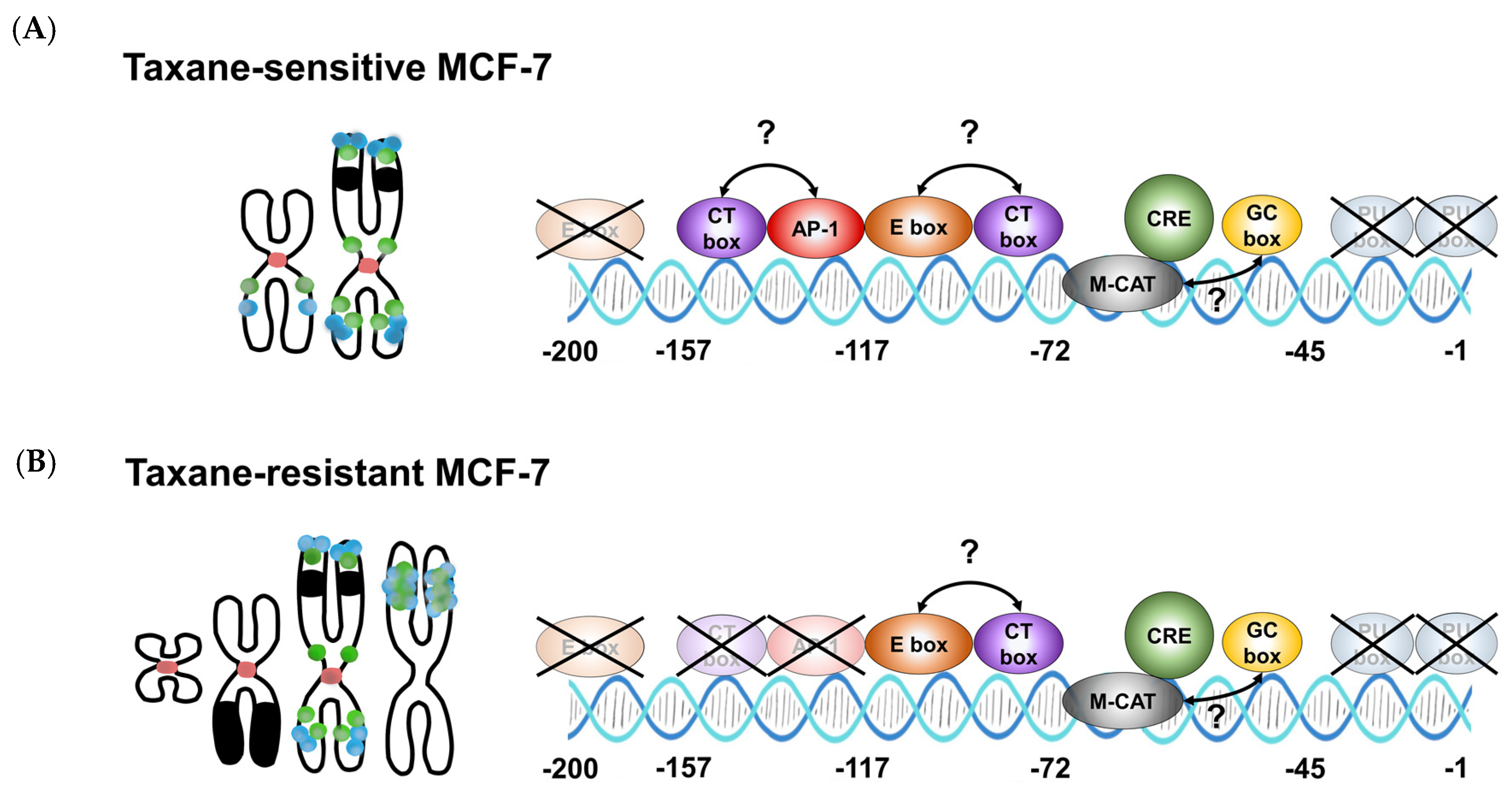
3.4. TRIP6 Expression Is Regulated by Cyclic AMP Response Element (CRE)
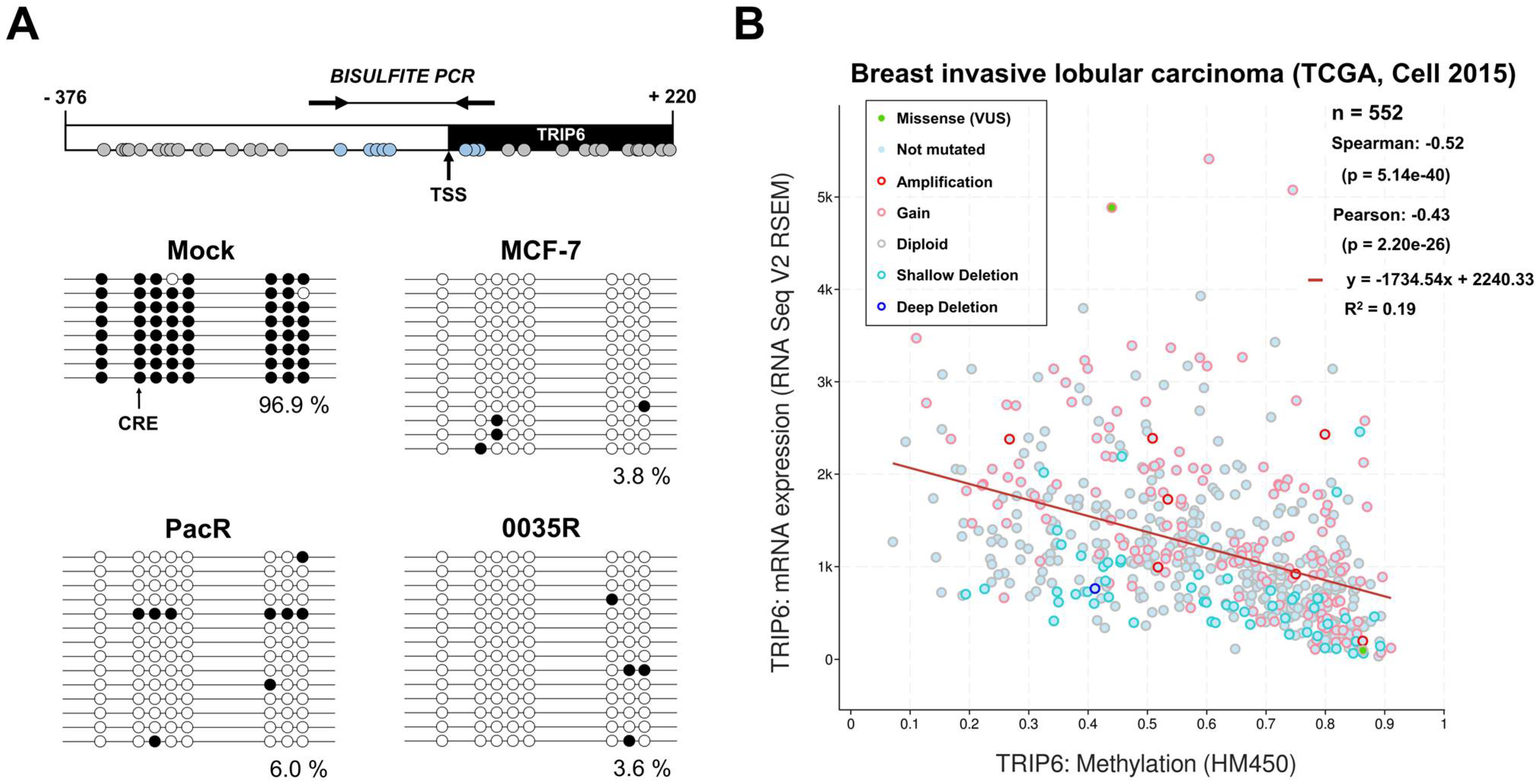
3.5. TRIP6 Proximal Promoter Is Hypomethylated in Taxane-Resistant MCF-7 Sublines
3.6. Associations of TRIP6 mRNA Level with Clinicopathological Features of Breast Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Yi, J.; Beckerle, M.C. The human TRIP6 gene encodes a LIM domain protein and maps to chromosome 7q22, a region associated with tumorigenesis. Genomics 1998, 49, 314–316. [Google Scholar] [CrossRef]
- Wang, Y.; Gilmore, T.D. LIM domain protein Trip6 has a conserved nuclear export signal, nuclear targeting sequences, and multiple transactivation domains. Biochim. Biophys. Acta 2001, 1538, 260–272. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, M.Q.; Badmalia, M.D.; Patel, T.R. Bioinformatic Analysis of Structure and Function of LIM Domains of Human Zyxin Family Proteins. Int. J. Mol. Sci. 2021, 22, 2647. [Google Scholar] [CrossRef] [PubMed]
- Lin, V.T.; Lin, F.T. TRIP6: An adaptor protein that regulates cell motility, antiapoptotic signaling and transcriptional activity. Cell Signal. 2011, 23, 1691–1697. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Lai, Y.J.; Lin, W.C.; Lin, F.T. TRIP6 enhances lysophosphatidic acid-induced cell migration by interacting with the lysophosphatidic acid 2 receptor. J. Biol. Chem. 2004, 279, 10459–10468. [Google Scholar] [CrossRef] [PubMed]
- Chastre, E.; Abdessamad, M.; Kruglov, A.; Bruyneel, E.; Bracke, M.; Di Gioia, Y.; Beckerle, M.C.; van Roy, F.; Kotelevets, L. TRIP6, a novel molecular partner of the MAGI-1 scaffolding molecule, promotes invasiveness. FASEB J. 2009, 23, 916–928. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Mana-Capelli, S.; Paramasivam, M.; Dasgupta, I.; Cirka, H.; Billiar, K.; McCollum, D. TRIP6 inhibits Hippo signaling in response to tension at adherens junctions. EMBO Rep. 2018, 19, 337–350. [Google Scholar] [CrossRef] [PubMed]
- Venkatramanan, S.; Ibar, C.; Irvine, K.D. TRIP6 is required for tension at adherens junctions. J. Cell Sci. 2021, 134, jcs247866. [Google Scholar] [CrossRef]
- Kassel, O.; Schneider, S.; Heilbock, C.; Litfin, M.; Göttlicher, M.; Herrlich, P. A nuclear isoform of the focal adhesion LIM-domain protein Trip6 integrates activating and repressing signals at AP-1- and NF-kappaB-regulated promoters. Genes Dev. 2004, 18, 2518–2528. [Google Scholar] [CrossRef]
- Kemler, D.; Dahley, O.; Roßwag, S.; Litfin, M.; Kassel, O. The LIM domain protein nTRIP6 acts as a co-repressor for the transcription factor MEF2C in myoblasts. Sci. Rep. 2016, 6, 27746. [Google Scholar] [CrossRef]
