Abstract
Family-1 UDP-glycosyltransferases (UGTs) are the most common and functional glycosyltransferases in the plant world. UGT is closely related to plant growth and the response to abiotic stress. However, despite systematic research, our understanding of potato UGT genes is still unclear. In this study, we identified 174 potato UGT proteins based on their conserved plant secondary product glycosyltransferase (PSPG) motifs. Phylogenetic analyses were used to compare these proteins with Arabidopsis UGTs and other plant UGTs, and it was found that they could be clustered into 18 distinct groups. Patterns of intron gain/loss and intron phases within potato UGTs revealed highly conserved intron insertion events. The promoter cis-elements of these 174 UGT genes were systematically investigated. The promoter regions of these UGT genes are known to contain various classes of cis-acting compounds. These include elements that are light-responsive, phytohormone-responsive, and stress-responsive. Transcriptome data analysis established that 25, 10, 6, and 4 of these 174 UGT genes were specifically expressed in leaves, roots, stolons, and young tubers, respectively. The mannitol-treated transcriptomic data showed thirty-eight UGT genes were significantly upregulated. The quantitative real-time PCR results showed that the four genes were all responsive to osmotic stress under a 10% PEG6000 treatment. The results of our study provide a basis for clarifying the molecular mechanism of potato osmotic stress resistance and better understanding its function in the future.
1. Introduction
Glycosylation is one of the most important protein translational modification reactions and is the last step in the synthesis of natural compounds, which promotes the diversity and complexity of plant secondary metabolic reactions [1]. Glycosylation is catalyzed by glycosyltransferase enzymes (GTs) and catalyzes the transfer of sugar moieties from donor molecules to specific acceptor molecules (sugars, lipids, etc.) [2]. The latest data from CAZy show that GTs are classified into 114 families in different species by their 3D structure (fold GT-A, GT-B, or predicted GT-C) and their mechanism (reversal or retention) [3]. Family-1 GTs comprise the largest family out of all subfamilies [4].
Family-1 GTs are the most common and functional glycosyltransferases in the plant world [5], in which the donor chemical is uridine 5′-diphosphate. Analysis of the UGTs’ sequences shows that their extremely varied N-terminal region is able to detect a range of substrates [6]. There is a motif in the C-terminal region termed PSPG-Box which is responsible for combining UDP sugars [7]. The PSPG-box is a highly conserved region comprising 44 amino acids in the UGT of all studied plants [8]. As their amino acid sequence and conserved domain have been identified, UGTs are no longer mysterious in many species. In Arabidopsis thaliana, there are a total of 120 UGT genes that were divided into 14 groups by phylogenetic analysis (groups A to N) [9]. Ninety-six UGTs were identified in Cicer arietinum, and their intron–exon structure was analyzed [10]. In Zea mays, 147 UGTs were identified, and then a phylogenetic tree was constructed with 18 UGTs of Arabidopsis and 2 UGTs of rice. The UGTs were divided into 17 groups, including 14 conserved groups in Arabidopsis and 3 new groups [11]. Most recently, in Gossypium hirsutum [12], Triticum aestivum L. [6], and Citrus grandis [13], 274, 179, and 145 UGTs were identified, respectively.
UGTs are present in many species and play important roles in the growth of plants [14]. Studies have shown that UGTs recognize different substrates, such as, for example, flavonoids, terpenoids, auxin, salicylic acid, and sterols [15]. These molecules can be glycosylated to enhance their activity, solubility, chemical stability, or biological activity and promote storage and accumulation in plant cells [16]. Moreover, recent studies have shown that many glycosyltransferase genes are related to abiotic stresses. For example, in Arabidopsis, overexpression of UGT75B1 genes increased the seed germination rate and seedling greening rate under salt and osmotic stresses [17]. Meanwhile, overexpression of UGT79B2/B3 greatly increased plant resistance to drought, low temperatures, and salt stressors; conversely, double mutants of UGT79B2 and B3 were more vulnerable to unfavorable circumstances [18]. Zhao et al.’s research demonstrated that inhibiting CsUGT78A14 reduced flavonoid accumulation and the reactive oxygen species scavenging ability, thus reducing the resistance of tea plants under cold stress. Inhibiting the expression of UGT91Q2 reduces cold stress by reducing accumulation of nerolidol glucoside and the reactive oxygen species (ROS) scavenging capacity in Camellia sinensis transgenic plants [19]. The involvement of AtUGT73C6 and AtUGT78D1 in the production of flavonoid glycosides in Arabidopsis has been verified [20]. Summarizing the above, flavonoids are decisive in abiotic stress. On the other hand, plant UGTs catalyze ABA glycosylation. For instance, UGT71B6 is able to identify (+)-ABA, the ABA enantiomer that occurs naturally [21]. Mutations in the UGT71C5 gene enhance drought tolerance in Arabidopsis [22]. UGT71B7 and UGT71B8 are induced by ABA, NaCl, and mannitol [23]. These results show that UGT genes are involved in responding to stress by altering ABA glycosylation.
Potato (Solanum tuberosum L.) is the world’s most important non-grain food crop and the fourth-largest crop in terms of production volume [24]. In previous studies, it was found that anthocyanin 5-O-glucosyltransferase (5-UGT) functions to modulate the process of potato tuber metabolism. The ectopic expression of 5-UGT enhances the pathogen infection resistance of potato to Erwinia [25]. Meanwhile, its overexpression increases the level of glycosylated anthocyanidins in transgenic potato plants. However, a systematic investigation of UGT genes under abiotic stress in potato still needs to be carried out. In the process of potato cultivation, abiotic stresses, especially osmotic stresses, are not conducive to potato growth, for example, in a water-scarce or high-salinity environment, resulting in ROS accumulation, cell death, and plant yield inhibition [26]. Studies have demonstrated that flavonoids are essential secondary metabolites (SMs) that are crucial in preventing and reducing the harm caused by osmotic stress [27].
In this study, we identified 174 UGT proteins from potato that could be divided into 18 groups. Transcriptome data showed that 38 UGT genes were significantly induced by a mannitol treatment. Four UGT genes were selected for further qRT-PCR verification. These genes were upregulated by a 10% PEG6000 stress treatment, which indicates that these genes might participate in the osmotic stress response. Overall, our findings provide candidate UGT genes involved in osmotic stress for further functional study.
