Identification of Genomic Regions Associated with Seedling Frost Tolerance in Sorghum
Abstract
1. Introduction
2. Materials and Methods
2.1. Genetic Material
2.2. Freezing Tolerance Assay
2.3. Cold Establishment
2.4. Growth Analysis
2.5. Evaluation of LT50
2.6. Freezing Treatment
2.7. Growth Analysis
2.8. Proline Content
2.9. DNA Extraction
2.10. DNA Quantification and Quality
2.11. Genotyping-by-Sequencing (GBS)
2.12. Linkage Map Construction
2.13. QTL Analysis
2.14. Statistical Analysis
3. Results
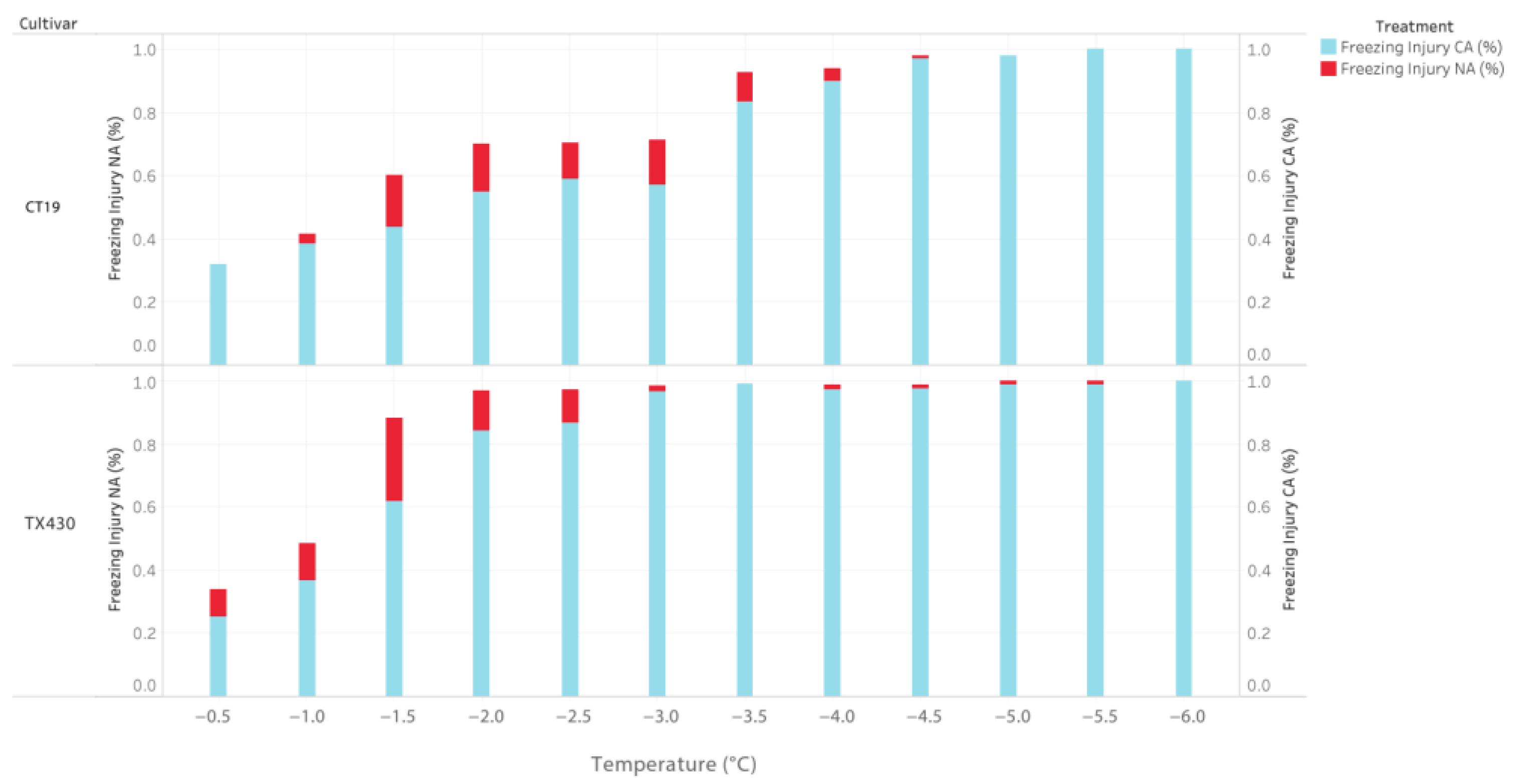
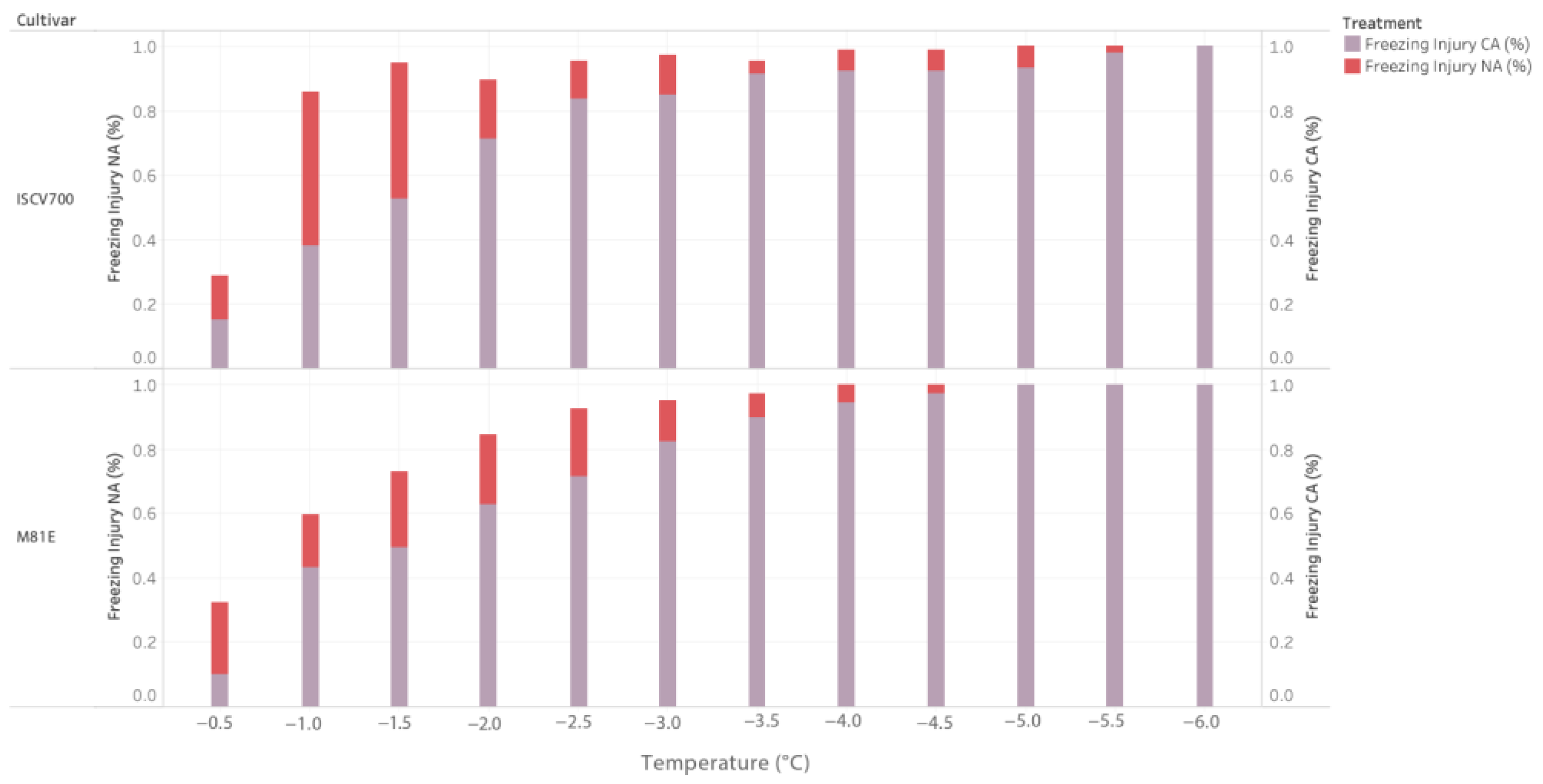
3.1. Determination of LT50
3.1.1. Effect of Cold Acclimation on Leaf Greenness and Growth
3.1.2. Changes in Leaf-Free Proline Content in Response to Cold-Acclimation
| Cultivar | Non-Acclimated (NA) | Chilling-Acclimated (CA) |
|---|---|---|
| CT19 | −1.44 a | −2.05 b |
| Tx430 | −1.02 c | −1.46 ad |
| M81E | −1.17 de | −1.75 f |