- Abbariki, T.N.; Gonda, Z.; Kemler, D.; Urbanek, P.; Wagner, T.; Litfin, M.; Wang, Z.Q.; Herrlich, P.; Kassel, O. The LIM domain protein nTRIP6 modulates the dynamics of myogenic differentiation. Sci. Rep. 2021, 11, 12904. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.; Haenold, R.; Urbánek, P.; Frappart, L.; Monajembashi, S.; Grigaravicius, P.; Nagel, S.; Min, W.K.; Tapias, A.; Kassel, O.; et al. TRIP6 functions in brain ciliogenesis. Nat. Commun. 2021, 12, 5887. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Kloeker, S.; Jensen, C.C.; Bockholt, S.; Honda, H.; Hirai, H.; Beckerle, M.C. Members of the Zyxin family of LIM proteins interact with members of the p130Cas family of signal transducers. J. Biol. Chem. 2002, 277, 9580–9589. [Google Scholar] [CrossRef]
- Takizawa, N.; Smith, T.C.; Nebl, T.; Crowley, J.L.; Palmieri, S.J.; Lifshitz, L.M.; Ehrhardt, A.G.; Hoffman, L.M.; Beckerle, M.C.; Luna, E.J. Supervillin modulation of focal adhesions involving TRIP6/ZRP-1. J. Cell Biol. 2006, 174, 447–458. [Google Scholar] [CrossRef]
- Lai, Y.J.; Lin, V.T.; Zheng, Y.; Benveniste, E.N.; Lin, F.T. The adaptor protein TRIP6 antagonizes Fas-induced apoptosis but promotes its effect on cell migration. Mol. Cell. Biol. 2010, 30, 5582–5596. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, X.M.; Wang, J.R.; Li, Y.; Ye, W.L.; Wang, Y.; Liu, Y.X.; Deng, Z.Y.; Gan, W.J.; Wu, H. TRIP6 promotes inflammatory damage via the activation of TRAF6 signaling in a murine model of DSS-induced colitis. J. Inflamm. 2022, 19, 1. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, S.A.; Loayza, D. LIM-domain proteins TRIP6 and LPP associate with shelterin to mediate telomere protection. Aging 2010, 2, 432–444. [Google Scholar] [CrossRef] [PubMed]
- Thul, P.J.; Åkesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Blal, H.A.; Alm, T.; Asplund, A.; Björk, L.; Breckels, L.M.; et al. A subcellular map of the human proteome. Science 2017, 356, eaal3321. [Google Scholar] [CrossRef] [PubMed]
- Gambardella, G.; Viscido, G.; Tumaini, B.; Isacchi, A.; Bosotti, R.; di Bernardo, D. A single-cell analysis of breast cancer cell lines to study tumour heterogeneity and drug response. Nat. Commun. 2022, 13, 1714. [Google Scholar] [CrossRef]
- Zhao, X.; Jiang, C.; Xu, R.; Liu, Q.; Liu, G.; Zhang, Y. TRIP6 enhances stemness property of breast cancer cells through activation of Wnt/β-catenin. Cancer Cell Int. 2020, 20, 51. [Google Scholar] [CrossRef]
- Wang, J.; Li, J.; Yang, J.; Zhang, L.; Gao, S.; Jiao, F.; Yi, M.; Xu, J. MicroRNA-138-5p regulates neural stem cell proliferation and differentiation in vitro by targeting TRIP6 expression. Mol. Med. Rep. 2017, 16, 7261–7266. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Shao, R.; Li, M.; Yan, Q.; Hu, H. MiR-485-3p modulates neural stem cell differentiation and proliferation via regulating TRIP6 expression. J. Cell. Mol. Med. 2020, 24, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Ling, Y.; Cao, C.; Li, S.; Qiu, M.; Shen, G.; Chen, Z.; Yao, F.; Chen, W. TRIP6, as a target of miR-7, regulates the proliferation and metastasis of colorectal cancer cells. Biochem. Biophys. Res. Commun. 2019, 514, 231–238. [Google Scholar] [CrossRef]