2. Materials and Methods
2.1. Identification of UGT Genes in Potato
The genome and genome annotation files (GFF3) of potato (estimated size: 785 MB with 56210 genes) were downloaded from Ensembl Plant (https://ensembl.gramene.org/Solanum_tuberosum/Info/Index, accessed on 17 October 2022). Using Tbtools (version No.2.007), a query was created using the 44 amino acid conserved sequences of the PSPG motif to search the potato genome database [28]. The expectation-value (E-value) cutoff was −5. The NCBI (https://www.ncbi.nlm.nih.gov/, accessed on 25 October 2022) was used to confirm each predicted potato UGT protein sequence.
2.2. Phylogenetic Analysis of Potato UGT Genes
Using MEGA-X’s ClustalW 10.0 software, multiple alignments of the potato UGT amino acid sequences were performed. With the bootstrap set to 1000 replicates and the neighbor-joining method enabled, the phylogenetic analysis was completed.
2.3. Chromosomal Locations
The chromosome position information of potato UGTs was obtained through the genome annotation files (GFF3). We also made use of Tbtools’ Drawing Atlas.
2.4. Intron Mapping
By identifying the intron splice locations, phases, and placements, the potato UGT intron map was created. Using CDSs and genomic sequences, the online Gene Structure Display Server 2.0 (http://gsds.gao-lab.org, accessed on 5 November 2022) was able to obtain the exon–intron structure and intron phases. After aligning every potato UGT, an amino acid sequence was created, and the introns were serially numbered in accordance with those places. The following were used to identify the intron phases: phase 0 corresponded to introns between two codons, phase 1 corresponded to introns after the first base in the codon, and phase 2 corresponded to introns after the second base in the codon [29].
2.5. Promoter Analysis of UGTs
The 1500 bp sequence before the CDS in UGTs, as the promoter region, was predicted using the PlantCARE website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 25 October 2023).
2.6. Plant Materials and Treatment
S. tuberosum L. Desiree (from the College of Agriculture, Yunnan University; it is a tetraploid) was used for analysis in this study. Four-week-old tissue culture plantlets’ stems were cut to a length of about 2 cm, and they were then placed in sterile glass jars filled with Murashige–Skoog (MS) liquid medium. They were placed in the culture room (light conditions for 16 h, 8 h of dark, and a temperature of 22 °C).
Four-week-old plants were cultivated in MS liquid medium containing 10% PEG6000 and were sampled at 0 h, 6 h, 12 h, and 24 h after treatment.
2.7. Expression Analysis and Quantitative Real-Time PCR
RNA-seq data of the potato UGT genes were obtained from the published sequencing file [30]. The generated heat maps use 7 distinct tissues of RH (RH89039-16—material in transcriptome sequencing; it is a diploid), as well as the control and mannitol treatments with DM (DM1-3 516 R44—material in transcriptome sequencing; it is a diploid).
For RNA extraction, the instructions provided in the reference kit (Takara, Dalian, China. code 9769) are used to isolate total RNA from a variety of potato tissues. Following the protocol, 0.5 μg RNA was used for reverse transcription reactions, and the Prime-Script RT reagent kit with a gDNA Eraser (Takara, Code No. 6110B) was used. Using a Bioer Technology FQD-96A and Tsingke 2×T5 Fast qPCR Mix (SYBR Green I), the qRT-PCR was performed, with three biological replicates. Elongation factor 1-α (Ef1α) served as the housekeeping gene in the qRT-PCR [31]. The 2−ΔΔCt technique was utilized to ascertain the relative expression levels [32]. GraphPad prism 8.0 (https://www.graphpad.com/, accessed on 4 March 2023) was used to analyze the data and present them in graphs. Tukey’s pairwise comparison was used to test the significance. The relevant assay primers are included in Supplementary Table S7.
3. Results
3.1. Identification and Characterization of the UGTs
Recently, the potato genome was sequenced to aid in the identification of potato gene families [30]. To identify the UGT family members in the potato, we used a conserved UGT domain of 44 amino acids called the PSPG-box to identify the UGT proteins in the potato genome. Blastp searches against the 56,210 potato protein models were conducted, and genes encoding identical proteins or sequences that were either too long or too short were eliminated [33]. A total of 174 UGT proteins were identified (Supplementary Table S1). Using the Ensemble potato sequence database and published sequencing files, detailed information was gathered about the potato UGT genes, such as the gene ID, transcript ID, length of the protein, molecular weight, isoelectric point, chromosome locations, splicing variants, and the putative function of the UGTs (Supplementary Table S1). The 174 UGT genes encoded proteins ranging in length from 352 (PGSC0003DMG400017249) to 506 (PGSC0003DMG400011740) amino acids, and the average length of the amino acids was 460. The gene molecular weight (MW) ranged from 39.428 to 57.037 kDa, and the isoelectric point (pI) ranged from 4.79 to 9.22. Their transcripts were encoded by 1-6 exons (PGSC0003DMG400025532 contains 6 exons), and they had from 1 to 3 splicing variants.
3.2. Phylogenetic Analysis of Potato UGT Genes
To study the evolutionary relationships between potato, Arabidopsis, maize, and rice UGT proteins, a phylogenetic tree was constructed with the four species (174 UGTs of potato, 17 UGTs (AtUGT79B2, AtUGT89B1, AtUGT90A1, AtUGT73B3, AtUGT71B1, AtUGT78D3, AtUGT85A1, AtUGT76B1, AtUGT83A1, AtUGT87A1, AtUGT86A1, AtUGT75C1, AtUGT92A1, AtUGT82A1, AtUGT73C6, AtUGT78D1, AtUGT76C2) of Arabidopsis, 3 UGTs (GRMZM2G075387, GRMZM5G834303, GRMZM2G067424) of maize, and 1 UGT (Oso2g0755500UGT85E1) of rice (Supplementary Table S2)). Phylogenetic analysis was used to divide the 174 UGT proteins into 18 groups (Figure 1). The distribution of all UGT proteins in each branch of the phylogenetic tree was uneven. It was found that 23 and 6 UGTs were clustered into two new groups, named groups O and P (Figure 1). In addition, we named a novel separate group R (Figure 1). There was only the S. tuberosum UGT (StUGT) protein in group C, and it was more abundant. Some of these groups have only StUGTs and Arabidopsis UGTs, where the specific distribution is as follows (total, potato, Arabidopsis): group D (26, 24, 2), group E (19, 18, 1), group L (19, 18, 1), group A (18, 17, 1), group I (3, 2, 1), group N (2, 1, 1), and so on. Group G also had the rice UGT (total, potato, Arabidopsis, rice): Group G (14, 12, 1, 1). Groups O and P contain potato, Arabidopsis, and rice UGTs. Group Q was not found to have potato UGTs.
