Exploring the Effect of High-Energy Heavy Ion Beam on Rice Genome: Transposon Activation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Whole-Genome Sequencing and Data Analysis
3. Results
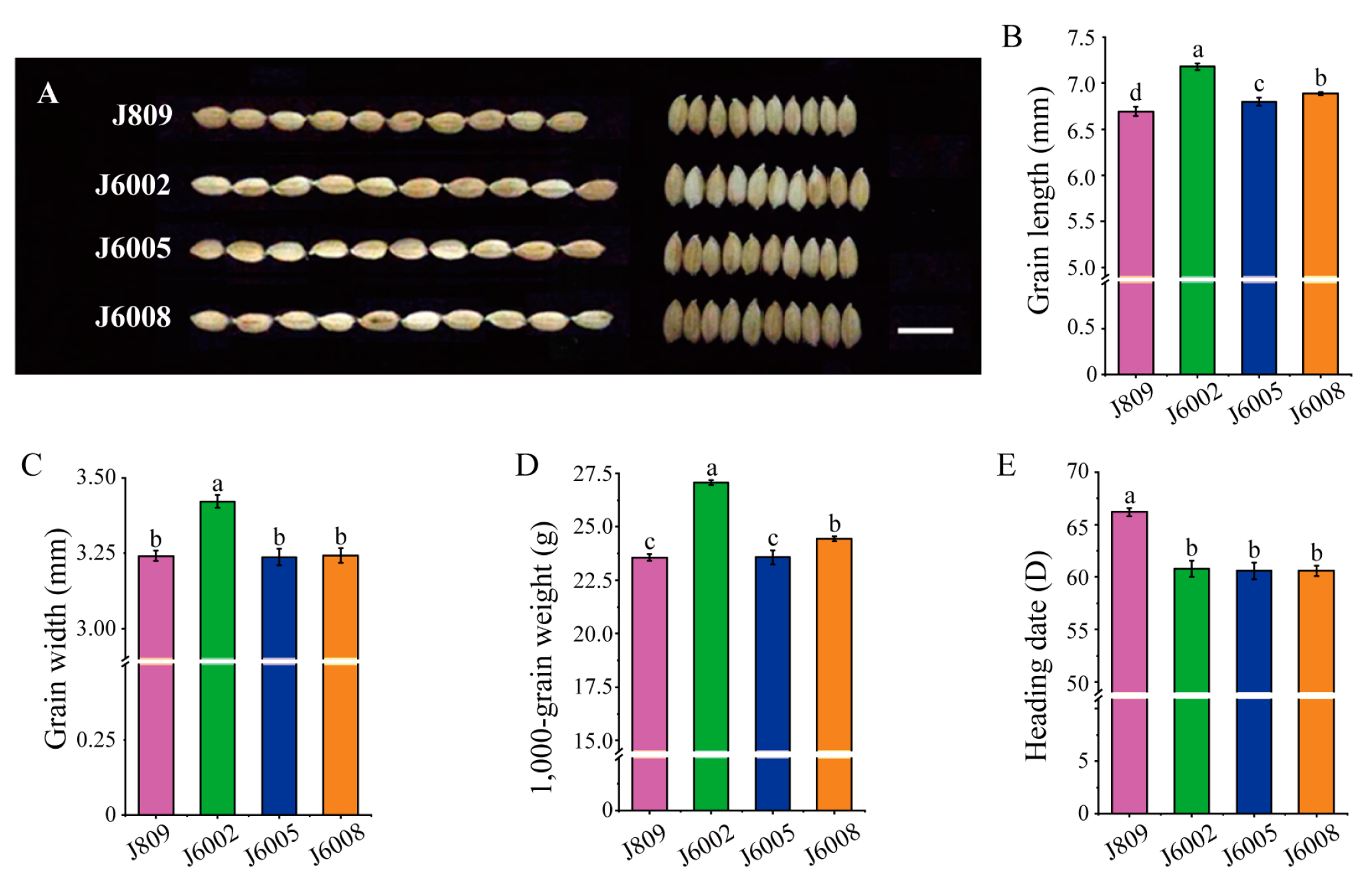
3.1. Phenotypic Variation in Carbon Ion Beam (CIB)-Induced Mutant Lines
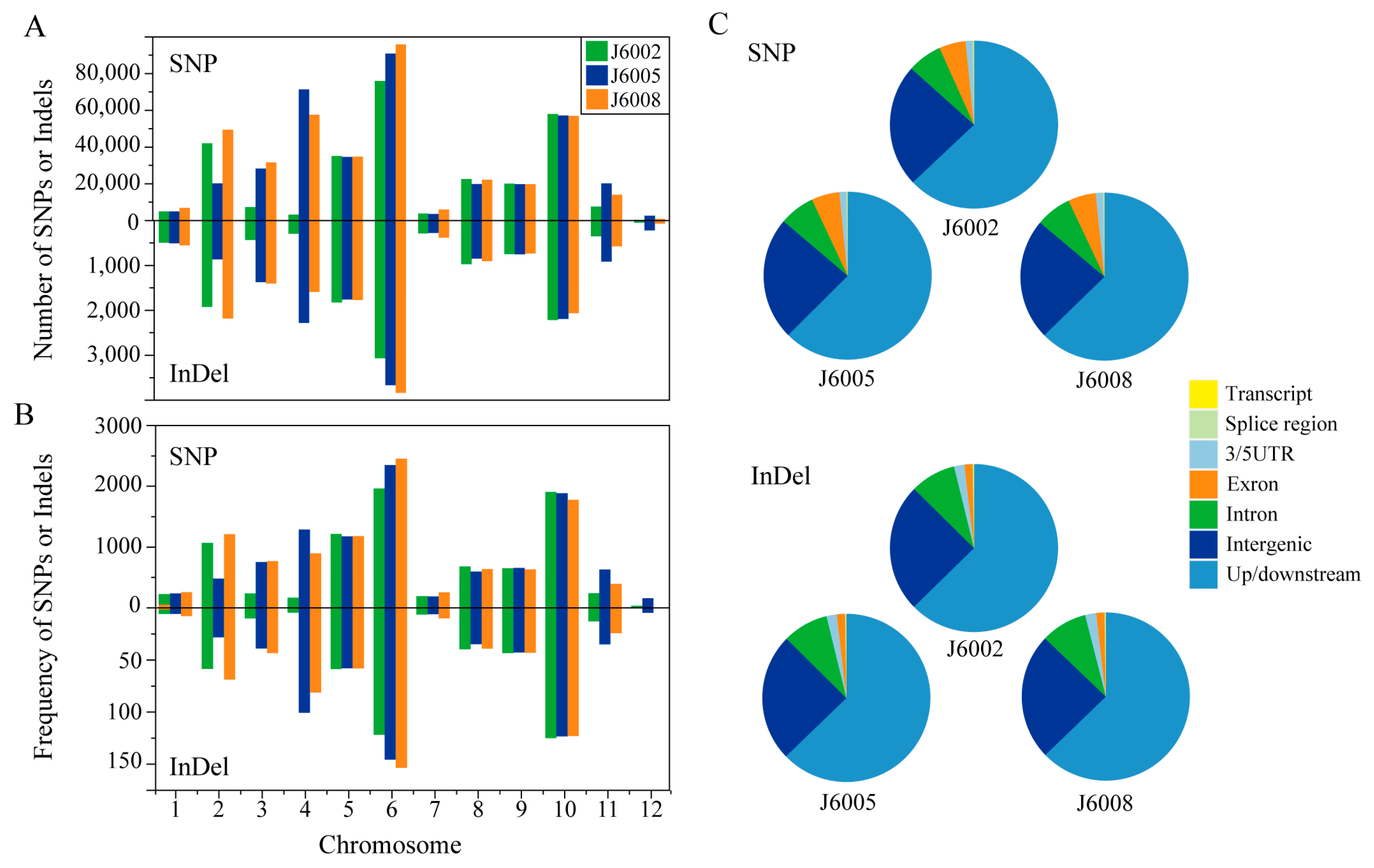
3.2. Genome-Wide Sequence Variations in Three CIB-Induced Mutant Lines
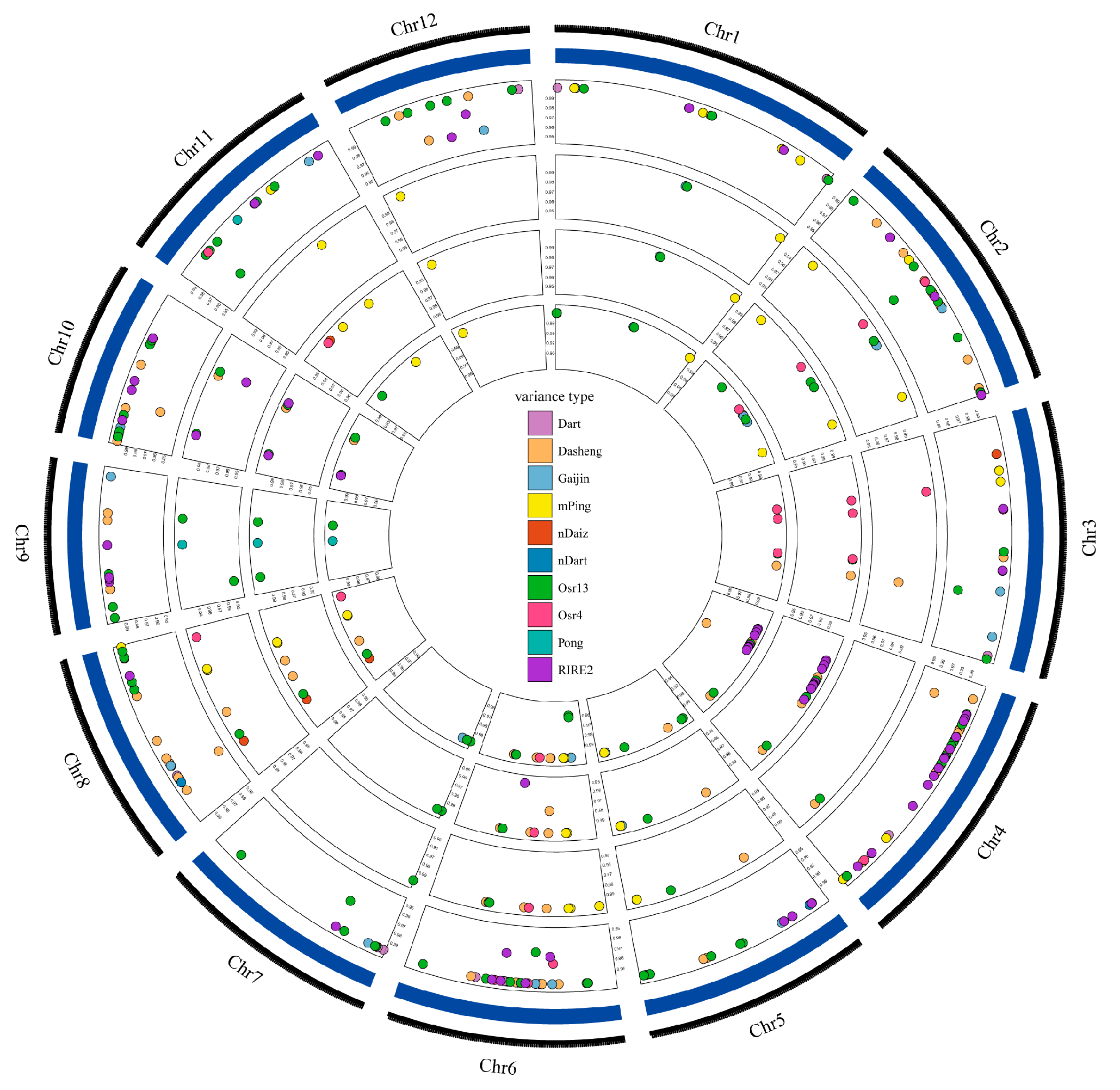
3.3. Transposon Activation in Three CIB-Induced Mutant Lines
3.4. Sequence Variations and Functional Genes Related to Phenotypic Changes in Mutants
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van Nguyen, N.; Ferrero, A. Meeting the challenges of global rice production. Paddy Water Environ. 2006, 4, 1–9. [Google Scholar] [CrossRef]
- Bind, D.; Dwivedi, V.K. Effect of mutagenesis on germination, plant survival and pollen sterility in M1 generation of in cowpea [Vigna unguiculata (L.) Walp]. Indian J. Agric. Res. 2014, 48, 398–401. [Google Scholar] [CrossRef]
- Oladosu, Y.; Rafii, M.Y.; Abdullah, N.; Hussin, G.; Ramli, A.; Rahim, H.A.; Miah, G.; Usman, M. Principle and application of plant mutagenesis in crop improvement: A review. Biotechnol. Biotechnol. Equip. 2016, 30, 1–16. [Google Scholar] [CrossRef]
- Shelake, R.M.; Pramanik, D.; Kim, J.Y. Evolution of plant mutagenesis tools: A shifting paradigm from random to targeted genome editing. Plant Biotechnol. Rep. 2019, 13, 423–445. [Google Scholar] [CrossRef]
- Shu, Q.Y.; Forster, B.P.; Nakagawa, H. Plant Mutation Breeding and Biotechnology; Cabi: Wallingford, UK, 2012. [Google Scholar]
- Yang, G.; Luo, W.; Zhang, J.; Yan, X.; Du, Y.; Zhou, L.; Li, W.; Wang, H.; Chen, Z.; Guo, T. Genome-wide comparisons of mutations induced by carbon-ion beam and gamma-rays irradiation in rice via resequencing multiple mutants. Front. Plant Sci. 2019, 10, 1514. [Google Scholar] [CrossRef] [PubMed]
- Kazama, Y.; Hirano, T.; Nishihara, K.; Ohbu, S.; Shirakawa, Y.; Abe, T. Effect of high-LET Fe-ion beam irradiation on mutation induction in Arabidopsis thaliana. Genes Genet. Syst. 2013, 88, 189–197. [Google Scholar] [CrossRef]
- Giap, H.; Giap, B. Historical perspective and evolution of charged particle beam therapy. AME Publ. Co. 2012, 1, 127–136. [Google Scholar]
- Joiner, M.C.; Burmeister, J.W.; Dörr, W. Linear energy transfer and relative biological effectiveness. In Basic Clinical Radiobiology; CRC Press: Boca Raton, FL, USA, 2018; pp. 54–60. [Google Scholar]
- Kazama, Y.; Hirano, T.; Saito, H.; Yang, L.; Abe, T. Characterization of highly efficient heavy-ion mutagenesis in Arabidopsis thaliana. BMC Plant Biol. 2011, 11, 161. [Google Scholar] [CrossRef]
- Tanaka, A.; Shikazono, N.; Hase, Y. Studies on biological effects of ion beams on lethality, molecular nature of mutation, mutation rate, and spectrum of mutation phenotype for mutation breeding in higher plants. J. Radiat. Res. 2010, 51, 223–233. [Google Scholar] [CrossRef]
