De Novo RNA Sequencing and Transcriptome Analysis of Sclerotium rolfsii Gene Expression during Sclerotium Development
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Specimen Collection
2.2. RNA Extraction
2.3. Library Construction and Sequencing
2.4. De Novo Assembly
2.5. Unigene Functional Analysis
2.6. Gene Quantification
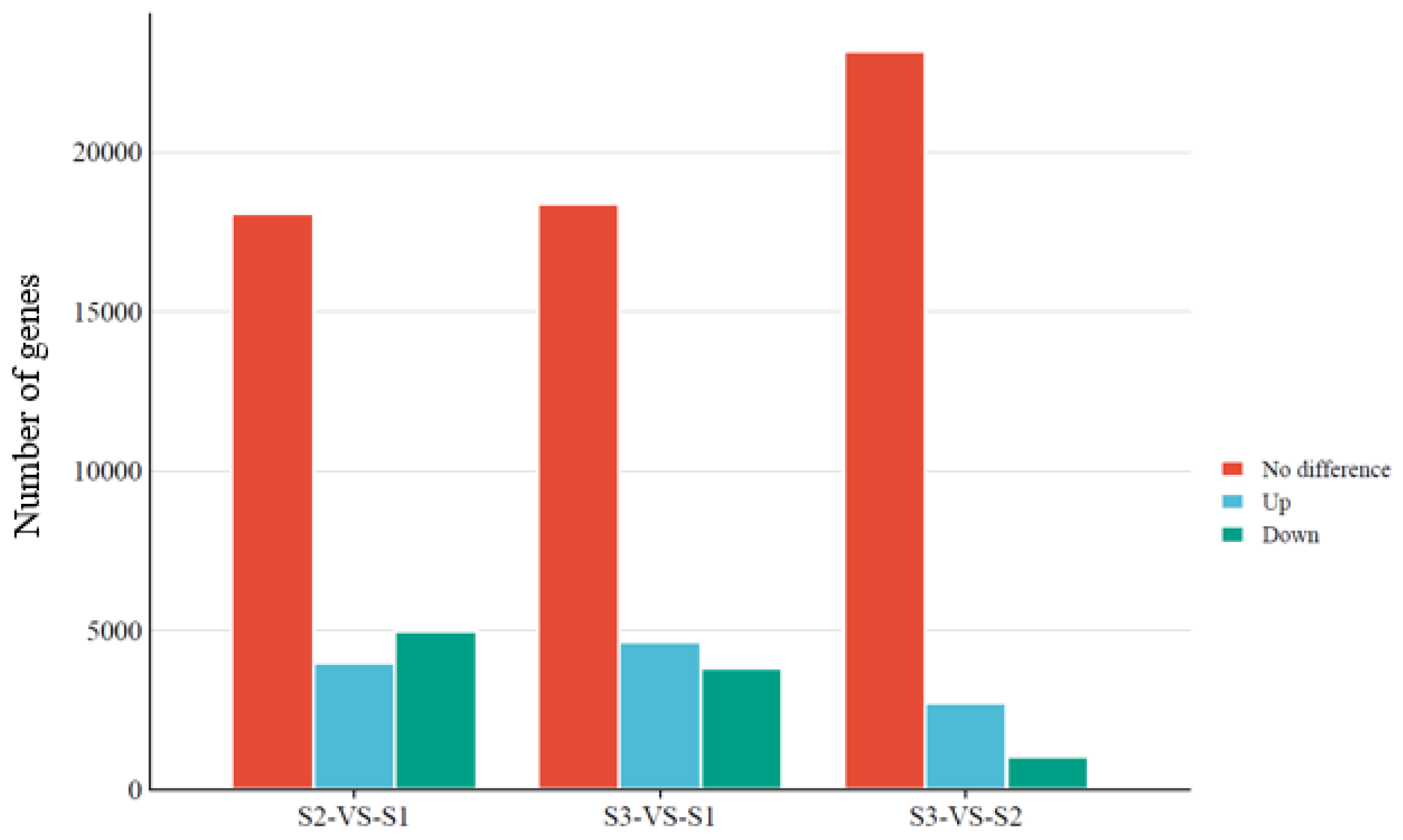
2.7. Analysis of Differentially Expressed Genes
2.8. Weighted Gene Co-Expression Network Analysis
2.9. Quantitative Real-Time PCR (qPCR) Validation of mRNA Expression
3. Results
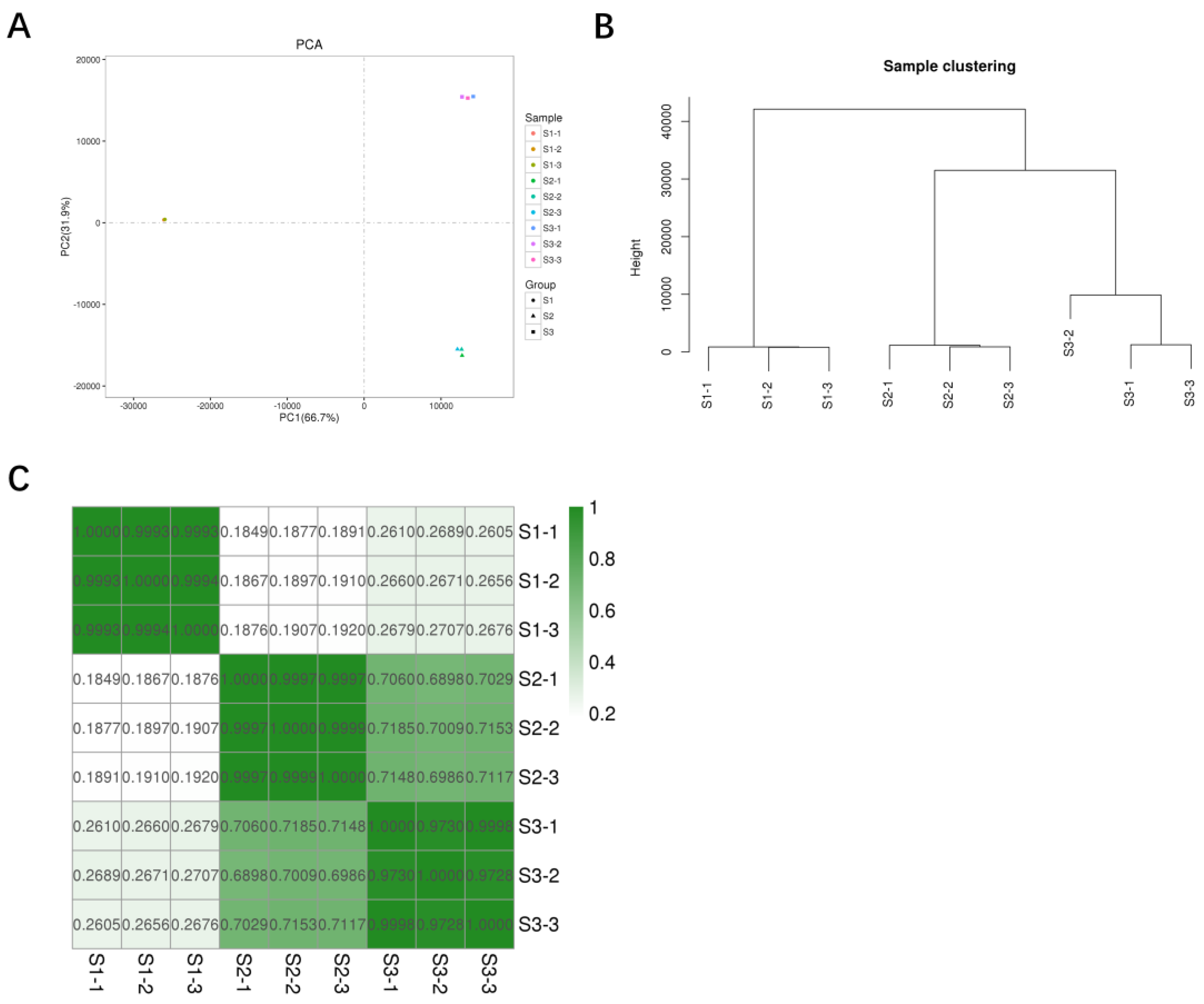
3.1. RNA-Seq of S. rolfsii during Sclerotial Development
3.2. Functional Annotation
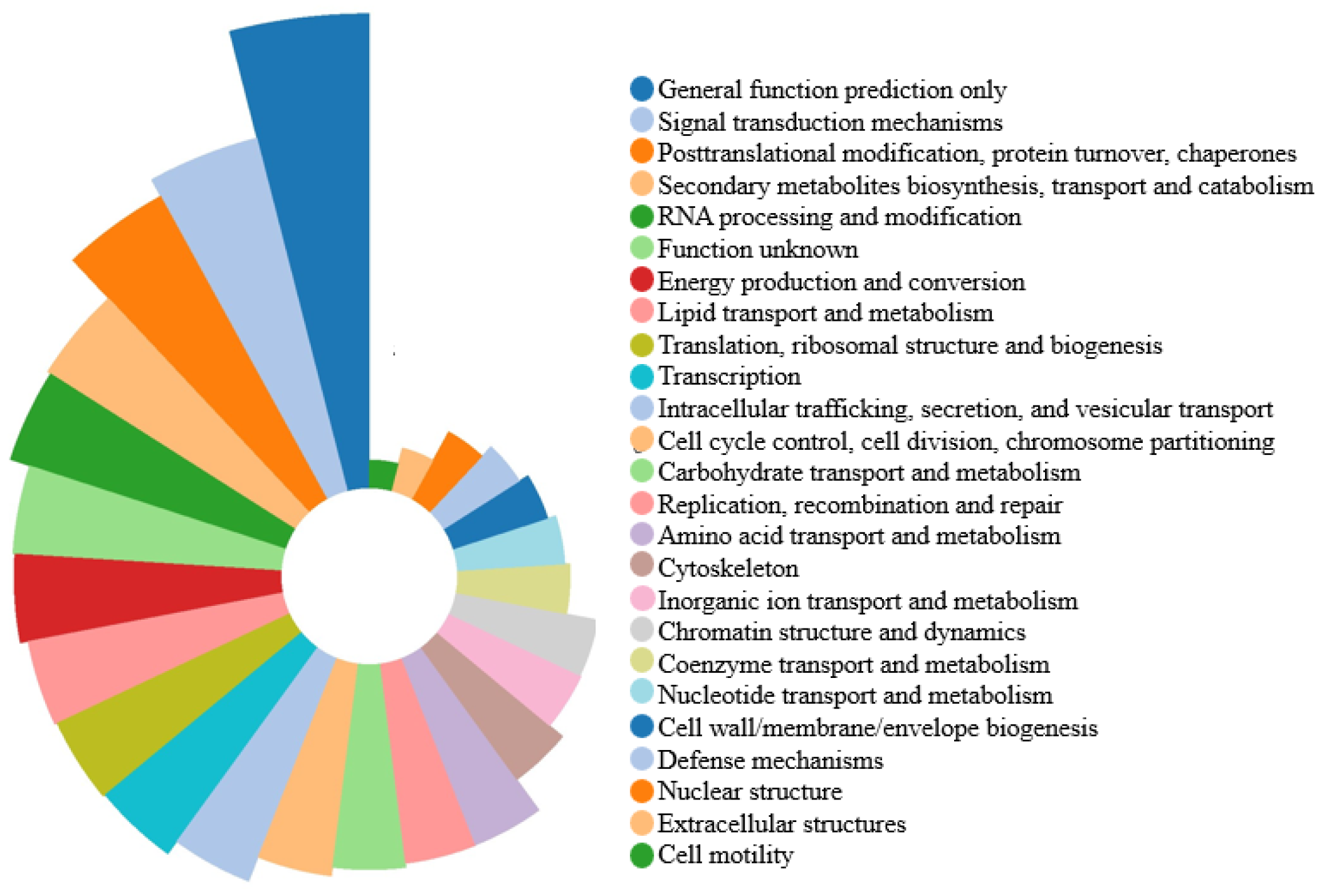
3.3. Classification of GO and KOG
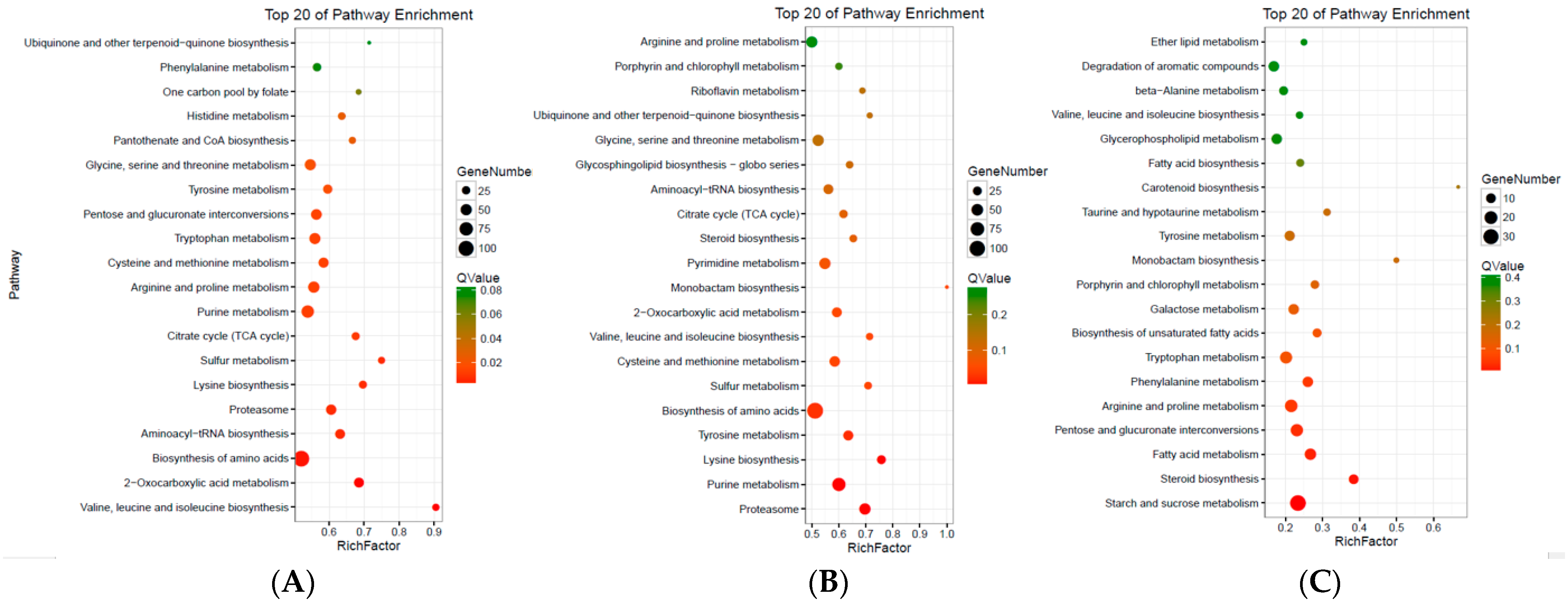
3.4. KEGG Pathway Analysis
3.5. Functional Distribution of DEGs between Mycelium and Sclerotial Developmental Stages
3.6. STEM Analysis of DEGs Involved in Sclerotium Formation
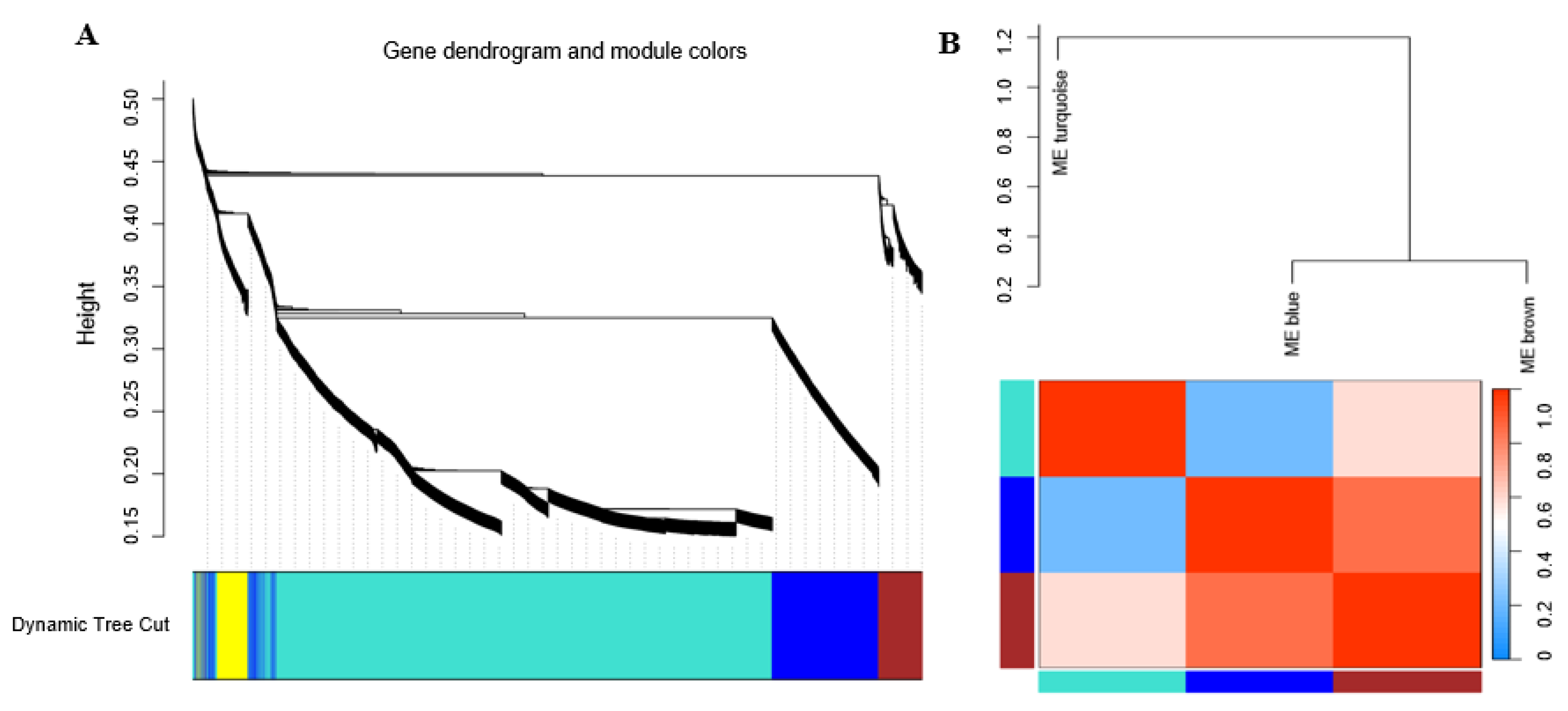
3.7. Correlation between Modules and Identification of Key Modules
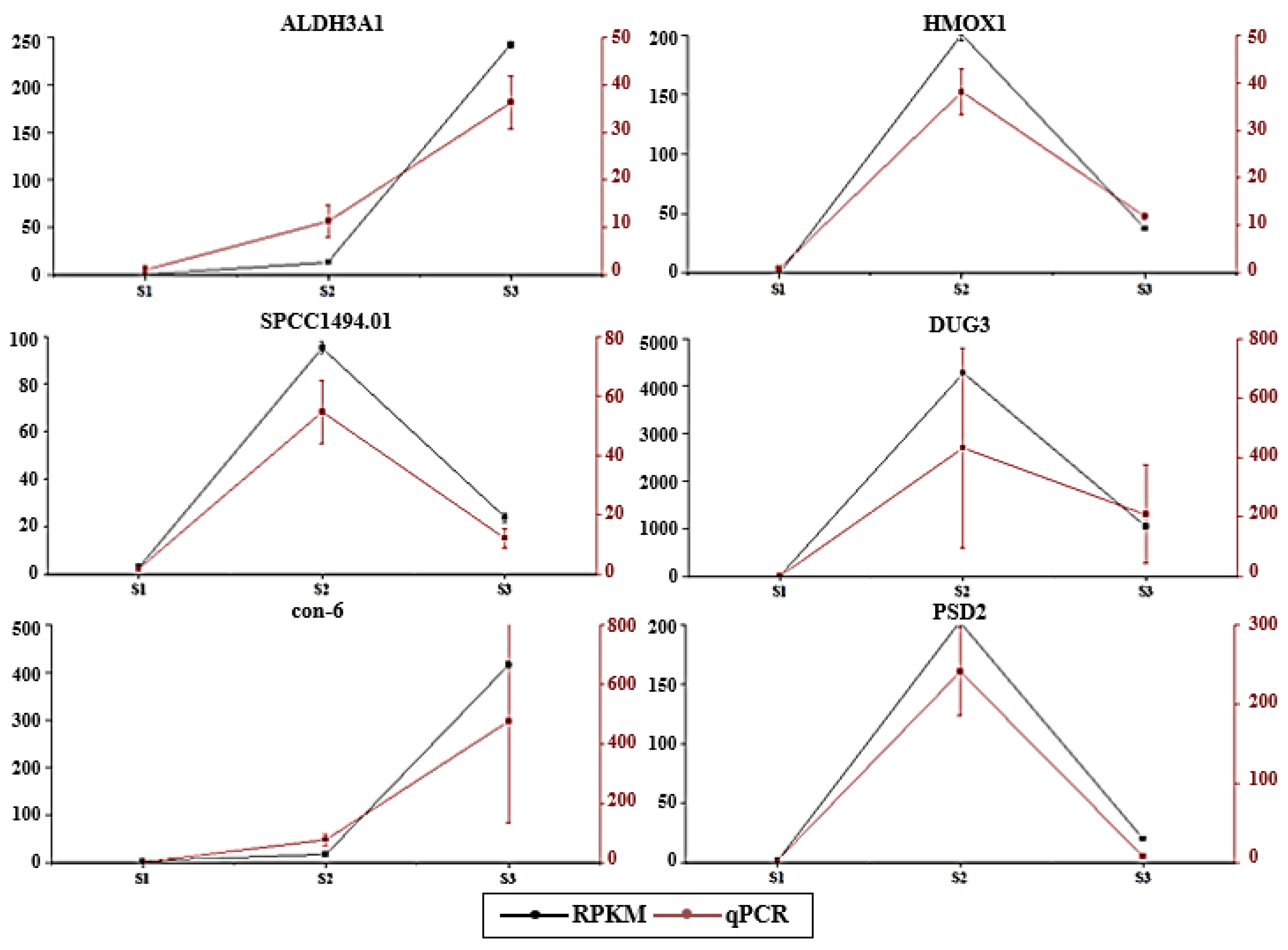
3.8. Validation of Differential Expression Profiles Using qPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dwivedi, S.K.; Prasad, G. Integrated management of Sclerotium rolfsii: An overview. Eur. J. Biomed. Pharm. Sci. 2016, 3, 137–146. [Google Scholar]
- Punja, Z.K.; Huang, J.S.; Jenkins, S.F. Relationship of mycelial growth and production of oxalic acid and cell wall degrading enzymes to virulence in Sclerotium rolfsii. Can. J. Plant Pathol. 1985, 7, 109–117. [Google Scholar] [CrossRef]