- Gou, H.; Liang, J.Q.; Zhang, L.; Chen, H.; Zhang, Y.; Li, R.; Wang, X.; Ji, J.; Tong, J.H.; To, K.F.; et al. TTPAL Promotes Colorectal Tumorigenesis by Stabilizing TRIP6 to Activate Wnt/β-Catenin Signaling. Cancer Res. 2019, 13, 3332–3346. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Turner, K.M.; Yeo, S.K.; Holm, T.M.; Shaughnessy, E.; Guan, J.L. Heterogeneity within molecular subtypes of breast cancer. Am. J. Physiol. Cell Physiol. 2021, 321, C343–C354. [Google Scholar] [CrossRef]
- Burstein, H.J.; Curigliano, G.; Thürlimann, B.; Weber, W.P.; Poortmans, P.; Regan, M.M.; Senn, H.J.; Winer, E.P.; Gnant, M.; Panelists of the St Gallen Consensus Conference. Customizing local and systemic therapies for women with early breast cancer: The St. Gallen International Consensus Guidelines for treatment of early breast cancer 2021. Ann. Oncol. 2021, 32, 1216–1235. [Google Scholar] [CrossRef]
- Lashen, A.; Toss, M.S.; Fadhil, W.; Oni, G.; Madhusudan, S.; Rakha, E. Evaluation Oncotype DX® 21-Gene Recurrence Score and Clinicopathological Parameters: A single institutional experience. Histopathology, 2023; accepted. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef]
- Pereira, B.; Chin, S.F.; Rueda, O.M.; Vollan, H.K.; Provenzano, E.; Bardwell, H.A.; Pugh, M.; Jones, L.; Russell, R.; Sammut, S.J.; et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 2016, 7, 11479. [Google Scholar] [CrossRef]
- Ojima, I.; Fumero-Oderda, C.L.; Kuduk, S.D.; Ma, Z.; Kirikae, F.; Kirikae, T. Structure-activity relationship study of taxoids for their ability to activate murine macrophages as well as inhibit the growth of macrophage-like cells. Bioorg. Med. Chem. 2003, 11, 2867–2888. [Google Scholar] [CrossRef]
- Calcagno, A.M.; Ambudkar, S.V. Molecular mechanisms of drug resistance in single-step and multi-step drug-selected cancer cells. Methods Mol. Biol. 2010, 596, 77–93. [Google Scholar] [CrossRef] [PubMed]
- Němcová-Fürstová, V.; Kopperová, D.; Balušíková, K.; Ehrlichová, M.; Brynychová, V.; Václavíková, R.; Daniel, P.; Souček, P.; Kovář, J. Characterization of acquired paclitaxel resistance of breast cancer cells and involvement of ABC transporters. Toxicol. Appl. Pharmacol. 2016, 310, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Jelínek, M.; Balušíková, K.; Daniel, P.; Němcová-Fürstová, V.; Kirubakaran, P.; Jaček, M.; Wei, L.; Wang, X.; Vondrášek, J.; Ojima, I.; et al. Substituents at the C3′ and C3′N positions are critical for taxanes to overcome acquired resistance of cancer cells to paclitaxel. Toxicol. Appl. Pharmacol. 2018, 347, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Brynychová, V.; Hlaváč, V.; Ehrlichová, M.; Václavíková, R.; Pecha, V.; Trnková, M.; Wald, M.; Mrhalová, M.; Kubáčková, K.; Pikus, T.; et al. Importance of transcript levels of caspase-2 isoforms S and L for breast carcinoma progression. Future Oncol. 2013, 9, 427–438. [Google Scholar] [CrossRef]