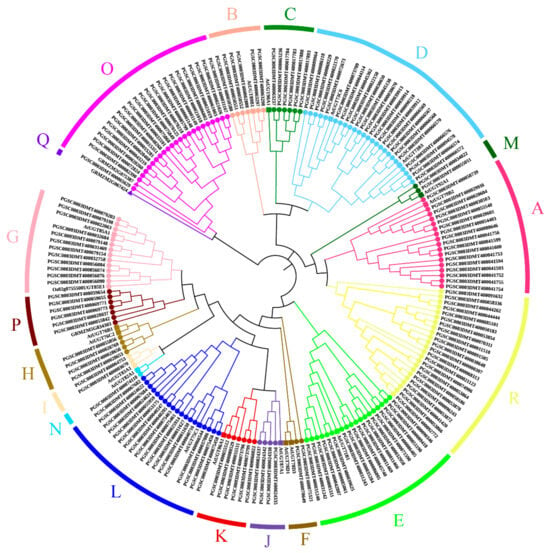
Figure 1.
Phylogenetic analysis of the UGT proteins from potato, Arabidopsis, maize, and rice. A total of 174 UGTs were divided into 18 groups (groups A–R). Bootstrap values over 50% are indicated above the nodes.
The distribution of plant UGTs in the phylogenetic groups is summarized in Table 1. In regard to potato, groups D, O, and R expanded more than the other groups. Group N has only one member. Group D is the largest group of potato UGTs as it has 24 members and is clustered with functionally characterized AtUGT73C6 and AtUGT73B3 (Figure 1). In group F and group H, there are also two Arabidopsis UGTs in each, but they only have two and four potato UGTs, respectively. This indicates that group D may play an important role in potato.

Table 1.
The number of UGTs in different phylogenetic groups.
3.3. Chromosomal Locations
Based on the existing annotation information of the potato genome, the genetic map of the UGT genes on the potato chromosome was further studied (Figure 2). Potato has 167 UGTs distributed across all 12 chromosomes, with 7 UGTs located on chromosome 00 (Figure 2). This chromosome is different from other chromosomes because it contains only one group (group R). There were 21 UGTs on chromosome 11, which mainly contains group A, group R, group K, and group R. They are concentrated in the range of 0 mb to 20 mb, followed by 19 UGTs on chromosome 2. Chromosome 06 had the smallest number (5) of UGTs.
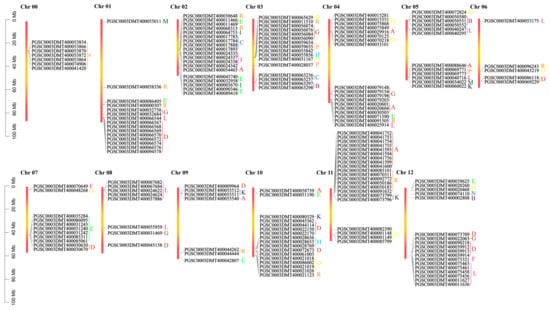
Figure 2.
Chromosome distribution of potato UGT genes. Each chromosome is shown with its chromosome number at the top. Different colored letters represent different phylogenetic groups of potato UGT genes.
Combined with the above phylogenetic tree, it was found that there were different groupings on each chromosome (except for chromosome 11). Chromosome 12 in particular contains eight groups (groups B, D, E, F, G, L, N, and O) (Figure 2). Group R, consisting of 23 potato UGTs, had 2, 2, 1, 1, 1, 2, 1, and 6 members located on chromosome 01, 02, 03, 04, 06, 09, 10, and 11. Group D, consisting of 24 potato UGTs, had 8 members located on chromosomes 01 and 2, 2, 1, 1, 5, and 5 members on chromosomes 06, 07, 08, 09, 10, and 12 (Figure 2).
3.4. Analysis of Intron Gain/Loss Events
To investigate the evolutionary relationships within the potato UGT gene family, the gene exon–intron structure was analyzed. Among the 174 sequences, 107 have no introns, while 61 and 4 have one and two introns each, respectively. In addition, PGSC0003DMT400031469 contains three introns and PGSC0003DMT400065629 has five introns (Figure 3). At least 10 intron insertion events occurred, numbered I-1 to I-10 according to their positions. Highly conserved introns were observed for intron I-5, which is contained in 39 (58%) potato UGTs belonging to groups A, E, F, G, I, J, K, N, and P (Figure 3). In addition, intron I-6 is concentrated in 12 sequences of a subgroup of group L. There were clearly intron gain/loss events that occurred in potato UGTs. Members from groups F, G, I, J, K, N, and P all gained intron 5, except PGSC0003DMT400079198, which lost intron 5 but gained intron 6. Meanwhile, for PGSC0003DMT400031469, it has intron 7 in addition to intron 5 and intron 6 (Figure 3).
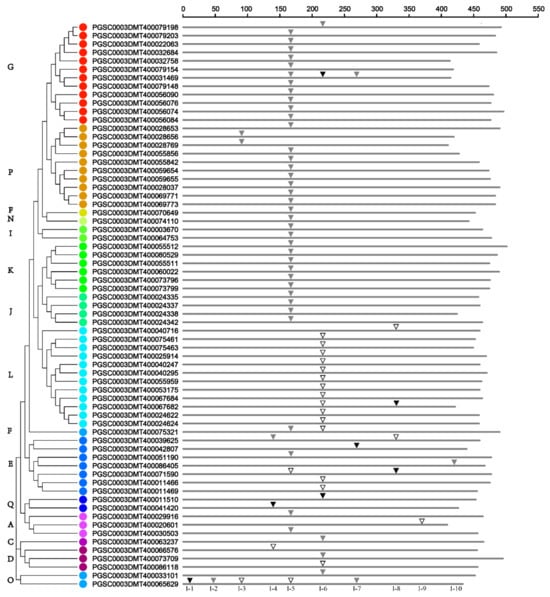
Figure 3.