| ISCV700 | −0.806 cg | −1.53 dh |
3.2. Correlation Analysis
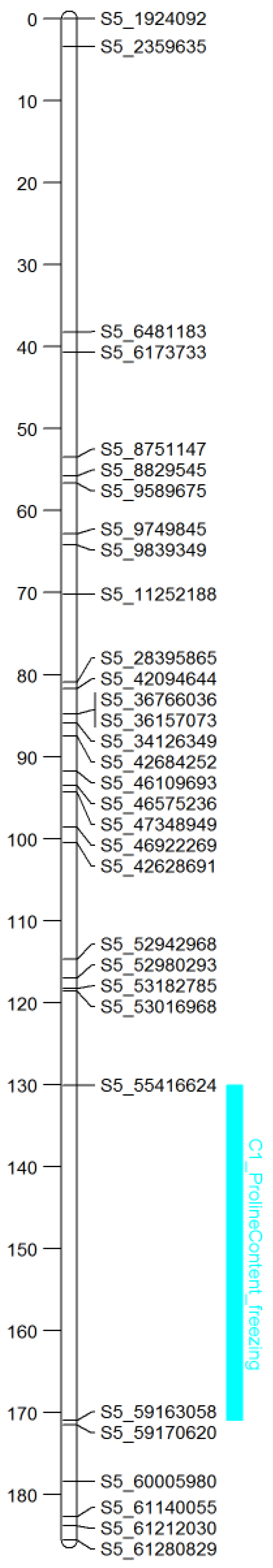
3.2.1. Linkage Map
(Frost Damage QTL)
4. Discussion
4.1. Genotyping and Linkage Map Development
4.2. Detection of Main Effect QTL
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kreyling, J.; Henry, H.A.L. Vanishing winters in Germany: Soil frost dynamics and snow cover trends, and ecological implications. Clim. Res. 2011, 46, 269–276. [Google Scholar] [CrossRef]
- Peng, Y.; Lin, W.; Cai, W.; Arora, R. Overexpression of a Panax Ginseng Tonoplast Aquaporin Alters Salt Tolerance, Drought Tolerance and Cold Acclimation Ability in Transgenic Arabidopsis plants. Planta 2007, 226, 729–740. [Google Scholar] [CrossRef]
- Warmund, M.R.; Guinan, P.; Fernandez, G. Temperatures and Cold Damage to Small Fruit Crops Across the Eastern United States Associated with the April 2007 Freeze. HortScience 2008, 43, 1643–1647. [Google Scholar] [CrossRef]
- Marino, G.P.; Kaiser, D.P.; Gu, L.; Ricciuto, D.M. Reconstruction of false spring occurrences over the southeastern United States, 1901–2007: An increasing risk of spring freeze damage? Environ. Res. Lett. 2011, 6, 024015. [Google Scholar] [CrossRef]
- Kistner, E.; Kellner, O.; Andresen, J.; Todey, D.; Morton, L.W. Vulnerability of specialty crops to short-term climatic variability and adaptation strategies in the Midwestern USA. Clim. Chang. 2018, 146, 145–158. [Google Scholar] [CrossRef]
- Smith, A.; Lott, N.; Houston, T.; Shein, K.; Crouch, J.; Enloe, J. US Billion-Dollar Weather & Climate Disasters 1980–2021; NOAA National Centers for Environmental Information: Asheville, NC, USA, 2021. [Google Scholar]
- Food and Agriculture Organization of the United Nations (FAO). 1997, pp. 1524–1527. Available online: https://www.fao.org/documents/card/en/c/0ee8bcbf-cdce-5644-afa4-979cc362235f (accessed on 20 November 2023).