- Hubackova, M.; Vaclavikova, R.; Ehrlichova, M.; Mrhalova, M.; Kodet, R.; Kubackova, K.; Vrána, D.; Gut, I.; Soucek, P. Association of superoxide dismutases and NAD(P)H quinone oxidoreductases with prognosis of patients with breast carcinomas. Int. J. Cancer 2012, 130, 338–348. [Google Scholar] [CrossRef]
- Tavassoli, F.A.; Devilee, P. International Agency for Research on Cancer; World Health Organization. In Pathology and Genetics of Tumours of the Breast and Female Genital Organs; IARC Press: Lyon, France, 2003; 432p. [Google Scholar]
- Cheang, M.C.; Chia, S.K.; Voduc, D.; Gao, D.; Leung, S.; Snider, J.; Watson, M.; Davies, S.; Bernard, P.S.; Parker, J.S.; et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J. Natl. Cancer Inst. 2009, 101, 736–750. [Google Scholar] [CrossRef]
- Goldhirsch, A.; Winer, E.P.; Coates, A.S.; Gelber, R.D.; Piccart-Gebhart, M.; Thürlimann, B.; Senn, H.J.; Panel members. Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann. Oncol. 2013, 24, 2206–2223. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Hlaváč, V.; Brynychová, V.; Václavíková, R.; Ehrlichová, M.; Vrána, D.; Pecha, V.; Koževnikovová, R.; Trnková, M.; Gatěk, J.; Kopperová, D.; et al. The expression profile of ATP-binding cassette transporter genes in breast carcinoma. Pharmacogenomics 2013, 14, 515–529. [Google Scholar] [CrossRef]
- Elsnerova, K.; Mohelnikova-Duchonova, B.; Cerovska, E.; Ehrlichova, M.; Gut, I.; Rob, L.; Skapa, P.; Hruda, M.; Bartakova, A.; Bouda, J.; et al. Gene expression of membrane transporters: Importance for prognosis and progression of ovarian carcinoma. Oncol. Rep. 2016, 35, 2159–2170. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Daniel, P.; Halada, P.; Jelínek, M.; Balušíková, K.; Kovář, J. Differentially Expressed Mitochondrial Proteins in Human MCF7 Breast Cancer Cells Resistant to Paclitaxel. Int. J. Mol. Sci. 2019, 20, 2986. [Google Scholar] [CrossRef] [PubMed]
- McGowan-Jordan, J.; Hastings, R.J.; Moore, S.; International Standing Committee on Human Cytogenomic nomenclature. ISCN 2020: An International System for Human Cytogenomic Nomenclature; Karger: Basel, Switzerland, 2020; 170p. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Touleimat, N.; Tost, J. Complete pipeline for Infinium® Human Methylation 450K BeadChip data processing using subset quantile normalization for accurate DNA methylation estimation. Epigenomics 2012, 4, 325–341. [Google Scholar] [CrossRef] [PubMed]
- Fleischer, T.; Frigessi, A.; Johnson, K.C.; Edvardsen, H.; Touleimat, N.; Klajic, J.; Riis, M.L.; Haakensen, V.D.; Wärnberg, F.; Naume, B.; et al. Genome-wide DNA methylation profiles in progression to in situ and invasive carcinoma of the breast with impact on gene transcription and prognosis. Genome Biol. 2014, 15, 435. [Google Scholar] [CrossRef]
- Fortin, J.P.; Triche, T.J., Jr.; Hansen, K.D. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics 2017, 33, 558–560. [Google Scholar] [CrossRef]
