Revisiting the Asian Buffalo Leech (Hirudinaria manillensis) Genome: Focus on Antithrombotic Genes and Their Corresponding Proteins
Abstract
:1. Introduction
2. Materials and Methods
2.1. The DNA and RNA Sequencing
2.2. Genome Assembling and Gene Prediction
2.3. Identification of Antithrombotic Genes
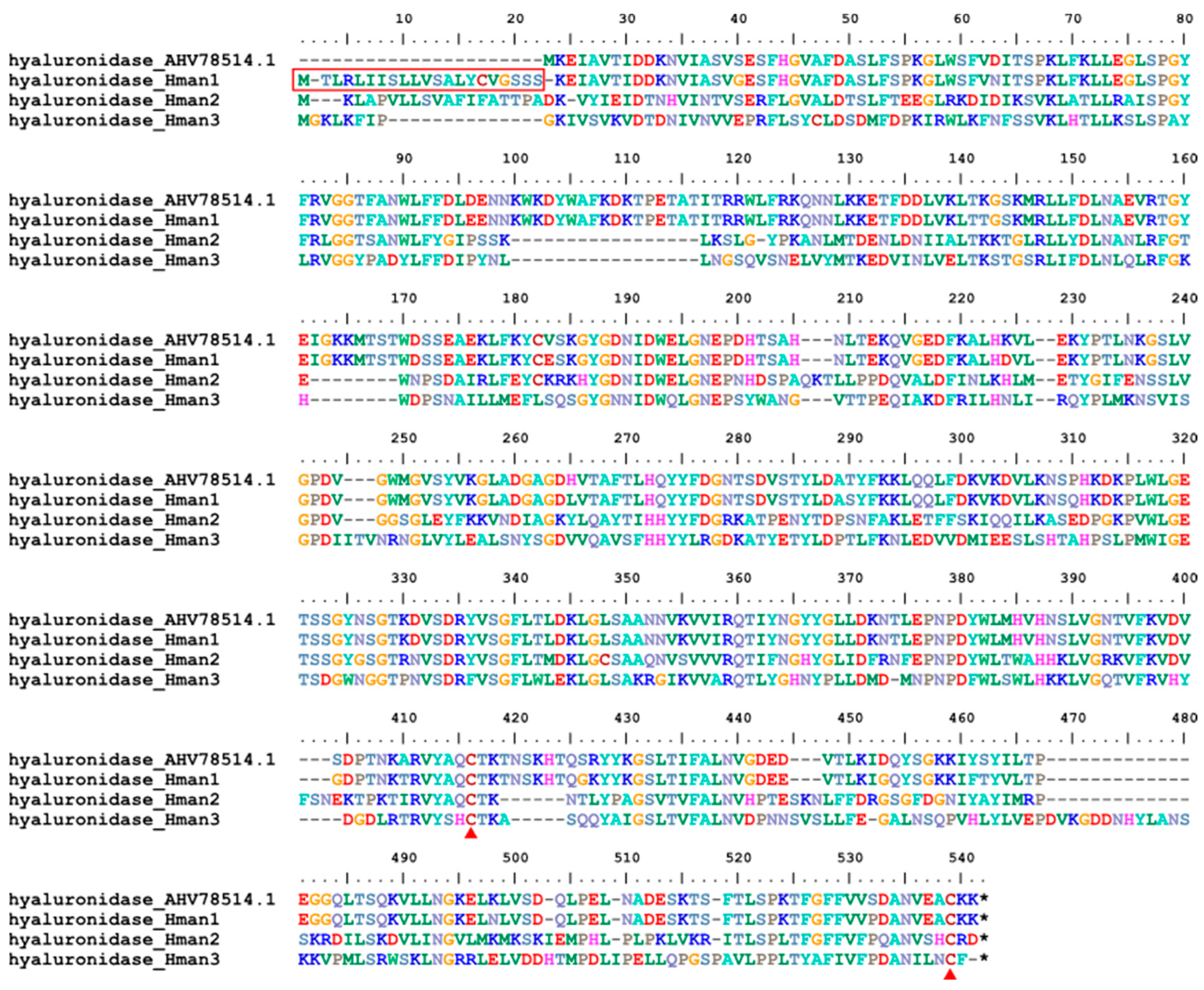
2.4. Variation Analysis