- Alizadeh, E.; Sanz, A.G.; García, G.; Sanche, L. Radiation Damage to DNA: The Indirect Effect of Low-Energy Electrons. J. Phys. Chem. Lett. 2013, 4, 820–825. [Google Scholar] [CrossRef]
- Ravanat, J.L.; Douki, T.; Cadet, J. Direct and indirect effects of UV radiation on DNA and its components. J. Photochem. Photobiol. B. Biol. 2001, 63, 88–102. [Google Scholar] [CrossRef] [PubMed]
- Tokuyama, Y.; Furusawa, Y.; Ide, H.; Yasui, A.; Terato, H. Role of isolated and clustered DNA damage and the post-irradiating repair process in the effects of heavy ion beam irradiation. J. Radiat. Res. 2015, 56, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Rodgers, K.; McVey, M. Error-prone repair of DNA double-strand breaks. J. Cell Physiol. 2016, 231, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Ling, A.P.K.; Ung, Y.C.; Hussein, S.; Harun, A.R.; Yoshihiro, H. Morphological and biochemical responses of Oryza sativa L. (cultivar MR219) to ion beam irradiation. J. Zhejiang Univ. Sci. B 2013, 14, 1132–1143. [Google Scholar] [CrossRef]
- Amoroso, A.; Concia, L.; Maggio, C.; Raynaud, C.; Maga, G. Oxidative DNA Damage Bypass in Arabidopsis thaliana Requires DNA Polymerase λ and Proliferating Cell Nuclear Antigen 2. Plant Cell 2011, 23, 806–822. [Google Scholar] [CrossRef]
- Thannickal, V.J.; Fanburg, B.L. Reactive oxygen species in cell signaling. Am. J. Physiol.-Lung Cell Mol. Physiol. 2000, 279, L1005–L1028. [Google Scholar] [CrossRef]
- Maekawa, M.; Hase, Y.; Shikazono, N.; Tanaka, A. Induction of somatic instability in stable yellow leaf mutant of rice by ion beam irradiation. Nucl. Instrum. Methods Phys. Res. Sect. B. Beam Interact. Mater. At. 2003, 206, 579–585. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. Mobile elements create structural variation: Analysis of a complete human genome. Genome Res. 2009, 19, 1516–1526. [Google Scholar]
- Ma, M.; Zhu, H.; Zhang, C.; Sun, X.; Gao, X.; Chen, G. “Liquid biopsy”—ctDNA detection with great potential and challenges. Ann. Transl. Med. 2015, 3, 235. [Google Scholar]
- Morita, R.; Ichida, H.; Ishii, K.; Hayashi, Y.; Abe, H.; Shirakawa, Y.; Ichinose, K.; Tsuneizumi, K.; Kazama, T.; Toriyama, K.; et al. LONG GRAIN 1: A novel gene that regulates grain length in rice. Mol. Breed. 2019, 39, 135. [Google Scholar] [CrossRef]