- Maksimovic, J.; Gordon, L.; Oshlack, A. SWAN: Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol. 2012, 13, R44. [Google Scholar] [CrossRef]
- Pidsley, R.; Zotenko, E.; Peters, T.J.; Lawrence, M.G.; Risbridger, G.P.; Molloy, P.; Van Djik, S.; Muhlhausler, B.; Stirzaker, C.; Clark, S.J. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 2016, 17, 208. [Google Scholar] [CrossRef]
- Kumaki, Y.; Oda, M.; Okano, M. QUMA: Quantification tool for methylation analysis. Nucleic Acids Res. 2008, 36, W170–W175. [Google Scholar] [CrossRef]
- Weissgerber, T.L.; Milic, N.M.; Winham, S.J.; Garovic, V.D. Beyond bar and line graphs: Time for a new data presentation paradigm. PLoS Biol. 2015, 13, e1002128. [Google Scholar] [CrossRef]
- Comşa, Ş.; Cîmpean, A.M.; Raica, M. The Story of MCF-7 Breast Cancer Cell Line: 40 years of Experience in Research. Anticancer Res. 2015, 35, 3147–3154. [Google Scholar]
- Fornes, O.; Castro-Mondragon, J.A.; Khan, A.; van der Lee, R.; Zhang, X.; Richmond, P.A.; Modi, B.P.; Correard, S.; Gheorghe, M.; Baranašić, D.; et al. JASPAR 2020: Update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020, 48, D87–D92. [Google Scholar] [CrossRef]
- Lutz, W.; Schwab, M. In vivo regulation of single copy and amplified N-myc in human neuroblastoma cells. Oncogene 1997, 15, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Ji, C.; Casinghino, S.; McCarthy, T.L.; Centrella, M. Multiple and essential Sp1 binding sites in the promoter for transforming growth factor-β type I receptor. J. Biol. Chem. 1997, 272, 21260–21267. [Google Scholar] [CrossRef] [PubMed]
- Mar, J.H.; Ordahl, C.P. M-CAT binding factor, a novel trans-acting factor governing muscle-specific transcription. Mol. Cell. Biol. 1990, 10, 4271–4283. [Google Scholar] [CrossRef] [PubMed]
- Iguchi-Ariga, S.M.; Schaffner, W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989, 3, 612–619. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.K.; Wang, Y.; Murphy, K.; Yi, J.; Beckerle, M.C.; Gilmore, T.D. LIM domain-containing protein trip6 can act as a coactivator for the v-Rel transcription factor. Gene Expr. 1999, 8, 207–217. [Google Scholar] [PubMed]
- Zhu, L.; Xu, X.; Tang, Y.; Zhu, X. TRIP6 functions as a potential oncogene and facilitated proliferation and metastasis of gastric cancer. Biologics 2019, 13, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Pavlíková, N.; Bartoňová, I.; Balušíková, K.; Kopperová, D.; Halada, P.; Kovář, J. Differentially expressed proteins in human MCF-7 breast cancer cells sensitive and resistant to paclitaxel. Exp. Cell Res. 2015, 333, 1–10. [Google Scholar] [CrossRef]
- Wang, Y.; Dong, L.; Liu, Y. Targeting Thyroid Receptor Interacting Protein 6 by MicroRNA-589-5p Inhibits Cell Proliferation, Migration, and Invasion in Endometrial Carcinoma. Cancer Biother. Radiopharm. 2019, 34, 529–536. [Google Scholar] [CrossRef]
- Heim, S.; Lage, H. Transcriptome analysis of different multidrug-resistant gastric carcinoma cells. In Vivo 2005, 19, 583–590. [Google Scholar]