Distribution of introns among 67 UGT genes in potato. According to the alignment of their amino acid sequences encoded by the UGT genes, the introns are mapped and numbered. Intron phases 0, 1, and 2 are indicated by open inverted triangles ( ), slash filled inverted triangles (▽), and black inverted triangles (▼), respectively. The phylogenetic relationships of potato UGT proteins can be seen on the left, and different phylogenetic groups are distinguished by colored dots.
), slash filled inverted triangles (▽), and black inverted triangles (▼), respectively. The phylogenetic relationships of potato UGT proteins can be seen on the left, and different phylogenetic groups are distinguished by colored dots.
 ), slash filled inverted triangles (▽), and black inverted triangles (▼), respectively. The phylogenetic relationships of potato UGT proteins can be seen on the left, and different phylogenetic groups are distinguished by colored dots.
), slash filled inverted triangles (▽), and black inverted triangles (▼), respectively. The phylogenetic relationships of potato UGT proteins can be seen on the left, and different phylogenetic groups are distinguished by colored dots.
Among the total 77 introns detected in potato UGT genes, 21, 49, and 7 were in phases 0, 1, and 2, respectively (Figure 3 and Supplementary Table S3). For the highly conserved I-5, only two were in phase 0, and phase 1 accounted for 94% of all introns. For introns I-6, 71% of UGTs were in phase 0. According to these results, the highly conserved introns were all in the same intron phase, which is in accordance with the view that most of the conserved introns are ancient elements, and their phases tend to be stable [36].
3.5. Expression Profiles of UGTs in Different Tissues
To determine the expression patterns of potato UGT genes in different tissues, we used published RH transcriptome sequencing data [30]. The seven different potato tissues analyzed were from the flower, leaf, stem, stolon, young tuber, mature tuber, and root (Figure 4). Out of the 174 potato UGT genes, 154 UGTs were expressed in potato tissues (accounting for 88.5%) (Supplementary Table S4). Additionally, 25 UGT genes (accounting for 14.3%), 10 UGT genes (5.4%), 6 UGT genes (3.4%), and 4 UGT genes (2.2%) were specifically expressed in the leaves, roots, stolons, and young tubers (Figure 4 and Supplementary Table S4). For example, in all UGTs, PGSC0003DMG400017508 was the most expressed in the roots but was not expressed in young tubers. PGSC0003DMG400011740 and PGSC0003DMG400000432 were expressed at high levels in the stolon and at low levels in other tissues. These results indicate that the special expressed genes might play an important role in leaves, roots, stolons, and young tubers, respectively.
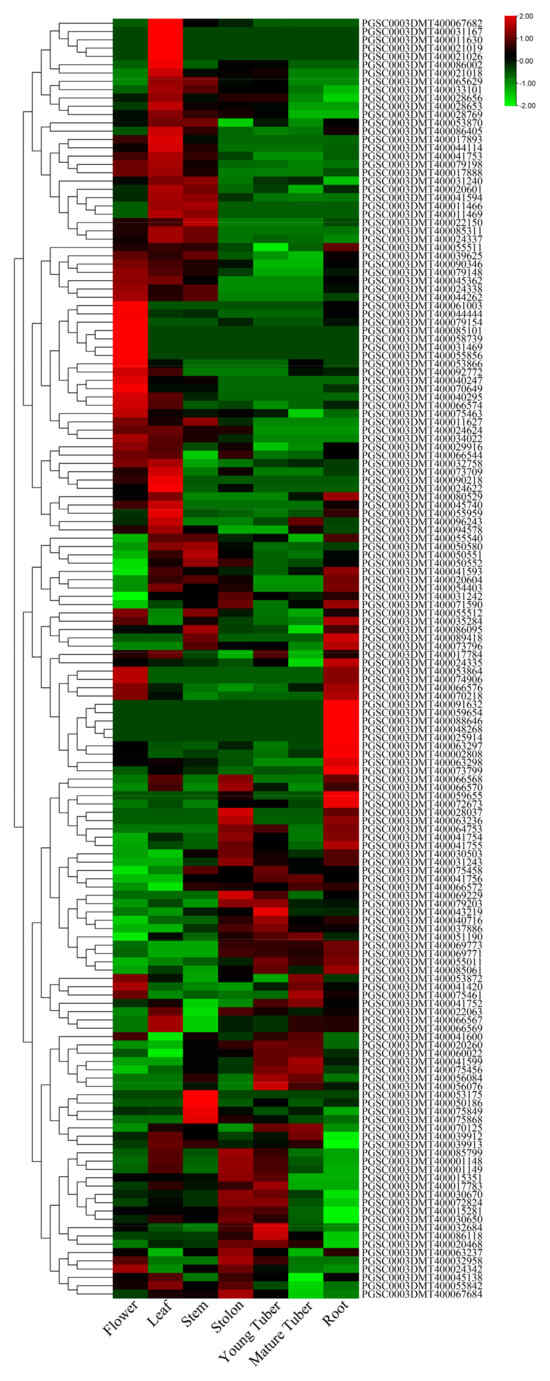
Figure 4.
Distribution of UGT genes in potato tissues. Genes with zero expression in all seven tissues were removed.
3.6. Promoter Cis-Elemental Analysis
A promoter sequence determines the spatiotemporal expression pattern and level of genes. A promoter analysis of genes can help to explore their potential functions. Therefore, a 1500 bp sequence prior to the CDS from the StUGT genes was used as a promoter region for cis-element analysis. We found that the promoters of 174 UGT genes contain many kinds of cis-responsive elements, including light-responsive elements (chs-CMA1a, GT1-motif, Box 4, G-box, AE-box, TCT-motif, AAGAA-motif, etc.), phytohormone-responsive elements (TCA, TATC-box, ABRE, CGTCA-motif, TGACG-motif, etc.), and stress-responsive elements (LTR, MBS, ARE TC-rich repeats, etc.). A total of 68 UGT genes contained 7–10 light-responsive elements, and 105 UGT genes contained 1–5 light-responsive elements. PGSC0003DMG400007981 did not contain any light-responsive elements. In total, 159 UGT genes contained 1–5 plant hormone-responsive elements, and 8 UGT genes contained 6–7 elements (7 genes did not contain these elements). And 140 UGT genes contained 1–4 stress-responsive elements, and 34 UGT genes did not contain these elements. The above results suggest that these UGT genes might be involved in the abiotic stress response (Supplementary Table S5).