- Ward, J.F.; Evans, J.W.; Limoli, C.L.; Calabro-Jones, P.M. Radiation and hydrogen peroxide induced free radical damage to DNA. Br. J. Cancer Suppl. 1987, 8, 105. [Google Scholar] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Lefouili, M.; Nam, K. The evaluation of Bcftools mpileup and GATK HaplotypeCaller for variant calling in non-human species. Sci. Rep. 2022, 12, 11331. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Cleary, J.G.; Braithwaite, R.; Gaastra, K.; Hilbush, B.S.; Inglis, S.; Irvine, S.A.; Jackson, A.; Littin, R.; Rathod, M.; Ware, D. Comparing variant call files for performance benchmarking of next-generation sequencing variant calling pipelines. BioRxiv 2015, 023754. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Adrion, J.R.; Song, M.J.; Schrider, D.R.; Hahn, M.W.; Schaack, S. Genome-wide estimates of transposable element insertion and deletion rates in Drosophila melanogaster. Genome Biol. Evol. 2017, 9, 1329–1340. [Google Scholar] [CrossRef]
- Li, S.W.; Li, M.; Song, H.P.; Feng, J.L.; Tai, X.S. Induction of a high-yield lovastatin mutant of Aspergillus terreus by 12 C 6+ heavy-ion beam irradiation and the influence of culture conditions on lovastatin production under submerged fermentation. Appl. Biochem. Biotechnol. 2011, 165, 913–925. [Google Scholar] [CrossRef]
- Ning, J.; Bao, Z.; Zhang, X.; Hirochika, H.; Wessler, S. An active DNA transposon in rice. Nature 2003, 421, 163–167. [Google Scholar]
- Ngezahayo, F.; Xu, C.; Wang, H.; Jiang, L.; Pang, J.; Liu, B. Tissue culture-induced transpositional activity of mPing is correlated with cytosine methylation in rice. BMC Plant Biol. 2009, 9, 91. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yu, Y.; Jiang, L.; Lin, X.; Zhang, C.; Ou, X.; Osabe, K.; Liu, B. Changes in DNA methylation and transgenerational mobilization of a transposable element (mPing) by the Topoisomerase II inhibitor, Etoposide, in rice. BMC Plant Biol. 2012, 12, 48. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Li, N.; Zhang, Z.; Meng, X.; Dong, Q.; Xu, C.; Gong, L.; Liu, B. CG hypomethylation leads to complex changes in DNA methylation and transpositional burst of diverse transposable elements in callus cultures of rice. Plant J. 2020, 101, 188–203. [Google Scholar] [CrossRef]
- Ohtsubo, H.; Kumekawa, N.; Ohtsubo, E. RIRE2, a novel gypsy-type retrotransposon from rice. Genes Genet. Syst. 1999, 74, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Fang, D.; Yang, R.; Gao, F.; An, X.; Zhou, X.; Li, Y.; Yi, C.; Zhang, T.; Liang, C.; et al. De novo genome assembly of Oryza granulata reveals rapid genome expansion and adaptive evolution. Commun. Biol. 2018, 1, 84. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N. Dasheng and RIRE2. A Nonautonomous Long Terminal Repeat Element and Its Putative Autonomous Partner in the Rice Genome. Plant Physiol. 2002, 130, 1697. [Google Scholar] [CrossRef] [PubMed]
- Kashkush, K.; Khasdan, V. Large-scale survey of cytosine methylation of retrotransposons and the impact of readout transcription from long terminal repeats on expression of adjacent rice genes. Genetics 2007, 177, 1975–1985. [Google Scholar] [CrossRef]