- Genovese, I.; Ilari, A.; Assaraf, Y.G.; Fazi, F.; Colotti, G. Not only P-glycoprotein: Amplification of the ABCB1-containing chromosome region 7q21 confers multidrug resistance upon cancer cells by coordinated overexpression of an assortment of resistance-related proteins. Drug Resist. Updat. 2017, 32, 23–46. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Juric, D.; Francisco, B.; Yu, R.X.; Duran, G.E.; Chen, K.G.; Chen, X.; Sikic, B.I. Regional activation of chromosomal arm 7q with and without gene amplification in taxane-selected human ovarian cancer cell lines. Genes Chromosom. Cancer 2006, 45, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Lombard, A.P.; Lou, W.; Armstrong, C.M.; D’Abronzo, L.S.; Ning, S.; Evans, C.P.; Gao, A.C. Activation of the ABCB1 Amplicon in Docetaxel- and Cabazitaxel-Resistant Prostate Cancer Cells. Mol. Cancer. Ther. 2021, 20, 2061–2070. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Nagpal, G.; Kumar, V.; Usmani, S.S.; Agrawal, P.; Raghava, G.P.S. HumCFS: A database of fragile sites in human chromosomes. BMC Genom. 2019, 19, 985. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Odom, D.T.; Koo, S.H.; Conkright, M.D.; Canettieri, G.; Best, J.; Chen, H.; Jenner, R.; Herbolsheimer, E.; Jacobsen, E.; et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl. Acad. Sci. USA 2005, 102, 4459–4464. [Google Scholar] [CrossRef]
- Tinti, C.; Yang, C.; Seo, H.; Conti, B.; Kim, C.; Joh, T.M.; Kim, K.S. Structure/function relationship of the cAMP response element in tyrosine hydroxylase gene transcription. J. Biol. Chem. 1997, 272, 19158–19164. [Google Scholar] [CrossRef]
- Seborova, K.; Kloudova-Spalenkova, A.; Koucka, K.; Holy, P.; Ehrlichova, M.; Wang, C.; Ojima, I.; Voleska, I.; Daniel, P.; Balusikova, K.; et al. The Role of TRIP6, ABCC3 and CPS1 Expression in Resistance of Ovarian Cancer to Taxanes. Int. J. Mol. Sci. 2021, 23, 73. [Google Scholar] [CrossRef]
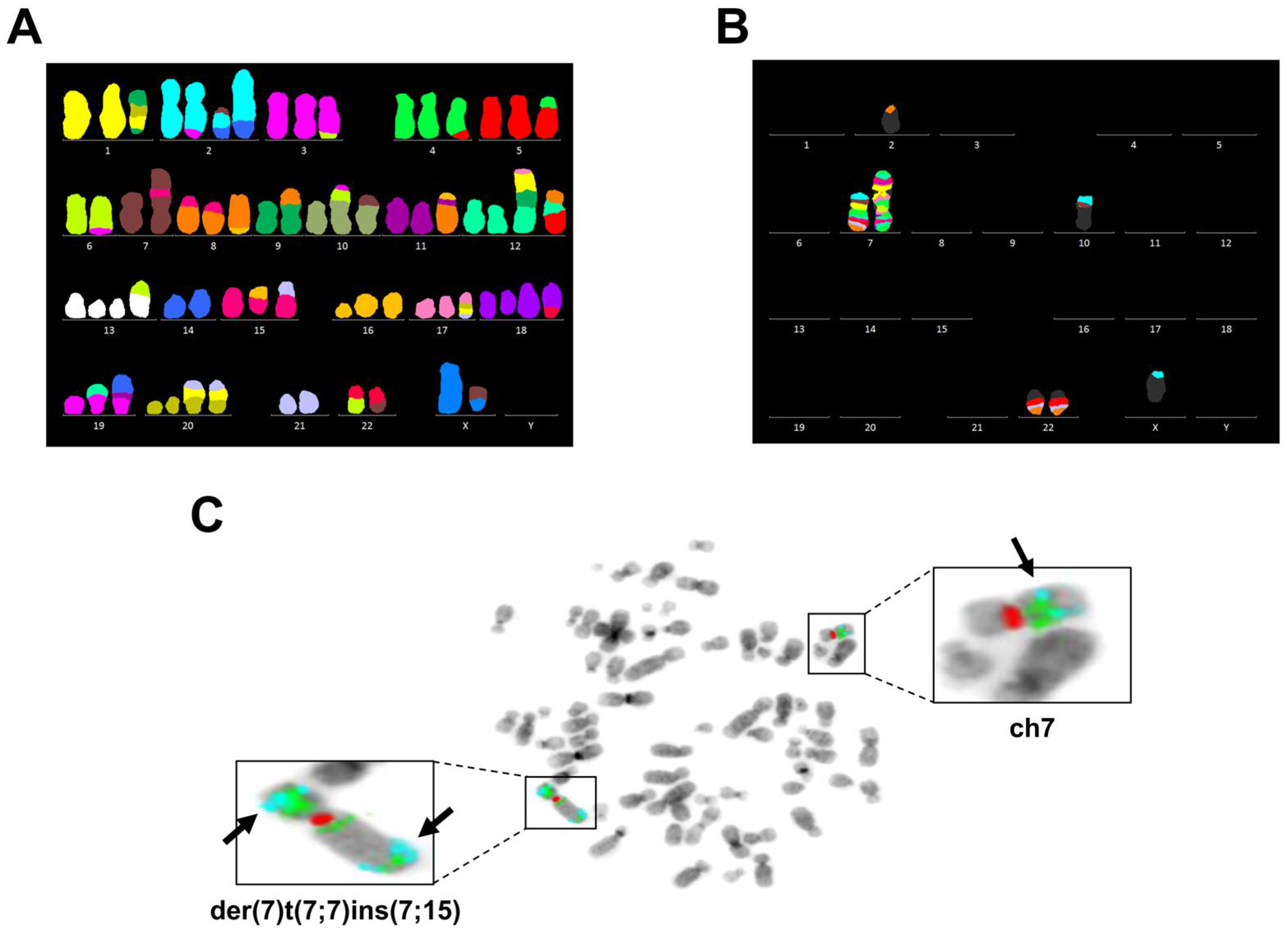
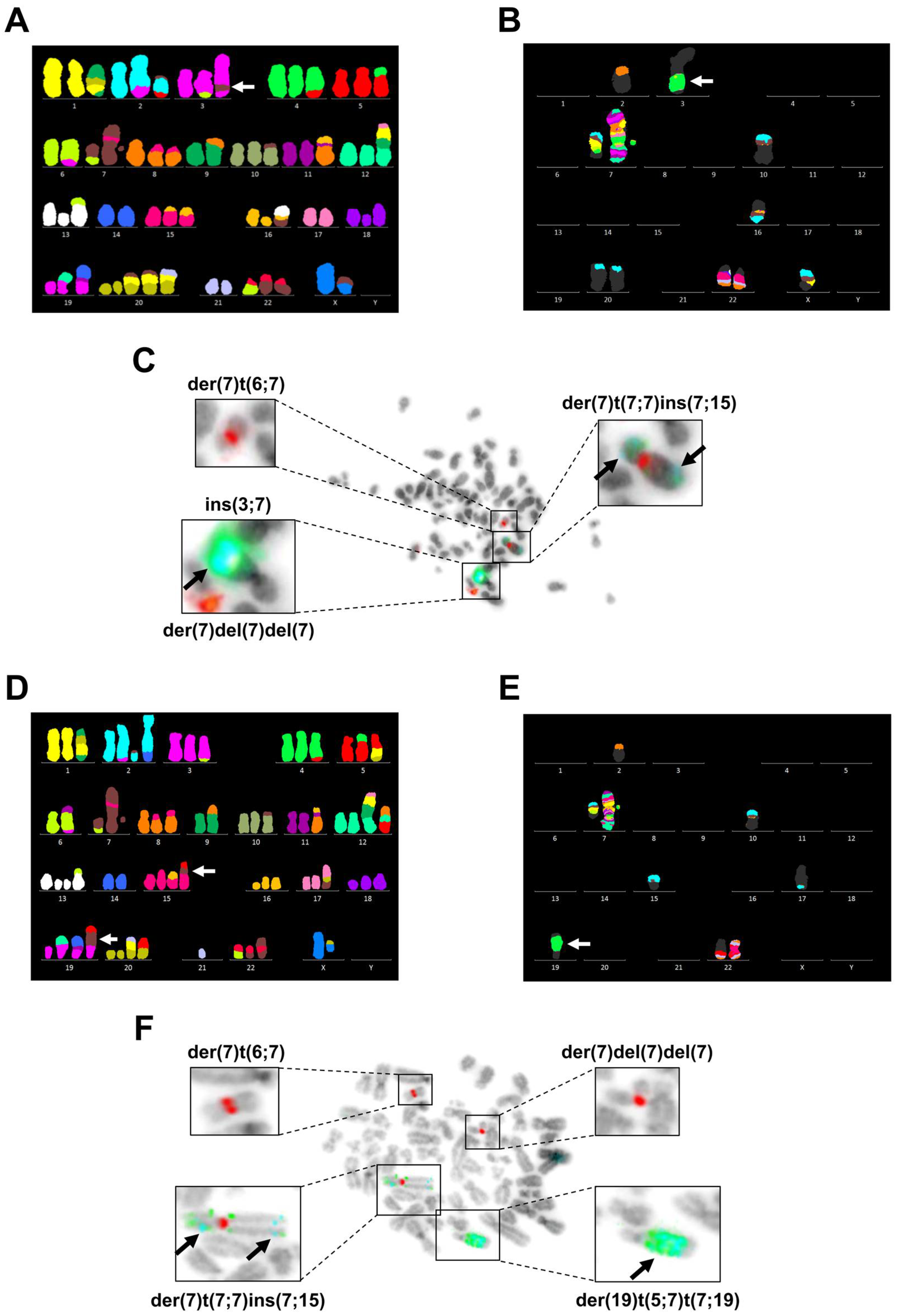
| Cell Line | Composite Karyotype |
|---|---|
| MCF-7 67~69 <3n> | X,-X,der(X)t(X;7)(p11.2;p15.3)del(X)(q21.3q28),der(1)t(9;20)(?;?)t(1;20)(p12;?)t(1;9)(q21;?),+2,der(2)t(2;3)(q?34;?),der(2)t(2;7)(p?;q32)t(2;14)(q36;q?),der(2)t(2;14)(q36;q?),der(3)del(3)(p?13p?26)t(3;6)(q27;q25),t(4;5)(q?31;p13),-6,der(6)t(3;6)(q27;q25),-7,der(7)t(7;7)(p15;?)ins(7;15)(p;?),der(8)t(8;15)(p11.2;q?)x2,der(8)t(8;16)(q?24;?),-9,der(9)t(8;9)(q?;p22),der(10)t(3;6)(?;?)t(6;10)(?;p14),der(10)t(7;10)(p21,p14),der(11)t(11;16)(p11.2;?)t(8;11)(q11.2;q13),+12,der(12)t(1;17)(?;?)t(1;9)(?;?)t(9;12)(?;p11.2),der(12)t(8;12)(?;p11.2)t(5;12)(?;q?21),+13,del(13)(q?22q?34)x2,der(13)t(6;13)(?;p11.2),-14,+15,der(15)t(15;16)(p11.2;q?)del(15)(q?22.3q?26),der(15)t(15;21)(p11.2;q?21),der(16)del(16)(p?13)del(16)(q?21),der(17)t(17;20)(q?24;?)t(1;20)(?;?)t(1;21)(?;?),+18,der(18)del(18)(p?11.2)del(18)(q?11.2),dup(18)(q?q?),der(18)t(18;22)(p11.2;q11.2),der(19)t(12;19)(q13;p13.3),der(19)t(11;14)(?;q?)t(11;19)(?;p12),+20,der(20)t(1;21)(?;q?)t(1;20)(?;p11.2)x2,-21,der(22)t(6;22)(?;q11.2),der(22)t(7;22)(q22;q11.2)x2[cp20] |