- Yu, D.Y.; Yan, L.Y.; Song, W.D.; Kang, Y.P.; Lei, Y.; Chen, Y.N.; Huai, D.X.; Wang, X.; Wang, Z.H.; Luo, H.Y.; et al. Progress on pathogenicity differentiation in Scletotium rolfsii isolates from peanut. Chin. J. Oil Crop Sci. 2022, 44, 930–936. [Google Scholar]
- Sun, S.; Sun, F.; Deng, D.; Zhu, X.; Zhu, Z. First report of southern blight of mung bean caused by Sclerotium rolfsii in China. Crop Prot. 2019, 130, 105055. [Google Scholar] [CrossRef]
- Yu, L.L.; Zhou, S.X.; Nie, Q.; Hsiang, T.; Zhou, Y. First report of Sclerotium rolfsii causing southern blight of Bletilla orchid in China. Plant Dis. 2018, 103, 762. [Google Scholar] [CrossRef]
- Yi, R.H.; Liao, H.Z.; Li, K.Y.; Feng, F. Fruit rot caused by Athelia rolfsii (Sclerotium rolfsii) on jackfruit (Artocarpus heterophyllus) in China. Plant Dis. 2023, 107, 3314. [Google Scholar]
- Ünal, F.; Aşkın, A.; Koca, E.; Yıldırır, M.; Bingöl, M.Ü. Mycelial compatibility groups, pathogenic diversity and biological control of Sclerotium rolfsii on turfgrass. Egypt. J. Biol. Pest Control. 2019, 29, 44. [Google Scholar] [CrossRef]
- Li, M.; Li, S.; Zhang, Z.X.; Du, P.Q.; Zhou, L.; Dong, W.S. Sensitivity to carboxin and population diversity of Sclerotium rolfsii from peanut in Henan province. J. Henan Agric. Sci. 2021, 50, 64–73. [Google Scholar]
- Georgiou, C.D.; Zervoudakis, G.; Tairis, N.; Kornaros, M. β-carotene production and its role in sclerotial differentiation of Sclerotium rolfsii. Fungal Genet. Biol. 2001, 34, 11–20. [Google Scholar] [CrossRef]