3.7. Expression Analysis under Abiotic Stresses
Water loss from plant cells under drought conditions leads to osmotic stress and oxidative stress. To identify osmotic stress-related UGT genes, we analyzed the expression pattern of UGTs by using the public transcriptome under 260 μM mannitol stress. A total of 38 of the 174 UGT genes were significantly upregulated (absolute log2 fold change > 1) (Figure 5 and Supplementary Table S6). The promoter cis-elements of these genes also had an abundant number of photo-responsive elements. A total of 32 UGT genes contained 4-10 light-responsive elements, 35 UGT genes contained 2-5 hormone-responsive elements, and 32 UGT genes contained 1-4 stress-responsive elements, while 8 UGT genes did not contain these elements (Figure 6 and Supplementary Table S5). The top two genes with the highest expression level and the two genes with anthocyanidine rhamnosy1-transferase function were selected for RT-qPCR verification. These genes included: PGSC0003DMT400041594, PGSC0003DMT400041753, PGSC0003DMT400017888, and PGSC0003DMT400044114. As shown in Figure 7, the expression of PGSC0003DMT400041594, PGSC0003DMT400041753, and PGSC0003DMT400017888 was significantly induced by 10% PEG6000, and peaked at 24 h, 6 h, and 6 h, respectively. Unexpectedly, the expression pattern of PGSC0003DMT400044114 was the reverse of what was anticipated. These results indicate that the four UGT genes selected might participate in the osmotic stress response.
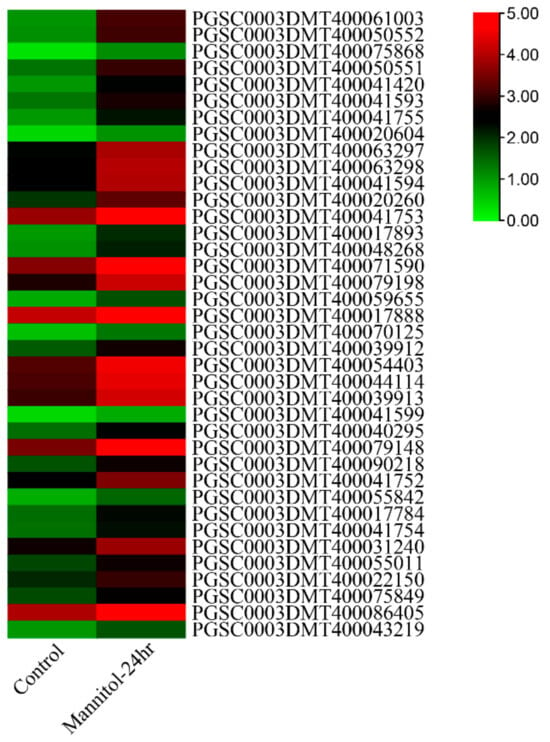
Figure 5.
Expression profiles of potato UGT genes after 24 h mannitol treatment.
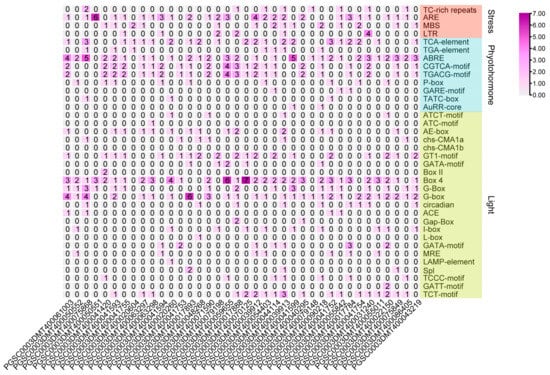
Figure 6.
Cis-element analysis of UGTs in potato—under mannitol treatment for 24 h, the expression level was upregulated (absolute log2 fold change > 1). Cis-elements are mainly divided into light, pressure, and hormone-responsive elements. The number represents the color intensity.
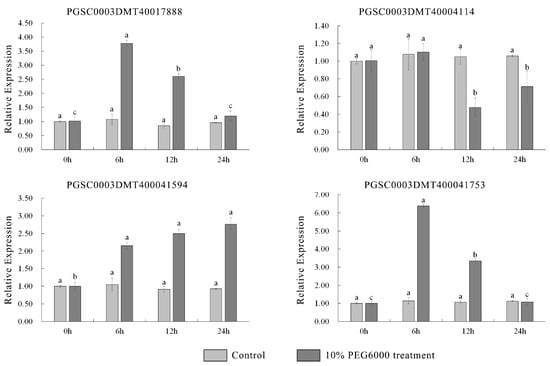
Figure 7.
qRT-PCR results under 10% PEG6000 treatment. The error bars indicate the standard errors of three biological replicates. Significant statistical differences (p < 0.05; Tukey’s) across different time points are denoted by the lowercase letter(s) above the vertical bars.
4. Discussion
Due to their important roles in SM biosynthesis, the regulation of cell homeostasis, and the detoxification of xenobiotics, UGTs in plants have been a source of great interest [37]. Studies have demonstrated that in A. thaliana and O. sativa, the UGT family accounts for the majority of GT genes (>25 and >35%, respectively) [9]. In addition, UGT has been identified in many other plants, including C. grandis [21], L. usitatissimum [29], Z. mays [11], V. vinifera, Malus domestica, Populus trichocarpa, Glycine max, and Mimulus guttatus [34]. Phylogenetic analysis shows that similar sequences probably have the same ancestor, share the same structure, and have a similar biological function. However, no clear analysis of StUGTs was previously conducted in potato.
This study identified 174 UGT genes in potato, accounting for approximately 0.3% of potato proteins. According to the phylogenetic analysis, the potato UGTs can be grouped into 18 groups, including 14 highly conserved groups (A to N) and 4 newly discovered groups (O, P, Q, and R) (Figure 1). By clustering the rice UGT of group Q with the dicotyledonous potato UGTs, it was found that there was no clustering in group Q. However, tomato UGTs were also not classified as group Q [35]. This is consistent with previous research that found that group Q may exist only in monocotyledons [13]. Compared to other plants, potato and tomato have more extended group O members. This suggests that group O members may have significant functions within the Solanaceae family. A novel group containing 23 UGTs was also identified in our analysis, and we named it group R. According to the previous studies, the number of UGT groups varied significantly among various plant species, and in most plant species, group E has the largest number of UGT members. However, in the potato genome, group E ranks third in the number of members. Group D, which now comprises 24 genes (13.7%) of the putative UGT genes, has grown to be the largest group. In group D, StUGTs were mostly clustered on chromosomes 1, 10, and 12. In addition, Several UGT73s (belonging to group D) were functional in catalyzing the glycosylation of (iso)flavonoids and in the biosynthesis of anthocyanins [37,38]. Previous studies demonstrated that flavonoid overaccumulation was essential to enhance tolerance to drought stress [39]. In tomatoes, group D is mainly involved in tomato detoxification [35]. Therefore, group D may be involved in the abiotic stress resistance of plants. This is consistent with priest DM’s research, where UGTs were found to be involved in the biosynthesis of plant natural products such as flavonoids, phenylpropanoids, terpenes, and steroids, as well as in the regulation of plant hormones [21].