| Cell Subline | Composite Karyotype |
|---|---|
| PacR 67-68 <3n> | X,-X,der(X)t(X;7)(p11.2;p15.3)del(X)(q21.3q28),der(1)t(9;20)(?;?)t(1;20)(p12;?)t(1;9)(q21;?),+2,der(2)t(2;3)(q?34;?),der(2)t(2;7)(p?;q32)t(2;5)(q?;?),der(2)t(2;14)(q36;q?),der(3)del(3)(p?13p?26)t(3;6)(q27;q25),ins(3;7)(q?23;q11.2q22),t(4;5)(q?31;p13),-6,der(6)t(3;6)(q27;q25),der(7)t(6;7)(?;q11.2),der(7)t(7;7)(p15;?)ins(7;15)(p;?),der(7)del(7)(p12)del(7)(q11.1),der(8)t(8;15)(p11.2;q?)x2,-9,der(9)t(8;9)(q?;p22),der(10)t(7;10)(p21;p14),der(11)t(11;16)(p11.2;?)t(8;11)(q11.2;q13),+12,der(12)t(1;17)(?;?)t(1;9)(?;?)t(9;12)(?;p11.2),der(12)t(8;12)(?;p11.2)t(5;12)(?;q?21),del(13)(q?22q?34),der(13)t(6;13)(?;p11.2),-14,der(15)t(15;16)(p11.2;q?),der(15)t(15;16)(p11.2;q?)del(15)(q?22.3q?26),+16,der(16)del(16)(p?13)del(16)(q?21)x2,der(16)t(13;16)(q?12;p11.2)t(7;16)(p15.3;q?13),-17,der(18)del(18)(p?11.2)del(18)(q?11.2),dup(18)(q?q?),der(19)t(12;19)(q13;p13.3),der(19)t(11;14)(?;q?)t(11;19)(?;p12),+20x2,der(20)t(1;7)(?;p21)t(1;20)(?;p11.2)x2,der(20)t(1;21)(?;q?)t(1;20)(?;p11.2),-21,der(22)t(6;22)(?;q11.2),der(22)t(7;22)(q22;q11.2)x2[cp8] |
| 0035R 67~70 <3n> | X,-X,der(X)t(X;20)(p11.2;?)del(X)(q21.3q28),der(1)t(9;20)(?;?)t(1;20)(p12;?)t(1;9)(q21;?),+2,der(2)t(2;3)(q?34;?),der(2)t(2;7)(p?12;q32)del(2)(q36),der(2)t(2;14)(q36;q?),der(3)del(3)(p?13p?26)t(3;6)(q27;q25),t(4;5)(q?31;p13),der(5)t(1;5)(q?13)t(1;20)(?;q?),-6,der(6)t(6;11)(p?21;q?)t(3;6)(q27;q25),der(7)t(6;7)(?;q11.2),der(7)t(7;7)(p15;?)ins(7;15)(p;?),der(7)del(7)(p12)del(7)(q11.1),der(8)t(8;15)(p11.2;q?)x2,-9,der(9)t(8;9)(q?;p22),der(10)t(7;10)(p21;p14),der(11)t(11;16)(p11.2;?)t(8;11)(q11.2;q13),+12,der(12)t(1;17)(?;?)t(1;9)(?;?)t(9;12)(?;p11.2),der(12)t(8;12)(?;p11.2)t(5;12)(?;q?21),+13,del(13)(q?22q?34)x2,der(13)t(6;13)(?;p11.2),-14,+15,der(15)t(15;16)(p11.2;q?)del(15)(q?22.3q?26),der(15)t(5;7)(?;p22)t(7;15)(p22;p11.2),der(16)del(16)(p?13)del(16)(q?21),der(17)t(17;20)(q?24;?)t(7;20)(p22;?),der(18)del(18)(p?11.2)del(18)(q?11.2),dup(18)(q?q?),+19,der(19)t(12;19)(q13;p13.3),der(19)t(11;14)(?;q?)t(11;19)(?;p12),der(19)t(5;7)(?;q21)t(7;19)(q21;p13.3),+20,der(20)t(1;21)(?;q?)t(1;20)(?;p11.2),der(20)t(5;20)(?;p?12),-21,der(22)t(6;22)(?;q11.2),der(22)t(7;22)(q22;q11.2),der(22)t(7;22)(q22;p11.2)t(7;22)(q22;q11.2)[cp12] |
| Region * | MCF-7 vs. PacR | MCF-7 vs. 0035R | ||||
|---|---|---|---|---|---|---|
| |Δβ| | p-Value | FDR | |Δβ| | p-Value | FDR | |
| Whole gene | 0.41 | 0.004 | 0.025 | 0.36 | 0.2 | ns |
| 1st exon | 0.23 | 0.121 | ns | 0.01 | 0.439 | ns |
| Body | 0.12 | 0.248 | ns | 0.11 | 0.248 | ns |
| TSS1500 | 0.49 | 0.02 | 0.025 | 0.36 | 0.002 | 0.008 |
| TSS200 | 0.40 | 0.05 | ns | 0.05 | 0.05 | ns |
| Characteristics | Breast Carcinoma Set |
|---|---|
| Mean age at diagnosis, years | 56.0 ± 10.7 |
| Menopausal status | N (%) |