- Xing, Y.M.; Yin, W.Q.; Liu, M.M.; Wang, C.L.; Guo, S.X. Oxalic acid and sclerotial differentiation of Polyporus umbellatus. Sci. Rep.-UK 2015, 5, 10759. [Google Scholar] [CrossRef]
- Singh, U.P.; Sarma, B.K.; Singh, D.P.; Amar, B. Studies on exudate-depleted sclerotial development in Sclerotium rolfsii and the effect of oxalic acid, sclerotial exudate, and culture filtrate on phenolic acid induction in chickpea (Cicer arietinum). Can. J. Microbiol. 2002, 48, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, U.; Dee, M.E.; Ownley, B.H.; Butler, D.M. Anaerobic soil disinfestation reduces germination and affects colonization of Sclerotium rolfsii sclerotia. Phytopathology 2018, 108, 342–351. [Google Scholar] [CrossRef]
- Cheng, X.H.; Guo, S.X.; Wang, C.L. Factors influencing formation of sclerotia in Grifola umbellate (Pers.) Pilát under artificial conditions. J. Integr. Plant Biol. 2010, 48, 1312–1317. [Google Scholar] [CrossRef]
- Xing, Y.M.; Zhang, L.C.; Liang, H.Q.; Lv, J.; Song, C.; Guo, S.X.; Wang, C.L.; Lee, T.S.; Lee, M.W. Sclerotial formation of Polyporus umbellatus by low temperature treatment under artificial conditions. PLoS ONE 2013, 8, e56190. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Zumaquero, A.; Kanematsu, S.; Nakayashiki, H.; Matas, A.; Martínez-Ferri, E.; Barceló-Muóz, A.; Pliego-Alfaro, F.; López-Herrera, C.; Cazorla, F.M.; Pliego, C.J. Transcriptome analysis of the fungal pathogen Rosellinia necatrix during infection of a susceptible avocado rootstock identifies potential mechanisms of pathogenesis. BMC Genom. 2019, 20, 1016. [Google Scholar] [CrossRef] [PubMed]