The occurrence of intron gain and loss events, as well as the phases and positions of introns in relation to protein sequences, provide key evolutionary clues [40]. Intron mapping of 174 peach UGTs revealed that 61% lacked introns, which is higher than the number of UGT genes lacking introns in maize (60%) [11] and Arabidopsis (58%) [41]. In potato UGT genes, ten intron positions have been identified, with I-5 being the most prevalent intron (Figure 3). Intron 5 was observed in most members of groups A, E, F, G, I, J, K, N, and P. In peach, maize, and black cottonwood [8,11,38], intron 5 is considered the oldest intron. The second most highly conserved intron was observed for I-6, which was mainly concentrated in group L of potato. A large number of introns in I-5 were in phase 1, while many introns in I-6 were in phase 0, and phase-0 and -1 introns outnumbered phase-2 introns. It appears that most conserved introns keep their phases stable [36].
Expression analysis was carried out with published RH/DM transcriptome sequencing data. It was found that 25, 10, 6, and 4 of these 174 UGTs were specifically expressed in leaves, roots, stolons, and young tubers, respectively. In other species, 1G091000 and 1G091100 are specifically expressed in peach blossoms and have been identified as playing important roles in the biosynthesis of anthocyanin [42]. These findings imply that genes expressed particularly in potatoes may be crucial to the growth and development of potatoes. In total, 62% were expressed in the stolon, 55% were expressed in the young tuber, and 48% were expressed in the mature tuber (Figure 4 and Supplementary Table S4). These observations suggest that there is a decline in secondary product metabolism during the maturation of potato tubers.
UGTs respond to abiotic stresses such as mannitol and PEG, thereby reducing damage caused by abiotic stresses. Under mannitol treatment, 38 UGTs were upregulated more than twofold (Supplementary Table S6). Analysis of the elements on the promoter of these genes showed that there were abundant light-, stress- and hormone-responsive elements. The presence of these elements further provides the basis for these genes to respond to stress. For example, the down-expression of UGT71C5 increased drought tolerance in transgenic plants, suggesting that UGT71C5 may play a major role in coping with hormonal stress [22]. Previous research has shown that ROS accumulation is one of the earliest cellular responses induced by osmotic stress [43], and anthocyanins are a type of important SM that helps eliminate and reduce the active oxygen damage caused by osmotic stress directly or indirectly [40]. It has been shown that all anthocyanins identified in Arabidopsis contain at least one glycan group, and glycosylation catalyzed by UGTs is the final step in anthocyanin biosynthesis [44,45]. Therefore, four genes were selected for qPCR validation. The results show that the expression levels of PGSC0003DMT400017888, PGSC0003DMT400041594, and PGSC0003DMT400041753 were significantly upregulated under the 10% PEG6000 treatment. Unexpectedly, the expression of PGSC0003DMT400044114 was significantly downregulated. Meanwhile, in the phylogenetic analysis, PGSC0003DMT400041594 and PGSC0003DMT400041753 were clustered with AtUGT79B2, which contribute to cold, salt, and drought stress tolerance via modulating anthocyanin accumulation [18]. PGSC0003DMT400044114 was clustered with AtUGT73B3, which participates in the regulation of redox status and general detoxification of ROS-reactive SMs [46]. Thus, we speculate that PGSC0003DMT400041594 and PGSC0003DMT400041753 may protect plants from osmotic stresses through glycosylation of flavonoids (e.g., anthocyanin), and thus the accumulation of products that scavenge excess ROS in the plant, with PGSC0003DMT400044114 being involved in the negative regulation of the redox state and general detoxification of ROS-reactive SMs.
5. Conclusions
In summary, we provided the first thorough account of the UGT gene family in potato. The phylogenetic analysis revealed that 18 subfamilies might be formed from the 174 StUGT proteins (groups A to R). We further analyzed through intron mapping the promoter cis-elements and the expression pattern of the UGTs in potato under osmotic treatment. Selected genes were able to respond to osmotic stress. The functions of PGSC0003DMT400017888, PGSC0003DMT400044114, PGSC0003DMT400041594, and PGSC0003DMT400041753 can be further investigated by genetic engineering under osmotic stress. The findings of this study contribute to furthering functional research and give insight into the history and possible uses of StUGT genes.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes14122144/s1; Table S1: Characteristics of Family-1 UGTs from S. tuberosum L.; Table S2: Arabidopsis and other UGT genes used for the identification of the phylogenetic groups; Table S3: Intron information of 67 potato UGT genes; Table S4: FPKM values in RH tissues; Table S5: Analysis of cis-elements of UGT gene promoters in potato; Table S6: Upregulated UGTs under mannitol treatment; Table S7: Information about primers used for qRT-PCR.