- Greaves, J.A. Improving Suboptimal Temperature Tolerance in Maize—The Search for Variation. J. Exp. Bot. 1996, 47, 307–323. [Google Scholar] [CrossRef]
- Singh, S.P. Sources of Cold Tolerance in Grain Sorghum. Can. J. Plant Sci. 1985, 65, 251–257. [Google Scholar] [CrossRef]
- Thomashow, M. Role of Cold-Responsive Genes in Plant Freezing Tolerance. Plant Physiol. 1998, 118, 1–8. Available online: http://www.plantphysiol.org/content/118/1/1.shor (accessed on 2 October 2016). [CrossRef]
- Lim, C.C.; Arora, R.; Townsend, E.C. Comparing Gompertz and Richards Functions to Estimate Freezing Injury in Rhododendron Using Electrolyte Leakage. J. Am. Soc. Hortic. Sci. 1998, 123, 246–252. [Google Scholar] [CrossRef]
- Gusta, L.V.; Wisniewski, M. Understanding Plant Cold Hardiness: An Opinion. Physiol. Plant 2013, 147, 4–14. Available online: http://www.ncbi.nlm.nih.gov/pubmed/22409670 (accessed on 25 January 2017). [CrossRef]
- Roberts, E.H.; Hadley, P.; Summerfield, R.J. Effects of Temperature and Photoperiod on Flowering in Chickpeas (Cicer arietinum L.). Ann. Bot. 1985, 55, 881–892. [Google Scholar] [CrossRef]
- Michel, S.; Löschenberger, F.; Hellinger, J.; Strasser, V.; Ametz, C.; Pachler, B.; Sparry, E.; Bürstmayr, H. Improving and Maintaining Winter Hardiness and Frost Tolerance in Bread Wheat by Genomic Selection. Front. Plant Sci. 2019, 10, 1195. [Google Scholar] [CrossRef] [PubMed]
- Slama, I.; Abdelly, C.; Bouchereau, A.; Flowers, T.; Savouré, A. Diversity, distribution and roles of osmoprotective compounds accumulated in halophytes under abiotic stress. Ann. Bot. 2015, 115, 433–447. [Google Scholar] [CrossRef] [PubMed]
- Meena, M.; Divyanshu, K.; Kumar, S.; Swapnil, P.; Zehra, A.; Shukla, V.; Yadav, M.; Upadhyay, R.S. Regulation of L-proline biosynthesis, signal transduction, transport, accumulation and its vital role in plants during variable environmental conditions. Heliyon 2019, 5, e02952. [Google Scholar] [CrossRef] [PubMed]
- Verbruggen, N.; Hermans, C. Proline accumulation in plants: A review. Amino Acids 2008, 35, 753–759. [Google Scholar] [CrossRef]
- Altuntaş, C.; Demiralay, M.; Muslu, A.S.; Terzi, R. Proline-stimulated signaling primarily targets the chlorophyll degradation pathway and photosynthesis associated processes to cope with short-term water deficit in maize. Photosynth. Res. 2020, 144, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Rydeen, A.E.; Brustad, E.M.; Pielak, G.J. Osmolytes and Protein–Protein Interactions. J. Am. Chem. Soc. 2018, 140, 7441–7444. [Google Scholar] [CrossRef]
- Govrin, R.; Obstbaum, T.; Sivan, U. Common Source of Cryoprotection and Osmoprotection by Osmolytes. J. Am. Chem. Soc. 2019, 141, 13311–13314. [Google Scholar] [CrossRef]
- Szabados, L.; Savouré, A. Proline: A Multifunctional Amino Acid. Trends Plant Sci. 2010, 15, 89–97. Available online: http://www.ncbi.nlm.nih.gov/pubmed/20036181 (accessed on 6 February 2017). [CrossRef]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid Determination of Free Proline for Water-Stress Studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Khedr, A.H.A.; Abbas, M.A.; Wahid, A.A.A.; Quick, W.P.; Abogadallah, G.M. Proline Induces the Expression of Salt-Stress-Responsive Proteins and May Improve the Adaptation of Pancratium maritimum L. to salt-stress. J. Exp. Bot. 2003, 54, 2553–2562. Available online: http://www.ncbi.nlm.nih.gov/pubmed/14512386 (accessed on 25 January 2017). [CrossRef] [PubMed]
- Xin, Z.; Chen, J. A high Throughput DNA Extraction Method with High Yield and Quality. Plant Methods 2012, 8, 26. Available online: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3441248&tool=pmcentrez&rendertype=abstract (accessed on 30 April 2015). [CrossRef] [PubMed]
- La Borde, N.; Rajewski, J.; Dweikat, I. Novel QTL for chilling tolerance at germination and early seedling stages in sorghum. Front. Genet. 2023, 14, 1129460. [Google Scholar] [CrossRef] [PubMed]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A Robust, Simple Genotyping-by-Sequencing (GBS) Approach for High Diversity Species. PLoS ONE 2011, 6, e19379. Available online: http://www.ncbi.nlm.nih.gov/pubmed/21573248 (accessed on 17 January 2017). [CrossRef] [PubMed]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.; Butler, D. ASMap: Linkage Map Construction Using the MSTmap Algorithm, R package version 0.4-5; R Core Team: Vienna, Austria, 2015. [Google Scholar]
- Kosambi, D.D. The estimation of map distances from recombination values. Ann. Eugen. 1943, 12, 172–175. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ye, G.; Wang, J. A modified Algorithm for the Improvement of Composite Interval Mapping. Genetics 2007, 175, 361–374. Available online: http://www.ncbi.nlm.nih.gov/pubmed/17110476 (accessed on 10 August 2016). [CrossRef]
- Doerge, R.W.; Churchill, G.A. Permutation Tests for Multiple Loci Affecting a Quantitative Character. Genetics 1996, 142, 285–294. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, J.; Bao, Y.; Wu, Y.; Zhang, H. Quantitative Trait Loci Controlling Rice Seed Germination under Salt Stress. Euphytica 2011, 178, 297–307. [Google Scholar] [CrossRef]
- Mendiburu, F.; Simon, R.; De Mendiburu, F. Agricolae-Ten years of an Open-source Statistical tool for experiments in Breeding, agriculture and biology. PeerJ PrePrints 2015. Available online: https://peerj.com/preprints/1404/ (accessed on 24 November 2022).