of Antithrombotic Proteins
2.5. Anticoagulation Analyses of Hirudins
3. Results
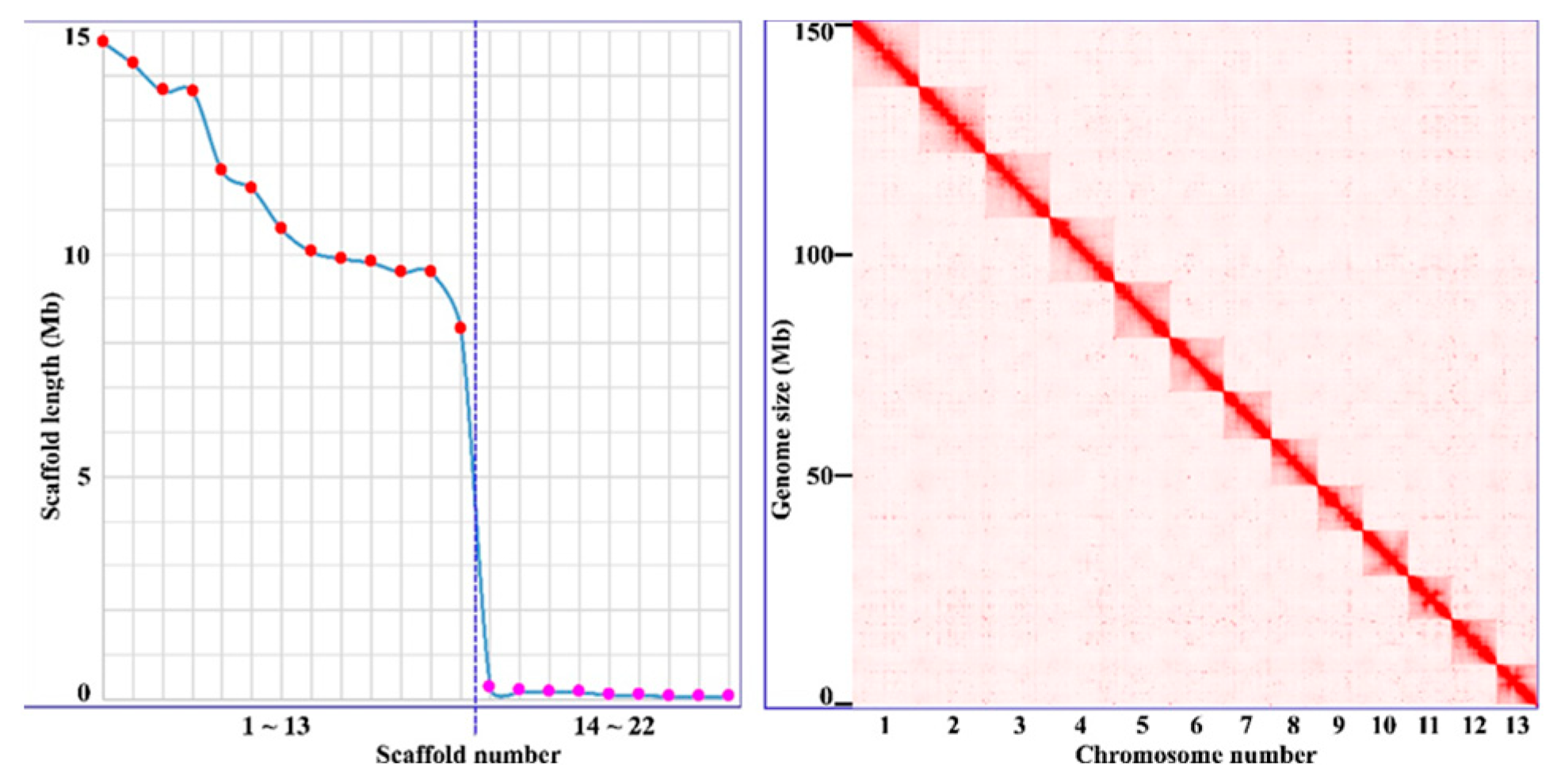