Author Contributions
Conceptualization, Y.W. and B.H.; methodology, S.S.; software, Y.W. and B.J.; validation, Y.W., S.S. and B.J.; formal analysis, Y.W. and J.L.; investigation, T.W.; resources, Y.W.; data curation, Y.W., J.L. and T.W.; writing—original draft preparation, Y.W., J.L. and B.J.; writing—review and editing, Y.W., B.J. and S.S.; visualization, Y.W.; supervision, B.H.; project administration, B.H.; funding acquisition, B.H. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Science and Technology Department of Yunnan Province (Grant No. 2019FY003015); Research Startup Funding of Yunnan University in China (Grant No. C176220100033); Science and Technology Major Project of Department of Science and Technology of Yunnan (Grant No. K204204210017); Yunnan Fundamental Research Projects (Grant No. 202301BF070001-026).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Acknowledgments
We thank Ping Wang of the Potato Research Group, College of Agriculture, Yunnan University, for help with data analysis and materials used for experiments.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ren, C.; Cao, Y.; Xing, M.; Guo, Y.; Li, J.; Xue, L.; Sun, C.; Xu, C.; Chen, K.; Li, X. Genome-wide analysis of UDP-glycosyltransferase gene family and identification of members involved in flavonoid glucosylation in Chinese bayberry (Morella rubra). Front. Plant Sci. 2022, 13, 998985. [Google Scholar] [CrossRef]
- Lairson, L.L.; Henrissat, B.; Davies, G.J.; Withers, S.G. Glycosyltransferases: Structures, functions, and mechanisms. Annu. Rev. Biochem. 2008, 77, 521–555. [Google Scholar] [CrossRef]
- Coutinho, P.M.; Stam, M.; Blanc, E.; Henrissat, B. Why are there so many carbohydrate-active enzyme-related genes in plants? Trends Plant Sci. 2003, 8, 563–565. [Google Scholar] [CrossRef]
- Liu, Q.; Chen, T.T.; Xiao, D.W.; Zhao, S.M.; Lin, J.S.; Wang, T.; Li, Y.J.; Hou, B.K. OsIAGT1 is a glucosyltransferase gene involved in the glucose conjugation of auxins in rice. Rice 2019, 12, 92. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.W.; Wang, W.C.; Jin, S.H.; Wang, J.; Wang, B.; Hou, B.K. Over-expression of a putative poplar glycosyltransferase gene, PtGT1, in tobacco increases lignin content and causes early flowering. J. Exp. Bot. 2012, 63, 2799–2808. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Ahmad, D.; Zhang, X.; Zhang, Y.; Wu, L.; Jiang, P.; Ma, H. Genome-wide analysis of family-1 UDP glycosyltransferases (UGT) and identification of UGT genes for FHB resistance in wheat (T. aestivum L.). BMC Plant Biol. 2018, 18, 67. [Google Scholar] [CrossRef]
- Speeckaert, N.; Adamou, N.M.; Hassane, H.A.; Baldacci-Cresp, F.; Mol, A.; Goeminne, G.; Boerjan, W.; Duez, P.; Hawkins, S.; Neutelings, G.; et al. Characterization of the UDP-glycosyltransferase UGT72 family in poplar and identification of genes involved in the glycosylation of monolignols. Int. J. Mol. Sci. 2020, 21, 5018. [Google Scholar] [CrossRef] [PubMed]
- Rehman, H.M.; Khan, U.M.; Nawaz, S.; Saleem, F.; Ahmed, N.; Rana, I.A.; Atif, R.M.; Shaheen, N.; Seo, H. Genome wide analysis of family-1 UDP glycosyltransferases in Populus trichocarpa specifies abiotic stress responsive glycosylation mechanisms. Genes 2022, 13, 1640. [Google Scholar] [CrossRef] [PubMed]
- Paquette, S.; Møller, B.L.; Bak, S. On the origin of family 1 plant glycosyltransferases. Phytochemistry 2003, 62, 399–413. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Rawat, V.; Suresh, C.G. Genome-wide identification and tissue-specific expression analysis of UDP-glycosyltransferases genes confirm their abundance in Cicer arietinum (Chickpea) genome. PLoS ONE 2014, 9, e109715. [Google Scholar] [CrossRef]
- Li, Y.; Li, P.; Wang, Y.; Dong, R.; Yu, H.; Hou, B. Genome-wide identification and phylogenetic analysis of Family-1 UDP glycosyltransferases in maize (Z. mays). Planta 2014, 239, 1265–1279. [Google Scholar] [CrossRef]
- Xiao, X.; Lu, Q.; Liu, R.; Gong, J.; Gong, W.; Liu, A.; Ge, Q.; Li, J.; Shang, H.; Li, P.; et al. Genome-wide characterization of the UDP-glycosyltransferase gene family in upland cotton. Biotechnology 2019, 9, 453. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Liu, X.; Xu, K.; Zhang, B. Genome-wide characterization, evolution and expression profiling of UDP-glycosyltransferase family in pomelo (C. grandis) fruit. BMC Plant Biol. 2020, 20, 459. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Vogt, T. Glycosyltransferases in secondary plant metabolism: Tranquilizers and stimulant controllers. Planta. 2001, 213, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Pang, C.; Fan, S.; Song, M.; Yu, J.; Wei, H.; Ma, Q.; Li, L.; Zhang, C.; Yu, S. Genome-wide analysis of the family 1 glycosyltransferases in cotton. Mol. Genet. Genom. 2015, 290, 1805–1818. [Google Scholar] [CrossRef]
- Wang, X. Structure, mechanism and engineering of plant natural product glycosyltransferases. FEBS Lett. 2009, 583, 3303–3309. [Google Scholar] [CrossRef]
- Chen, T.T.; Liu, F.F.; Xiao, D.W.; Jiang, X.Y.; Li, P.; Zhao, S.M.; Hou, B.K.; Li, Y.J. The Arabidopsis UDP-glycosyltransferase75B1, conjugates abscisic acid and affects plant response to abiotic stresses. Plant Mol. Biol. 2020, 102, 389–401. [Google Scholar] [CrossRef]
- Li, P.; Li, Y.J.; Zhang, F.J.; Zhang, G.Z.; Jiang, X.Y.; Yu, H.M.; Hou, B.K. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 2017, 89, 85–103. [Google Scholar] [CrossRef]
- Zhao, M.; Zhang, N.; Gao, T.; Jin, J.; Jing, T.; Wang, J.; Wu, Y.; Wan, X.; Schwab, W.; Song, C. Sesquiterpene glucosylation mediated by glucosyltransferase UGT91Q2 is involved in the modulation of cold stress tolerance in tea plants. New Phytol. 2020, 226, 362–372. [Google Scholar] [CrossRef]