- Yilmaz, S.; Marakli, S.; Yuzbasioglu, G.; Gozukirmizi, N. Short-term mutagenicity test by using IRAP molecular marker in rice grown under herbicide treatment. Biotechnol. Biotechnol. Equip. 2018, 32, 923–928. [Google Scholar] [CrossRef]
- Zhang, Q.J.; Gao, L.Z. Rapid and recent evolution of LTR retrotransposons drives rice genome evolution during the speciation of AA-genome Oryza species. G3 Genes Genomes Genet. 2017, 7, 1875–1885. [Google Scholar] [CrossRef]
- Monden, Y.; Naito, K.; Okumoto, Y.; Saito, H.; Oki, N.; Tsukiyama, T.; Ideta, H.; Nakazaki, T.; Wessler, S.R. High potential of a transposon mPing as a marker system in japonica× japonica cross in rice. DNA Res. 2009, 16, 131–140. [Google Scholar] [CrossRef]
- Weng, J.; Gu, S.; Wan, X.; Jiang, L.; Gao, H.; Zhai, H.; Su, N.; Cheng, Z.; Guo, T. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 2008, 18, 1199–1209. [Google Scholar] [CrossRef] [PubMed]
- Qiao, J.; Jiang, H.; Lin, Y.; Shang, L.; Qian, Q. A novel miR167a-OsARF6-OsAUX3 module regulates grain length and weight in rice. Mol. Plant 2021, 14, 1683–1698. [Google Scholar] [CrossRef] [PubMed]
- Si, L.; Chen, J.; Huang, X.; Gong, H.; Luo, J.; Hou, Q.; Zhou, T.; Liu, T.; Zhu, J.; Shangguan, Y.; et al. OsSPL13 controls grain size in cultivated rice. Nat. Genet. 2016, 48, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.L.; Dong, N.Q.; Guo, T.; Ye, W.W.; Shan, J.X.; Lin, H.X. A quantitative trait locus GW6 controls rice grain size and yield through the gibberellin pathway. Plant J. 2020, 103, 1174–1188. [Google Scholar] [CrossRef] [PubMed]
- Yi, C.D.; Wang, D.R.; Jiang, W.; Li, W.; Cheng, X.; Wang, Y.; Zhou, Y.; Liang, G.; Gu, M. Development of functional markers and identification of haplotypes for rice grain shape gene GW8. Acta Agron. Sin. 2016, 42, 1291–1297. [Google Scholar] [CrossRef]
- Kim, S.L.; Lee, S.; Kim, H.J.; Nam, H.G.; An, G. OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol. 2007, 145, 1484–1494. [Google Scholar] [CrossRef] [PubMed]
- Sui, P.; Shi, J.; Gao, X.; Shen, W.H.; Dong, A. H3K36 methylation is involved in promoting rice flowering. Mol. Plant 2013, 6, 975–977. [Google Scholar] [CrossRef]
- Kojima, S.; Takahashi, Y.; Kobayashi, Y.; Monna, L.; Sasaki, T.; Araki, T.; Yano, M. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 2002, 43, 1096–1105. [Google Scholar] [CrossRef]
- Nakagawa, M.; Shimamoto, K.; Kyozuka, J. Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J. 2002, 29, 743–750. [Google Scholar] [CrossRef]
- Xue, W.; Xing, Y.; Weng, X.; Zhao, Y.; Tang, W.; Wang, L.; Zhou, H.; Yu, S.; Xu, C.; Li, X. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 2008, 40, 761–767. [Google Scholar] [CrossRef]