- Nagalakshmi, U.; Waern, K.; Snyder, M. RNA-Seq: A method for comprehensive transcriptome analysis. Curr. Protoc. Mol. Biol. 2010, 89, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, C.; Folly, Y.M.E.; Chang, J.; Wang, Y.; Zhou, L.; Zhang, H.; Liu, Y. Morphological and transcriptomic analysis of the inhibitory effects of Lactobacillus plantarum on Aspergillus flavus growth and aflatoxin production. Toxins 2019, 11, 636. [Google Scholar] [CrossRef] [PubMed]
- Gkarmiri, K.; Finlay, R.D.; Alström, S.; Thomas, E.; Cubeta, M.A.; Högberg, N. Transcriptomic changes in the plant pathogenic fungus Rhizoctonia solani AG-3 in response to the antagonistic bacteria Serratia proteamaculans and Serratia plymuthica. BMC Genom. 2015, 16, 630. [Google Scholar] [CrossRef]
- Wu, Y.; Zhu, W.; Wei, W.; Zhao, X.; Wang, Q.; Zeng, W.; Zheng, Y.; Chen, P.; Zhan, S. De novo assembly and transcriptome analysis of sclerotial development in Wolfiporia cocos. Gene 2016, 588, 149–155. [Google Scholar] [CrossRef]
- Schmid, J.; Müller-Hagen, D.; Bekel, T.; Funk, L.; Stahl, U.; Sieber, V.; Meyer, V. Transcriptome sequencing and comparative transcriptome analysis of the scleroglucan producer Sclerotium rolfsii. BMC Genom. 2010, 11, 329. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Tang, T.; Mao, T.; Duan, Y.; Guo, X.; You, J. Development of EST-SSR primers and genetic diversity analysis of the southern blight pathogen Sclerotium rolfsii using transcriptome data. Front. Microbiol. 