- R Core Team, R. A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2017. [Google Scholar]
- Patton, A.J.; Reicher, Z.J. Zoysiagrass Species and Genotypes Differ in Their Winter Injury and Freeze Tolerance. Crop Sci. 2007, 47, 1619–1627. [Google Scholar] [CrossRef]
- Taylor, A.O.; Rowley, J.A. Plants under Climatic Stress: I. Low Temperature, High Light Effects on Photosynthesis. Plant Physiol. 1971, 47, 713–718. [Google Scholar] [CrossRef] [PubMed]
- Ercoli, L.; Mariotti, M.; Masoni, A.; Arduini, I. Growth responses of sorghum plants to chilling temperature and duration of exposure. Eur. J. Agron. 2004, 21, 93–103. [Google Scholar] [CrossRef]
- Renaut, J.; Hoffmann, L.; Hausman, J.-F. Biochemical and Physiological Mechanisms Related to Cold Acclimation and Enhanced Freezing Tolerance in Poplar plantlets. Physiol. Plant 2005, 125, 82–94. [Google Scholar] [CrossRef]
- Ensminger, I.; Busch, F.; Huner, N.P.A. Photostasis and cold acclimation: Sensing low temperature through photosynthesis. Physiol. Plant. 2006, 126, 28–44. [Google Scholar] [CrossRef]
- Hayat, S.; Hayat, Q.; Alyemeni, M.N.; Wani, A.S.; Pichtel, J.; Ahmad, A. Role of Proline under Changing Environments: A Review. Plant Signal. Behav. 2012, 7, 1456–1466. Available online: http://www.tandfonline.com/doi/full/10.4161/psb.21949#abstract (accessed on 20 November 2023). [CrossRef] [PubMed]
- Claussen, W. Proline as a Measure of Stress in Tomato Plants. Plant Sci. 2005, 168, 241–248. Available online: http://linkinghub.elsevier.com/retrieve/pii/S0168945204003723 (accessed on 20 November 2023). [CrossRef]
- Su, M.; Li, X.-F.; Ma, X.-Y.; Peng, X.-J.; Zhao, A.-G.; Chen, S.-Y.; Liu, G.-S. Cloning two P5CS Genes from Bioenergy Sorghum and Their Expression Profiles under Abiotic Stresses and MeJA Treatment. Plant Sci. 2011, 181, 652–659. Available online: http://www.ncbi.nlm.nih.gov/pubmed/21958707 (accessed on 6 February 2017). [CrossRef]
- Gore , M.A.; Fang, D.D.; Poland, J.A.; Zhang, J.; Percy, R.G.; Cantrell, R.G.; Chen, S.Y.; Liu, G.S. Linkage Map Construction and Quantitative Trait Locus Analysis of Agronomic and Fiber Quality Traits in Cotton. Plant Genome 2014, 7, 1–62. Available online: https://www.crops.org/publications/tpg/abstracts/7/1/plantgenome2013.07.0023 (accessed on 8 January 2015).