3.1. The Genome Sequencing and Assembling
3.2. Gene Prediction and Annotation
3.3. Identification of Antithrombotic Genes
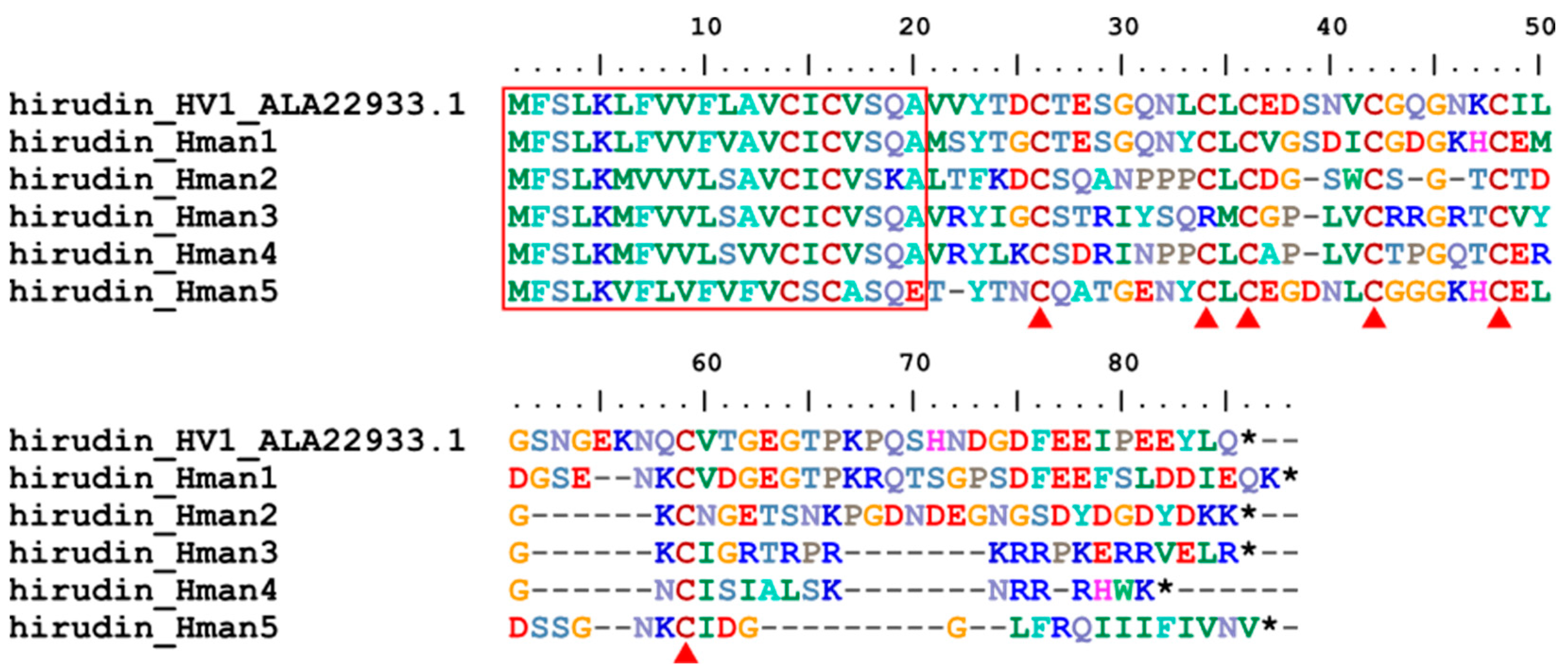
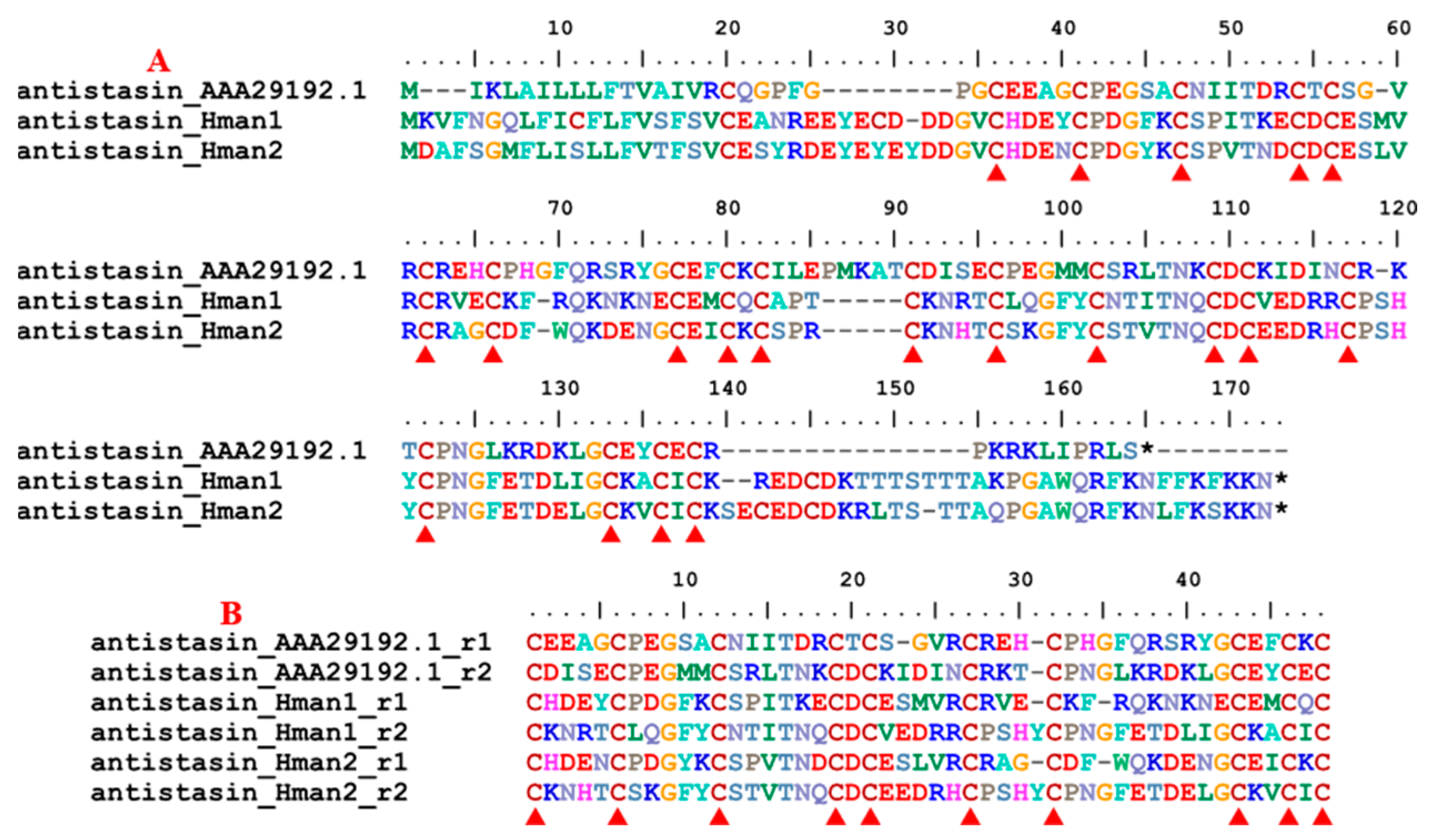
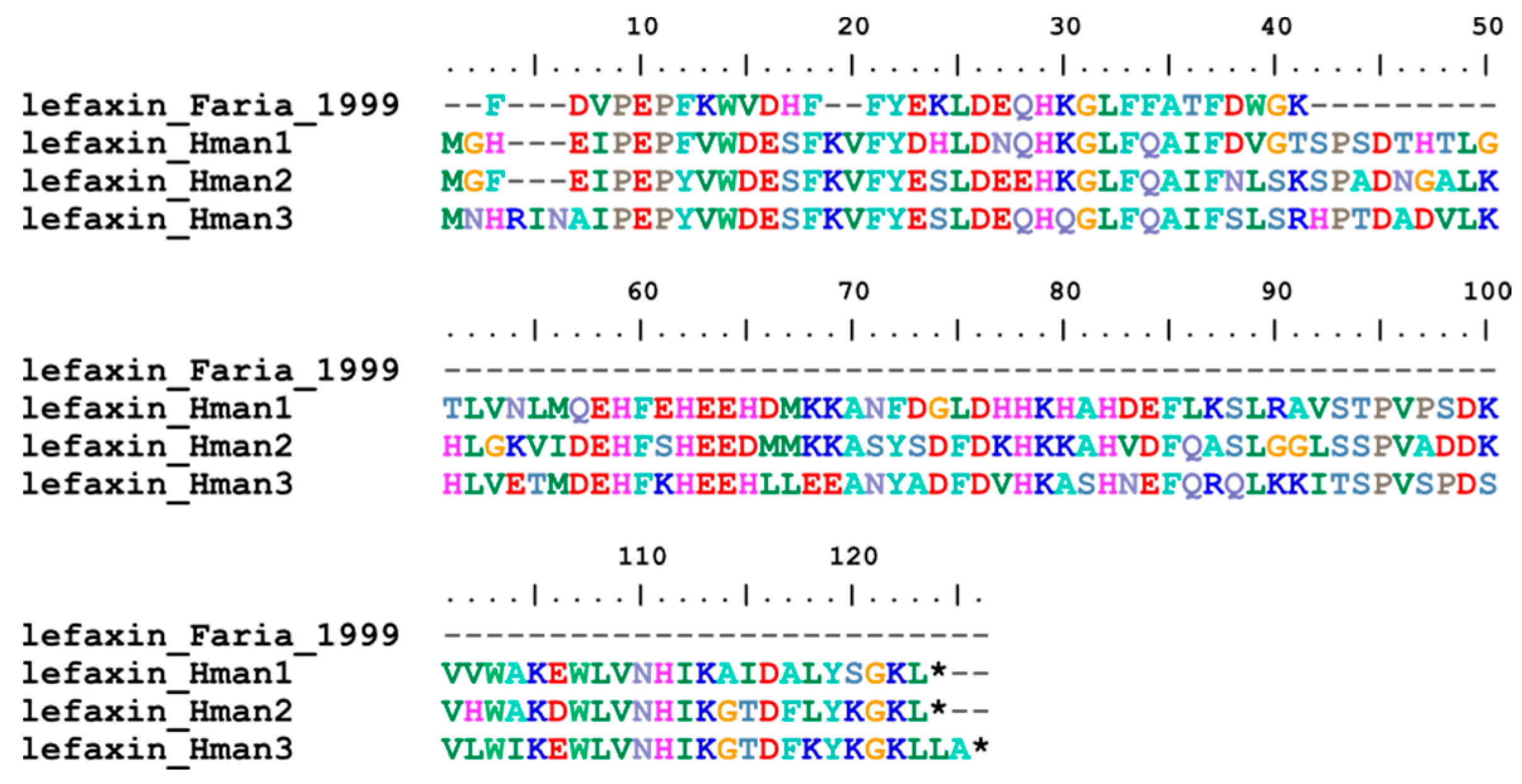