2023, 14, 1152865. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.J. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; Mccue, K.; Schaeffer, L.; Wold, B.J. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Ernst, J.; Bar-Joseph, Z. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinform. 2006, 7, 191. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S.J.B.B.; Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2009, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Qiu, Y.; Zhao, R.; Hou, J.; Tu, L.; Nie, Z.; Wang, J.; Zheng, Y.; Wang, M. Transcriptomics and metabolomics analysis of Sclerotium rolfsii fermented with differential carbon sources. Foods 2022, 11, 3706. [Google Scholar] [CrossRef]
- Yu, J.J.; Yu, M.N.; Nie, Y.F.; Sun, W.X.; Yin, X.L.; Zhao, J.; Wang, Y.H.; Ding, H.; Qi, Z.Q.; Du, Y.; et al. Comparative transcriptome analysis of fruiting body and sporulating mycelia of Villosiclava virens reveals genes with putative functions in sexual reproduction. Curr. Genet. 2016, 62, 575–584. [Google Scholar] [CrossRef]
- Liu, X.; Xie, J.; Fu, Y.; Jiang, D.; Cheng, J. The subtilisin-like protease Bcser2 affects the sclerotial formation, conidiation and virulence of Botrytis cinerea. Int. J. Mol. Sci. 2020, 21, 603. [Google Scholar] [CrossRef]
- Shu, C.; Zhao, M.; Anderson, J.P.; Garg, G.; Zhou, E. Transcriptome analysis reveals molecular mechanisms of sclerotial development in the rice sheath blight pathogen Rhizoctonia solani AG1-IA. Funct. Integr. Genom. 2019, 19, 743–758. [Google Scholar] [CrossRef]