| Premenopausal | 27 (28.4) |
| Postmenopausal | 66 (69.5) |
| Not available | 2 (2.1) |
| Histological type | N (%) |
| Invasive ductal carcinoma | 80 (84.2) |
| Others | 15 (15.8) |
| Histological grade | N (%) |
| G1 | 13 (13.7) |
| G2 | 59 (62.1) |
| G3 | 22 (23.1) |
| Not available | 1 (1.1) |
| Stage | N (%) |
| I | 31 (32.6) |
| II | 59 (62.1) |
| III | 4 (4.2) |
| IV | 1 (1.1) |
| Estrogen receptor status | N (%) |
| Positive | 86 (90.5) |
| Negative | 9 (9.5) |
| Progesterone receptor status | N (%) |
| Positive | 70 (73.7) |
| Negative | 25 (26.3) |
| ERBB2 status | N (%) |
| Positive | 27 (28.4) |
| Negative | 68 (71.6) |
| Ki67 status1 | N (%) |
| Positive | 70 (73.7) |
| Negative | 13 (13.7) |
| Unknown | 12 (12.6) |
| Molecular subtype | N (%) |
| Luminal A | 60 (63.2) |
| Luminal B | 26 (27.3) |
| ERBB2 | 7 (7.4) |
| Triple negative | 2 (2.1) |
| Therapeutic regimens | N (%) |
| Neoadjuvant (NACT) 2 | 13 (13.7) |
| Adjuvant (ACT) | 82 (86.3) |
| Relapse | N (%) |
| Yes | 9 (9.5) |
| No | 86 (90.5) |
| Overall survival (OS) | |
| Mean (months) ± SD | 70.9 ± 28.0 |
| Disease-free survival (DFS) | |
| Mean (months) ± SD | 61.1 ± 28.4 |
| Characteristics | TRIP6 Expression Relative to IPO8 and MRPL19 | Significance (Mann–Whitney) |
|---|---|---|
| Premenopausal | 1.05 ± 0.59 | 0.033 |
| Postmenopausal | 0.77 ± 0.49 | |
| Progesterone receptor positive | 0.93 ± 0.56 | 0.020 |
| Progesterone receptor negative | 0.61 ± 0.34 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Daniel, P.; Balušíková, K.; Václavíková, R.; Šeborová, K.; Ransdorfová, Š.; Valeriánová, M.; Wei, L.; Jelínek, M.; Tlapáková, T.; Fleischer, T.; et al. ABCB1 Amplicon Contains Cyclic AMP Response Element-Driven TRIP6 Gene in Taxane-Resistant MCF-7 Breast Cancer Sublines. Genes 2023, 14, 296. https://doi.org/10.3390/genes14020296
Daniel P, Balušíková K, Václavíková R, Šeborová K, Ransdorfová Š, Valeriánová M, Wei L, Jelínek M, Tlapáková T, Fleischer T, et al. ABCB1 Amplicon Contains Cyclic AMP Response Element-Driven TRIP6 Gene in Taxane-Resistant MCF-7 Breast Cancer Sublines. Genes. 2023; 14(2):296. https://doi.org/10.3390/genes14020296
Chicago/Turabian StyleDaniel, Petr, Kamila Balušíková, Radka Václavíková, Karolína Šeborová, Šárka Ransdorfová, Marie Valeriánová, Longfei Wei, Michael Jelínek, Tereza Tlapáková, Thomas Fleischer, and et al. 2023. "ABCB1 Amplicon Contains Cyclic AMP Response Element-Driven TRIP6 Gene in Taxane-Resistant MCF-7 Breast Cancer Sublines" Genes 14, no. 2: 296. https://doi.org/10.3390/genes14020296
APA StyleDaniel, P., Balušíková, K., Václavíková, R., Šeborová, K., Ransdorfová, Š., Valeriánová, M., Wei, L., Jelínek, M., Tlapáková, T., Fleischer, T., Kristensen, V. N., Souček, P., Ojima, I., & Kovář, J. (2023). ABCB1 Amplicon Contains Cyclic AMP Response Element-Driven TRIP6 Gene in Taxane-Resistant MCF-7 Breast Cancer Sublines. Genes, 14(2), 296. https://doi.org/10.3390/genes14020296