- Li, C.; Zhu, S.; Zhang, H.; Wang, J.; Chai, J.; Wu, F.; Cheng, Z.; Guo, X. OsLBD37 and OsLBD38, two class II type LBD proteins, are involved in the regulation of heading date by controlling the expression of Ehd1 in rice. Biochem. Biophys. Res. Commun. 2017, 486, 720–725. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Wang, J.; Cai, M.; Zhang, H.; Wu, F.; Xu, Y.; Li, C.; Cheng, Z.; Zhang, X.; Guo, X. The OsHAPL1-DTH8-Hd1 complex functions as the transcription regulator to repress heading date in rice. JEB 2017, 68, 553–568. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhou, C.; Zhao, Y.; Zhou, S.; Wang, W.; Zhou, D. The rice enhancer of zeste [E(z)] genes SDG711 and SDG718 are respectively involved in long day and short day signaling to mediate the accurate photoperiod control of flowering time. Front. Plant Sci. 2014, 5, 591. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Zhang, Y.; Zhang, M.; Zhan, X.; Cao, L. The rice CONSTANS-like protein OsCOL15 suppresses flowering by promoting Ghd7 and repressing RID1. Biochem. Biophys. Res. Commun. 2018, 495, 1349–1355. [Google Scholar] [CrossRef] [PubMed]
- Yano, K.; Yamamoto, E.; Aya, K.; Takeuchi, H.; Lo, P.C.; Hu, L.; Yamasaki, M.; Yoshida, S.; Kitano, H.; Hirano, K. Genome-wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice. Nat. Genet. 2016, 48, 927–934. [Google Scholar] [CrossRef]
- Yano, K.; Morinaka, Y.; Wang, F.; Huang, P.; Takehara, S.; Hirai, T.; Ito, A.; Koketsu, E.; Kawamura, M.; Kotake, K. GWAS with principal component analysis identifies a gene comprehensively controlling rice architecture. PNAS 2019, 116, 21262–21267. [Google Scholar] [CrossRef]
- Ying, J.Z.; Gao, J.P.; Shan, J.X.; Zhu, M.Z.; Shi, M.; Lin, H.X. Dissecting the genetic basis of extremely large grain shape in rice cultivar ‘JZ1560’. J. Genet. Genom. 2012, 39, 325–333. [Google Scholar] [CrossRef]
- Yang, Z.; Jin, L.; Zhu, H.; Wang, S.; Liu, G. Analysis of epistasis among QTLs on heading date based on single segment substitution lines in rice. Sci. Rep. 2018, 8, 3059. [Google Scholar] [CrossRef]
| Category | Type | Super Family | J809 | J6008 | J6005 | J6002 |
|---|---|---|---|---|---|---|
| Retrotransposon | Gypsy | Dart | 90 | 80 | 80 | 78 |
| Dasheng | 674 | 661 | 662 | 675 | ||
| Gaijin | 184 | 183 | 180 | 181 | ||
| Jing | 37 | 38 | 38 | 38 | ||
| Karma | 4 | 4 | 4 | 5 | ||
| mGing | 16 | 17 | 17 | 17 | ||
| nDaiz | 25 | 25 | 27 | 25 | ||
| nDart | 14 | 13 | 14 | 14 | ||
| RIRE2 | 606 | 605 | 606 | 603 | ||
| Copia | Lullaby | 5 | 5 | 5 | 5 | |
| Osr13 | 478 | 477 | 473 | 477 | ||
| Osr4 | 85 | 87 | 85 | 83 | ||
| Tos17 | 8 | 7 | 7 | 8 | ||
| DNA transposon | DNA | mPing | 42 | 40 | 47 | 44 |
| Pong | 5 | 6 | 5 | 5 | ||
| Total | 2273 | 2248 | 2250 | 2258 |
| Super Family | Type | J6002 | J6005 | J6008 |
|---|---|---|---|---|
| Dart | (+) | 0 | 0 | 1 |
| (−) | 8 | 9 | 10 | |
| Dasheng | (+) | 10 | 20 | 20 |
| (−) | 16 | 23 | 21 | |
| Gaijin | (+) | 3 | 3 | 8 |
| (−) | 4 | 4 | 7 | |
| Jing | (+) | 0 | 0 | 0 |
| (−) | 0 | 0 | 0 | |
| Lullaby | (+) | 0 | 0 | 0 |