- Hiremath, P.J.; Kumar, A.; Penmetsa, R.V.; Farmer, A.; Schlueter, J.A.; Chamarthi, S.K.; Whaley, A.M.; Carrasquilla-Garcia, N.; Gaur, P.M.; Upadhyaya, H.D.; et al. Large-scale development of cost-effective SNP marker assays for diversity assessment and genetic mapping in chickpea and comparative mapping in legumes. Plant Biotechnol. J. 2012, 10, 716–732. [Google Scholar] [CrossRef]
- Zheng, Y.; Wu, L.; Xia, X.; Peng, Y.; Zhu, H.; Liu, Y.; Wu, Y.; Li, S.; He, Z. QTL Mapping of Adult-Plant Resistance to Stripe Rust in Chinese Wheat Cultivar Chuanyu 16. J. Agric. Sci. 2011, 4, 57–70. Available online: http://www.ccsenet.org/journal/index.php/jas/article/view/11304 (accessed on 20 November 2023). [CrossRef][Green Version]
- Thu, T.T.; Mai, T.T.X.; Dewaele, E.; Farsi, S.; Tadesse, Y.; Angenon, G.; Jacobs, M. Molecular mapping of QTLs for plant type and earliness traits in pigeonpea (Cajanus cajan L. Millsp.). Mol. Breed. 2003, 11, 159–168. [Google Scholar] [CrossRef]
- Ejeta, G.; Knoll, J.E. Marker-Assisted Selection in Sorghum. In Genomics-Assisted Crop Improvement; Springer: Dordrecht, The Netherlands, 2007; pp. 187–205. [Google Scholar]
- Burow, G.; Burke, J.J.; Xin, Z.; Franks, C.D. Genetic Dissection of Early-Season Cold Tolerance in Sorghum (Sorghum bicolor (L.) Moench). Mol. Breed. 2010, 28, 391–402. [Google Scholar] [CrossRef]
- Kumar, S.; Banks, T.W.; Cloutier, S. SNP Discovery through Next-Generation Sequencing and Its Applications. Int. J. Plant Genom. 2012, 2012, 831460. Available online: http://www.ncbi.nlm.nih.gov/pubmed/23227038 (accessed on 17 January 2017). [CrossRef]
- Gelli, M.; Mitchell, S.; Liu, K.; Clemente, T.; Weeks, D.; Zhang, C.; Holding, D.; Dweikat, I. Mapping QTLs and association of differentially expressed gene transcripts for multiple agronomic traits under different nitrogen levels in sorghum. BMC Plant Biology 2016, 16, 16. [Google Scholar] [CrossRef] [PubMed]
- Bekele, W.A.; Fiedler, K.; Shiringani, A.; Schnaubelt, D.; Windpassinger, S.; Uptmoor, R.; Friedt, W.; Snowdon, R.J. Unravelling the genetic complexity of sorghum seedling development under low-temperature conditions. Plant Cell Environ. 2014, 37, 707–723. [Google Scholar] [CrossRef] [PubMed]
- Kubala, S.; Wojtyla, Ł.; Quinet, M.; Lechowska, K.; Lutts, S.; Garnczarska, M. Enhanced expression of the proline synthesis gene P5CSA in relation to seed osmopriming improvement of Brassica napus germination under salinity stress. J. Plant Physiol. 2015, 183, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Davidson, R.M.; Gowda, M.; Moghe, G.; Lin, H.; Vaillancourt, B.; Shiu, S.H.; Jiang, N.; Buell, C.R. Comparative Transcriptomics of three Poaceae Species Reveals Patterns of Gene Expression Evolution. Plant J. 2012, 71, 492–502. [Google Scholar] [CrossRef]

| Population C1 | Population C2 | |||||||
|---|---|---|---|---|---|---|---|---|
| Parents | RILs | Parents | RILs | |||||
| Traits | CT19 | Tx430 | Mean ± SD | Range | M81E | ISCV700 | Mean ± SD | Range |