- Jones, P.; Messner, B.; Nakajima, J.; Schäffner, A.R.; Saito, K. UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in A. thaliana. J. Biol. Chem. 2003, 278, 43910–43918. [Google Scholar] [CrossRef]
- Priest, D.M.; Ambrose, S.J.; Vaistij, F.E.; Elias, L.; Higgins, G.S.; Ross, A.R.; Abrams, S.R.; Bowles, D.J. Use of the glucosyltransferase UGT71B6 to disturb abscisic acid homeostasis in A. thaliana. Plant J. 2006, 46, 492–502. [Google Scholar] [CrossRef]
- Liu, Z.; Yan, J.P.; Li, D.K.; Luo, Q.; Yan, Q.; Liu, Z.B.; Ye, L.M.; Wang, J.M.; Li, X.F.; Yang, Y. UDP-glucosyltransferase71c5, a major glucosyltransferase, mediates abscisic acid homeostasis in Arabidopsis. Plant Physiol. 2015, 167, 1659–1670. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.; Xu, Z.Y.; Park, Y.; Kim, D.H.; Lee, Y.; Hwang, I. Abscisic acid uridine diphosphate glucosyltransferases play a crucial role in abscisic acid homeostasis in Arabidopsis. Plant Physiol. 2014, 165, 277–289. [Google Scholar] [CrossRef]
- Mu, T.; Sun, H. Progress in research and development of potato staple food processing technology. J. Appl. Glycosci. 2017, 64, 51–64. [Google Scholar] [CrossRef]
- Lorenc-Kukuła, K.; Jafra, S.; Oszmiański, J.; Szopa, J. Ectopic expression of anthocyanin 5-o-glucosyltransferase in potato tuber causes increased resistance to bacteria. J. Agric. Food Chem. 2005, 53, 272–281. [Google Scholar] [CrossRef]
- Bundy, J.G.; Davey, M.P.; Viant, M.R. Environmental metabolomics: A critical review and future perspectives. Metabolomics 2009, 5, 3–21. [Google Scholar] [CrossRef]
- Dias, M.C.; Pinto, D.C.G.A.; Silva, A.M.S. Plant flavonoids: Chemical characteristics and biological activity. Molecules 2021, 26, 5377. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Barvkar, V.T.; Pardeshi, V.C.; Kale, S.M.; Kadoo, N.Y.; Gupta, V.S. Phylogenomic analysis of UDP glycosyltransferase 1 multigene family in Linum usitatissimum identified genes with varied expression patterns. BMC Genomics 2012, 13, 175. [Google Scholar] [CrossRef]
- Xu, X.; Pan, S.; Cheng, S.; Zhang, B.; Mu, D.; Ni, P.; Zhang, G.; Yang, S.; Li, R.; Wang, J.; et al. Genome sequence and analysis of the tuber crop potato. Nature 2011, 475, 189–195. [Google Scholar] [CrossRef]
- Nicot, N.; Hausman, J.F.; Hoffmann, L.; Evers, D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef]
- Derveaux, S.; Vandesompele, J.; Hellemans, J. How to do successful gene expression analysis using real-time PCR. Methods 2010, 50, 227–230. [Google Scholar] [CrossRef] [PubMed]
- Vogt, T.; Jones, P. Glycosyltransferases in plant natural product synthesis: Characterization of a supergene family. Trends Plant Sci. 2000, 5, 380–386. [Google Scholar] [CrossRef]
- Caputi, L.; Malnoy, M.; Goremykin, V.; Nikiforova, S.; Martens, S. A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 2012, 69, 1030–1042. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Chen, Q.; Chen, F.; Liu, H.; Lin, J.; Chen, R.; Ren, C.; Wei, J.; Zhang, Y.; Yang, F.; et al. Glutathione promotes degradation and metabolism of residual fungicides by inducing udp-glycosyltransferase genes in tomato. Front. Plant Sci. 2022, 13, 893508. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.W.; Gilbert, W. Rates of intron loss and gain: Implications for early eukaryotic evolution. Proc. Natl. Acad. Sci. USA 2005, 102, 5773–5778. [Google Scholar] [CrossRef]
- Song, C.; Hong, X.; Zhao, S.; Liu, J.; Schulenburg, K.; Huang, F.C.; Franz-Oberdorf, K.; Schwab, W. Glucosylation of 4-hydroxy-2,5-dimethyl-3(2h)-furanone, the key strawberry flavor compound in strawberry fruit. Plant Physiol. 2016, 171, 139–157. [Google Scholar] [CrossRef]
- Wu, B.; Gao, L.; Gao, J.; Xu, Y.; Liu, H.; Cao, X.; Zhang, B.; Chen, K. Genome-wide identification, expression patterns, and functional analysis of UDP glycosyltransferase family in peach (Prunus persica L. Batsch). Front. Plant Sci. 2017, 8, 389. [Google Scholar] [CrossRef]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K.; et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014, 77, 367–379. [Google Scholar] [CrossRef]
- Rogozin, I.B.; Lyons-Weiler, J.; Koonin, E.V. Intron sliding in conserved gene families. Trends Genet. 2000, 16, 430–432. [Google Scholar] [CrossRef]
- Li, Y.; Baldauf, S.; Lim, E.K.; Bowles, D.J. Phylogenetic analysis of the UDP-glycosyltransferase multigene family of A. thaliana. J. Biol. Chem. 2001, 276, 4338–4343. [Google Scholar] [CrossRef]
- Cheng, J.; Wei, G.; Zhou, H.; Gu, C.; Vimolmangkang, S.; Liao, L.; Han, Y. Unraveling the mechanism underlying the glycosylation and methylation of anthocyanins in peach. Plant Physiol. 2014, 166, 1044–1058. [Google Scholar] [CrossRef] [PubMed]
- Naing, A.H.; Kim, C.K. Abiotic stress-induced anthocyanins in plants: Their role in tolerance to abiotic stresses. Physiol. Plant 2021, 172, 1711–1723. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.Z.; Xie, D.Y. Biosynthesis and metabolic engineering of anthocyanins in A. thaliana. Recent. Pat. Biotechnol. 2014, 8, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Simon, C.; Langlois-Meurinne, M.; Didierlaurent, L.; Chaouch, S.; Bellvert, F.; Massoud, K.; Garmier, M.; Thareau, V.; Comte, G.; Noctor, G.; et al. The secondary metabolism glycosyltransferases UGT73B3 and UGT73B5 are components of redox status in resistance of Arabidopsis to Pseudomonas syringae pv. tomato. Plant Cell Environ. 2014, 37, 1114–1129. [Google Scholar] [CrossRef]
- Tohge, T.; Nishiyama, Y.; Hirai, M.Y.; Yano, M.; Nakajima, J.; Awazuhara, M.; Inoue, E.; Takahashi, H.; Goodenowe, D.B.; Kitayama, M.; et al. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 2005, 42, 218–235. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).