| (−) | 0 | 0 | 0 | |
| mGing | (+) | 0 | 0 | 0 |
| (−) | 0 | 0 | 0 | |
| mPing | (+) | 12 | 12 | 10 |
| (−) | 6 | 9 | 12 | |
| nDaiz | (+) | 1 | 3 | 2 |
| (−) | 0 | 0 | 0 | |
| nDart | (+) | 0 | 0 | 0 |
| (−) | 2 | 2 | 1 | |
| Osr13 | (+) | 12 | 20 | 24 |
| (−) | 7 | 14 | 14 | |
| Osr4 | (+) | 4 | 7 | 7 |
| (−) | 6 | 6 | 6 | |
| Pong | (+) | 1 | 1 | 1 |
| (−) | 0 | 1 | 0 | |
| RIRE2 | (+) | 2 | 14 | 12 |
| (−) | 9 | 12 | 15 | |
| Tos17 | (+) | 0 | 0 | 0 |
| (−) | 0 | 0 | 0 |
| Mutant Gene | Line | Variant Information |
|---|---|---|
| OsMADS65 (LOC_Os01g69850) | J6002 | Chr1,40346050, G/A; 40346099, T/C; 40346701, A/C; 40348092, A/C; 40353298, T/C; 40354446, T/C; 40358884, G/A; 40364205, A/G; 40347761, DEL:1;40353958, INS:30;40354148, INS:22 |
| J6005 | Chr1,40346050, G/A; 40346099, T/C; 40346701, A/C; 40348092, A/C; 40353298, T/C; 40354446, T/C; 40358884, G/A; 40364205, A/G; 40347761, DEL:1;40353958, INS:30;40354148, INS:22 | |
| J6008 | Chr1,40346050, G/A; 40346099, T/C; 40346701, A/C; 40348092, A/C; 40353298, T/C; 40354446, T/C; 40358884, G/A; 40364205, A/G; 40347761, DEL:1;40353958, INS:30;40354148, INS:22 | |
| RCN2 (LOC_Os02g32950) | J6008 | Chr2,19567866, C/T; 19567924, A/T; 19568531, C/T; 19568568, C/G; 19568704, G/A; 19568752, G/A; 19568814, A/T; 19567960, DEL:13 |
| SDG725 (LOC_Os02g34850) | J6002 | Chr2, 20900113, C/T; 20901575, A/G;20902659, T/C;20902806, G/A;20902923, A/G;20902938, C/T;20903136, A/T;20903813, A/C;20904000, A/G;20904272, C/T;20904360, A/G;20905262, T/C;20905318, T/A;20905609, A/T;20905719, T/C;20905892, C/T;20906077, G/A;20907028, T/G;220902714, DEL:13 |
| J6008 | Chr2,20900113, C/T;20901575, A/G;20902659, T/C;20902806, G/A;20902923, A/G;20902938, C/T; 20903136, A/T;20903813, A/C;20904000, A/G;20904272, C/T;20904360, A/G;20905262, T/C; 20905318, T/A;20905609, A/T;20905719, T/C;20905892, C/T;20906077, G/A;20907028, T/G; 220902714, DEL:18 | |
| Ghd2 (LOC_Os02g49880) | J6002 | Chr2,30473609, G/A;30474736, T/C |
| J6005 | Chr2,30473609, G/A; 30474736, T/C | |
| J6008 | Chr2,30473609, G/A; 30474736, T/C | |
| OsLBD38 (LOC_Os03g41330) | J6005 | Chr3,22977039, T/C;22977138, A/T;22977558, C/G;22977961, T/G;22978006, G/A;22978008, G/A;22978082, C/G;22978142, A/G;22978156, A/C;22978174, G/A;22979442, A/G;22979762, A/C; 22977280, INS:9; 22977453, DEL14; 22978092, DEL12 |
| J6008 | Chr3, 22977039, T/C;22977138, A/T;22977558, C/G;22977961, T/G;22978006, G/A;22978008, G/A;22978082, C/G;22978142, A/G;22978156, A/C;22978174, G/A;22979442, A/G;22979762, A/C; 22977280, INS:9; 22977453, DEL14; 22978092, DEL12 | |
| OsHAPL1 (LOC_Os05g41450) | J6002 | Chr5,24277541, T/C;24277683, T/A;24278012, A/G;24278551, T/C |
| J6005 | Chr5,24277541, T/C; 24277683, T/A; 24278012, A/G; 24278551, T/C; 24280474, A/G; 24280680, T/C | |
| J6008 | Chr5,24277541, T/C; 24277683, T/A; 24278012, A/G; 24278551, T/C | |