- Vijayakumar, R.; Zhao, C.X.; Gopal, R.; Jaleel, C.A. Non-enzymatic and enzymatic antioxidant variations in tender and mature leaves of Strychnos nux-vomica L. (Family: Loganiaceae). C. R. Biol. 2009, 332, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, J. DHN melanin biosynthesis in the plant pathogenic fungus botrytis cinerea is based on two developmentally regulated key enzyme (PKS)-encoding genes. Mol. Microbiol. 2016, 99, 729–748. [Google Scholar] [CrossRef] [PubMed]
- Maiuri, M.C.; Zalckvar, E.; Kimchi, A.; Kroemer, G. Self-eating and self-killing: Crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2007, 8, 741–752. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Tan, J.; Miao, Y.; Lei, P.; Zhang, Q. Ros and autophagy: Interactions and molecular regulatory mechanisms. Cell Mol. Neurobiol. 2015, 35, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Baruah, K.; Norouzitallab, P.; Linayati, L.; Sorgeloos, P.; Bossier, P. Reactive oxygen species generated by a heat shock protein (Hsp) inducing product contributes to hsp70 production and hsp70-mediated protective immunity in artemia franciscana against pathogenic vibrios. Dev. Comp. Immunol. 2014, 46, 470–479. [Google Scholar] [CrossRef] [PubMed]
- Rollins, J.A.; Dickman, M.B. Increase in endogenous and exogenous cyclic AMP levels inhibits sclerotial development in Sclerotinia sclerotiorum. Appl. Environ. Microb. 1998, 64, 2539–2544. [Google Scholar] [CrossRef]