| SPADNA * | 24.8 | 17.6 | 19.57 ± 4.63 | 8.9–33.6 | 24.2 | 17.3 | 18.55 ± 3.83 | 8.2–32.1 |
| SPADCA * | 23.6 | 13.0 | 14.69 ± 3.6 | 7.2–27.0 | 18.5 | 10.6 | 13.6 ± 3.6 | 4.2–24.8 |
| SPADF * | 15.7 | 9.2 | 10.54 ± 3.74 | 1.8–22.5 | 13.7 | 8.2 | 10.0 ± 3.77 | 1.8–22.5 |
| ProNA * | 0.499 | 0.4434 | 0.52 ± 0.0296 | 0.411–0.599 | 0.064 | 0.051 | 0.0517 ± 0.03 | 0.004–0.17 |
| ProCA * | 0.5689 | 0.5403 | 0.560 ± 0.038 | 0.481–0.707 | 0.147 | 0.08 | 0.115 ± 0.017 | 0.15–0.91 |
| ProF * | 0.6531 | 0.6149 | 0.622 ± 0.057 | 0.502–0.817 | 0.233 | 0.117 | 0.164 ± 0.087 | 0.27–0.52 |
| StmNA * | 17.9 | 14.14 | 20.8 ± 4.3 | 9.62–24.2 | 23.58 | 17.3 | 18.11 ± 4.4 | 0.00–29.0 |
| StmCA * | 11.7 | 16.32 | 12.4 ± 3.5 | 3.66–20.5 | 15.16 | 9.62 | 14.17 ± 3.34 | 0.0–22.9 |
| SPADNA | SPADCA | SPADF | StmNa | ProNa | ProCA | ProF | |
|---|---|---|---|---|---|---|---|
| SPADNA | 1 | ||||||
| SPADCA | 0.1126 | 1 | |||||
| SPADF | 0.0504 | 0.6076 *** | 1 | ||||
| StmNA | 0.126 * | 0.2359 *** | −0.0697 | 1 | |||
| ProNA | −0.0161 | −0.0788 | 0.0365 | 0.0178 | 1 | ||
| ProCA | 0.0626 | −0.0477 | −0.0795 | −0.0619 | 0.6056 *** | 1 | |
| ProF | 0.0611 | −0.0678 | −0.1117 | −0.0351 | 0.3311 *** | 0.6731 *** | 1 |
| SPADNA | SPADCA | SPADF | StmNA | StmCA | ProNA | ProCA | ProF | |
|---|---|---|---|---|---|---|---|---|
| SPADNA | 1 | |||||||
| SPADCA | 0.676 *** | 1 | ||||||
| SPADF | 0.0511 *** | 0.692 *** | 1 | |||||
| StmNA | 0.0078 | 0.0646 | 0.1978 | 1 | ||||
| StmCA | 0.0095 | −0.104 | −0.0747 | 0.1216 | 1 | |||
| ProNA | 0.0499 | 0.0247 | −0.0087 | −0.0643 | 0.0059 | 1 | ||
| ProCA | 0.0717 | 0.1095 | 0.0074 | −0.0092 | −0.0565 | 0.0194 * | 1 | |
| ProF | 0.3167 ** | 0.2628 ** | 0.1396 | −0.0847 | 0.0789 | 0.1715 | 0.2548 ** | 1 |
| Population | QTL | LG | Peak cM | Marker Interval | AE | LOD | PVE |
|---|---|---|---|---|---|---|---|
| C1 | PROF | SB_05 | 145 | S5_55416624/S5_59163058 | −0.48 | 33.85 | 98.12 |
| C2 | SPADCA | SB_10.2 | 11 | S10_58412586/S10_59030486 | −1.51 | 3.86 | 17.75 |
| SPADF | SB_04 | 5 | S4_61780584/S4_61537581 | −1.68 | 4.68 | 21.93 | |
| PRONA | SB_03 | 223 | S3_2604980/S3_2556101 | −0.02 | 9.22 | 54.21 | |
| PRONA | SB_03 | 230 | S3_1472653/S3_555829 | 0.01 | 5.07 | 26.13 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borde, N.L.; Dweikat, I. Identification of Genomic Regions Associated with Seedling Frost Tolerance in Sorghum. Genes 2023, 14, 2117. https://doi.org/10.3390/genes14122117
Borde NL, Dweikat I. Identification of Genomic Regions Associated with Seedling Frost Tolerance in Sorghum. Genes. 2023; 14(12):2117. https://doi.org/10.3390/genes14122117
Chicago/Turabian StyleBorde, Niegel La, and Ismail Dweikat. 2023. "Identification of Genomic Regions Associated with Seedling Frost Tolerance in Sorghum" Genes 14, no. 12: 2117. https://doi.org/10.3390/genes14122117
APA StyleBorde, N. L., & Dweikat, I. (2023). Identification of Genomic Regions Associated with Seedling Frost Tolerance in Sorghum. Genes, 14(12), 2117. https://doi.org/10.3390/genes14122117