| Hd3a/FT/Ehd3 (LOC_Os06g06320) | J6008 | Chr6,2942192, C/G |
| SDG711 (LOC_Os06g16390) | J6005 | Chr6,9355225, T/C |
| J6008 | Chr6,9355225, T/C | |
| OsCOL15 (LOC_Os08g42440) | J6002 | Chr8,26793385, T/C; 26793749, T/C; 26794802, A/C; 26795049, A/C; 26795325, T/A; 26795632, A/C; 26795721, C/T; 26796722, C/G; 26792973, DEL,14; 26794149; INS:1; 26797108, DEL:26 |
| J6005 | Chr8,26793385, T/C; 26793749, T/C; 26794802, A/C; 26795049, A/C; 26795325, T/A; 26795632, A/C; 26795721, C/T; 26796722, C/G; 26792973, DEL,26; 26794149; INS:1; 26797108, DEL:6 | |
| J6008 | Chr8,26793385, T/C; 26793749, T/C; 26794802, A/C; 26795049, A/C; 26795325, T/A; 26795632, A/C; 26795721, C/T; 26796722, C/G; 26792973, DEL,26; 26794149; INS:1; 26797108, DEL:25 | |
| OsGATA28 (LOC_Os11g08410) | J6002 | Chr11,4433156, C/T; 4433087, INS:6, 4433137, DEL:11 |
| J6008 | Chr11,4433156, C/T; 4434073, T/A; 4433087, INS:6, 4433137, DEL:19 |
| Mutant Gene | Lines | Variant Information |
|---|---|---|
| GW5 (LOC_Os05g09520) | J6002 | Chr5, 5365256, G/A; 5366491, INS:20 |
| J6005 | Chr5, 5365256, G/A; 5366491, INS:20 | |
| J6008 | Chr5, 5365256, G/A; 5366491, INS:20 | |
| qGL5 (LOC_Os05g37470) | J6002 | Chr5, 21926820, T/C; 21926832, G/A; 21926911, A/T; 21927295, G/A; 21928349, T/C; 21928809, T/A; 21929398, C/T; 21929564, C/T; 21930387, A/G; 21930730, G/A; 21930884, G/A; 21930900, T/C; 21929789, INS:12 |
| J6005 | Chr5, 21926820, T/C; 21926832, G/A; 21926911, A/T; 21927295, G/A; 21928349, T/C; 21928809, T/A; 21929398, C/T; 21929564, C/T; 21930387, A/G; 21930730, G/A; 21930884, G/A; 21930900, T/C; 21929789, INS:12 | |
| J6008 | Chr5, 21926820, T/C; 21926832, G/A; 21926911, A/T; 21927295, G/A; 21928349, T/C; 21928809, T/A; 21929398, C/T; 21929564, C/T; 21930387, A/G; 21930730, G/A; 21930884, G/A; 21930900, T/C; 21929789, INS:12 | |
| GW6 (LOC_Os06g15620) | J6005 | Chr6, 8847736, A/G; 8847914, A/T; 8848057, A/T; 8848148, G/A; 8848223, A/C; 8848234, A/T; 8848326, C/G; 8848531, T/G; 8848532, C/G; 8848614, C/T; 8848639, C/T; 8848711, G/A; 8848740, A/C; 8848752, T/C; 8847997, INS:2; 8848285, DEL:20; 8848295, DEL:22; 8848308, INS:3; 8848330, DEL:12; 8848601, INS:6; 8848645,DEL:27; 8848655, DEL:23; 8848689, DEL:18; 8848837, DEL:18 |
| J6008 | Chr6, 8847736, A/G; 8847914, A/T; 8848057, A/T; 8848148, G/A; 8848223, A/C; 8848234, A/T; 8848326, C/G; 8848531, T/G; 8848532, C/G; 8848614, C/T; 8848639, C/T; 8848711, G/A; 8848740, A/C; 8848752, T/C; 8847997, INS:2; 8848285, DEL:22; 8848295, DEL:22; 8848308, INS:3; 8848330, DEL:13; 8848601, INS:6; 8848645, DEL:24; 8848654, DEL:23;8848689, DEL:24; 8848837, DEL:18 | |
| GLW7 (LOC_Os07g32170) | J6002 | Chr7, 19100099, INS:2 |
| J6005 | Chr7, 19100099, INS:2 | |
| J6008 | Chr7, 19100099, INS:2 | |