- Harel, A.; Gorovits, R.; Yarden, O. Changes in protein kinase a activity accompany sclerotial development in Scierotinia sclerotiorum. Phytopathology 2005, 95, 397–404. [Google Scholar] [CrossRef]
- Schlicker, C.; Hall, R.A.; Vullo, D.; Middelhaufe, S.; Gertz, M.; Supuran, C.T.; Mühlschlegel, F.A.; Steegborn, C. Structure and inhibition of the CO2-sensing carbonic anhydrase Can2 from the pathogenic fungus Cryptococcus neoformans. J. Mol. Biol. 2009, 385, 1207–1220. [Google Scholar] [CrossRef]
- Teh, A.H.; Sim, P.F.; Hisano, T. Structural basis for binding uronic acids by family 32 carbohydrate-binding modules-ScienceDirect. Biochem. Biophys. Res. Commun. 2020, 533, 257–261. [Google Scholar] [CrossRef]



| Annotation Database | No. of Annotation | Percent of Annotation (%) |
|---|---|---|
| Total Unigenes | 28,158 | 100% |
| NR | 14,763 | 52.43% |
| SwissProt | 9188 | 32.63% |
| GO | 10,275 | 36.49% |
| KEGG | 3096 | 11% |
| Annotation genes | 15,322 | 54.41% |
| Without annotation gene | 12,836 | 45.59% |
| Sample Name | Raw Reads | Clean Reads | Clean Bases | Q20 (%) | Q30 (%) | Content (%) |
|---|---|---|---|---|---|---|
| S1-1 | 27,778,288 | 27,173,090 | 2.72G | 98.56 | 95.5 | 52.17 |
| S1-2 | 27,374,584 | 26,896,318 | 2.69G | 98.7 | 95.8 | 52.25 |
| S1-3 | 23,420,774 | 22,913,500 | 2.29G | 98.58 | 95.5 | 52.3 |
| S2-1 | 25,401,562 | 24,936,756 | 2.49G | 98.69 | 95.83 | 51.62 |
| S2-2 | 23,896,106 | 23,481,220 | 2.35G | 98.69 | 95.77 | 51.59 |
| S2-3 | 27,575,880 | 27,038,696 | 2.70G | 98.61 | 95.59 | 51.67 |
| S3-1 | 27,697,540 | 27,164,438 | 2.72G | 98.6 | 95.55 | 52.14 |