| GW8 (LOC_Os08g41940) | J6002 | Chr8, 26501258, C/T; 26501458, C/T; 26502275, A/G; 26502571, C/G; 26503084, C/T; 26503280, T/A; 26503323, A/G; 26503397, T/C; 26504208, G/A; 26504323, T/A; 26504422, G/C; 26504599, A/G; 26505008, C/T; 26505025, G/A; 26505046, A/G; 26505387, C/A; 26505658, G/T; 8848285, DEL:20; 26501190, INS:10; 26501542, DEL:16; 26501777, DEL:5; 26501945, INS:15; 26502637, INS:15; 26503416, DEL:13; 26504889, INS:15; 26504922, DEL:14; 26506051, INS:15; |
| J6005 | Chr8, 26501258, C/T; 26501458, C/T; 26502275, A/G; 26502571, C/G; 26503084, C/T; 26503280, T/A; 26503323, A/G; 26503397, T/C; 26504208, G/A; 26504323, T/A; 26504422, G/C; 26504599, A/G; 26505008, C/T; 26505025, G/A; 26505046, A/G; 26505387, C/A; 26505658, G/T; 26501190, INS:10; 26501542, DEL:17;26501777, DEL:3; 26501945, INS:19; 26502637, INS:12; 26503416, DEL:13; 26504889, INS:17; 26504922, DEL:22; 26506051, INS:16; | |
| J6008 | Chr8, 26501258, C/T; 26501458, C/T; 26502275, A/G; 26502571, C/G; 26503084, C/T; 26503280, T/A; 26503323, A/G; 26503397, T/C; 26504208, G/A; 26504323, T/A; 26504422, G/C; 26504599, A/G; 26505008, C/T; 26505025, G/A; 26505046, A/G; 26505387, C/A; 26505658, G/T; 26501190, INS:10; 26501542, DEL:23; 26501777, DEL:9; 26501945, INS:19; 26502637, INS:18; 26503416, DEL:18; 26504889, INS:14; 26504922, DEL:14; 26506051, INS:18; | |
| WTG1 (LOC_Os08g42540) | J6002 | Chr8, 26883121, C/T; 26883146, G/C; 26883932, T/A; 26884169, T/G; 26885532, G/A; 26885992, G/A; 26886283, C/A; 26886650, C/T; 26886715, G/A; 26886719, C/A; 26887098, C/T; 26887145, T/A; 26884796, INS:30; 26886522, DEL:16 |
| J6005 | Chr8, 26883121, C/T; 26883146, G/C; 26883932, T/A; 26884169, T/G; 26885532, G/A; 26885992, G/A; 26886283, C/A; 26886715, G/A; 26886719, C/A; 26884796, INS:25; 26886522, DEL:12 | |
| J6008 | Chr8, 26883121, C/T; 26883146, G/C; 26883932, T/A; 26884169, T/G; 26885532, G/A; 26885992, G/A; 26886283, C/A; 26886650, C/T; 26886715, G/A; 26886719, C/A; 26887098, C/T; 26887145, T/A; 26884796, INS:21; 26886522, DEL:13 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, X.; Li, J.; Yang, F.; Zhang, X.; Li, Y. Exploring the Effect of High-Energy Heavy Ion Beam on Rice Genome: Transposon Activation. Genes 2023, 14, 2178. https://doi.org/10.3390/genes14122178
Wen X, Li J, Yang F, Zhang X, Li Y. Exploring the Effect of High-Energy Heavy Ion Beam on Rice Genome: Transposon Activation. Genes. 2023; 14(12):2178. https://doi.org/10.3390/genes14122178
Chicago/Turabian StyleWen, Xiaoting, Jingpeng Li, Fu Yang, Xin Zhang, and Yiwei Li. 2023. "Exploring the Effect of High-Energy Heavy Ion Beam on Rice Genome: Transposon Activation" Genes 14, no. 12: 2178. https://doi.org/10.3390/genes14122178
APA StyleWen, X., Li, J., Yang, F., Zhang, X., & Li, Y. (2023). Exploring the Effect of High-Energy Heavy Ion Beam on Rice Genome: Transposon Activation. Genes, 14(12), 2178. https://doi.org/10.3390/genes14122178