| S3-2 | 29,499,244 | 28,988,848 | 2.90G | 98.69 | 95.82 | 51.74 |
| S3-3 | 25,507,578 | 25,025,724 | 2.50G | 98.62 | 95.57 | 52.21 |
| Total | 238,151,556 | 233,618,590 | 23.36 | 887.74 | 860.93 | 467.69 |
| Genes Num | GC Percentage | N50 | Max Length | Min Length | Average Length | Total Assembled Bases |
|---|---|---|---|---|---|---|
| 28,158 | 48.29% | 2379 | 16,402 | 201 | 1284 | 36,165,475 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, F.; Wang, X.; Tang, T.; Duan, Y.; Mao, T.; Guo, X.; Wang, Q.; You, J. De Novo RNA Sequencing and Transcriptome Analysis of Sclerotium rolfsii Gene Expression during Sclerotium Development. Genes 2023, 14, 2170. https://doi.org/10.3390/genes14122170
Wang F, Wang X, Tang T, Duan Y, Mao T, Guo X, Wang Q, You J. De Novo RNA Sequencing and Transcriptome Analysis of Sclerotium rolfsii Gene Expression during Sclerotium Development. Genes. 2023; 14(12):2170. https://doi.org/10.3390/genes14122170
Chicago/Turabian StyleWang, Fanfan, Xiaoyue Wang, Tao Tang, Yuanyuan Duan, Ting Mao, Xiaoliang Guo, Qingfang Wang, and Jingmao You. 2023. "De Novo RNA Sequencing and Transcriptome Analysis of Sclerotium rolfsii Gene Expression during Sclerotium Development" Genes 14, no. 12: 2170. https://doi.org/10.3390/genes14122170
APA StyleWang, F., Wang, X., Tang, T., Duan, Y., Mao, T., Guo, X., Wang, Q., & You, J. (2023). De Novo RNA Sequencing and Transcriptome Analysis of Sclerotium rolfsii Gene Expression during Sclerotium Development. Genes, 14(12), 2170. https://doi.org/10.3390/genes14122170