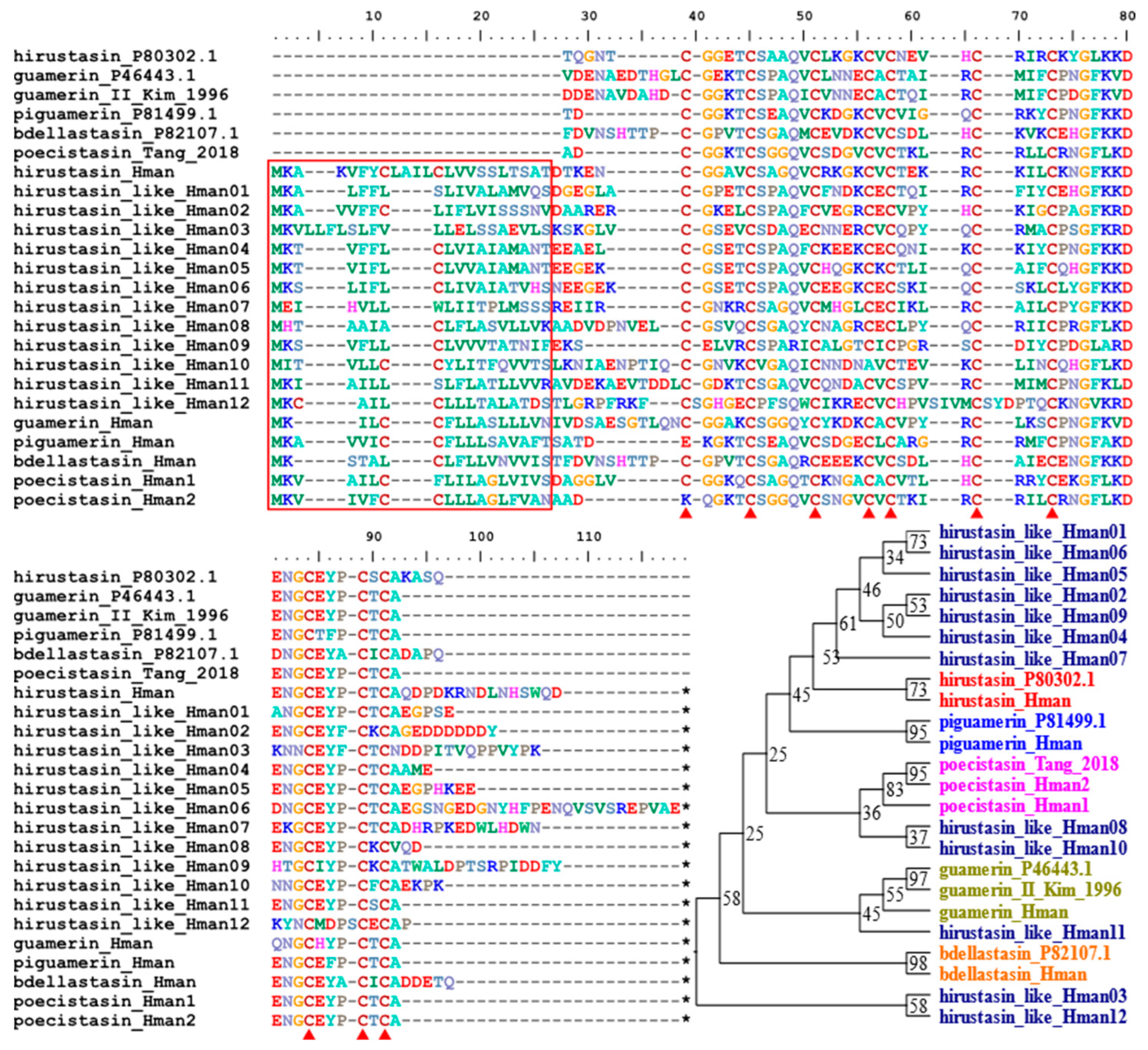
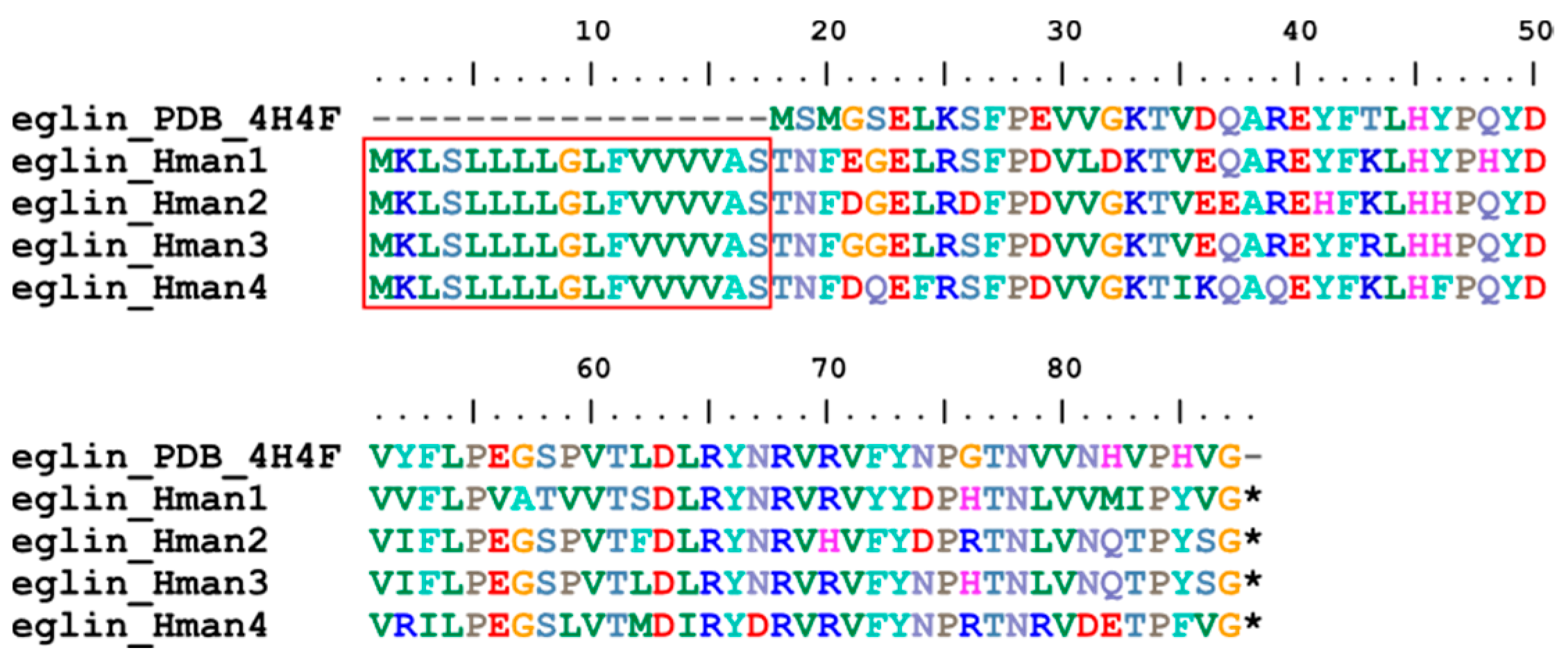
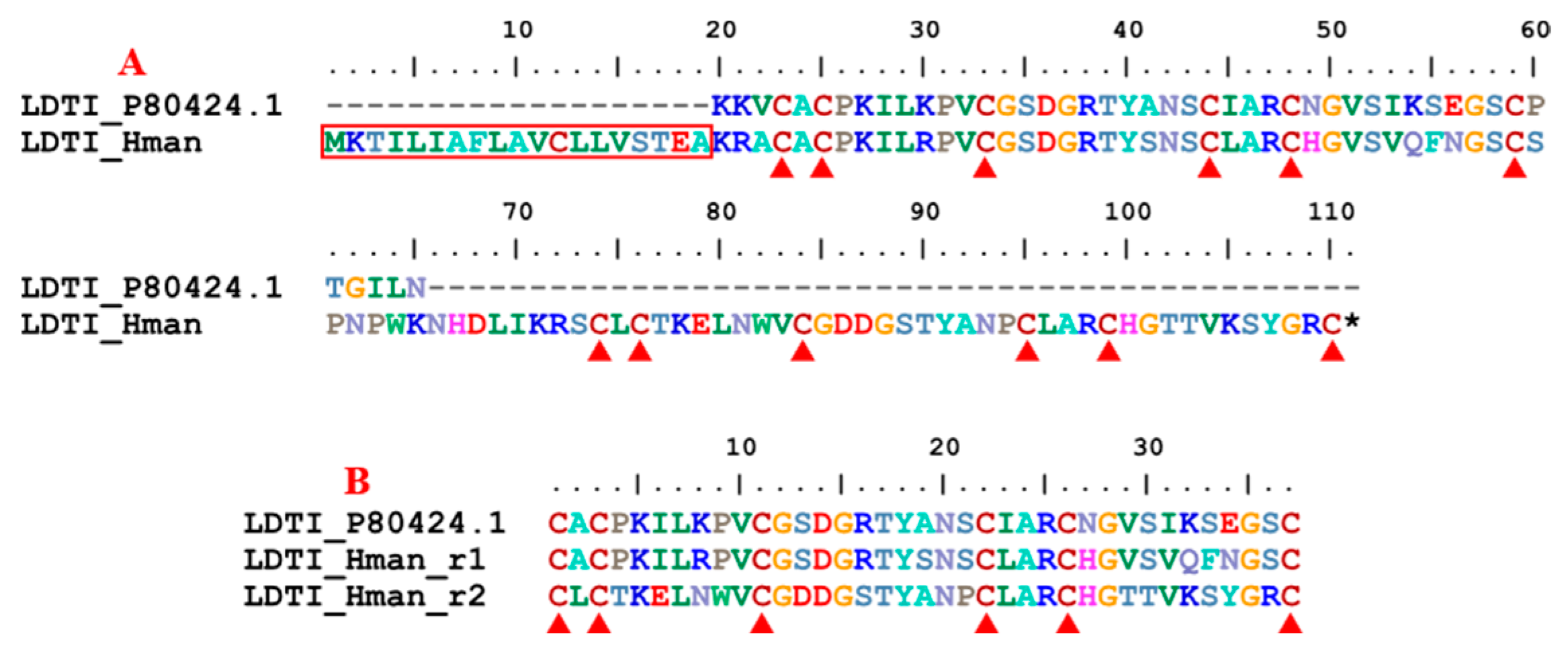
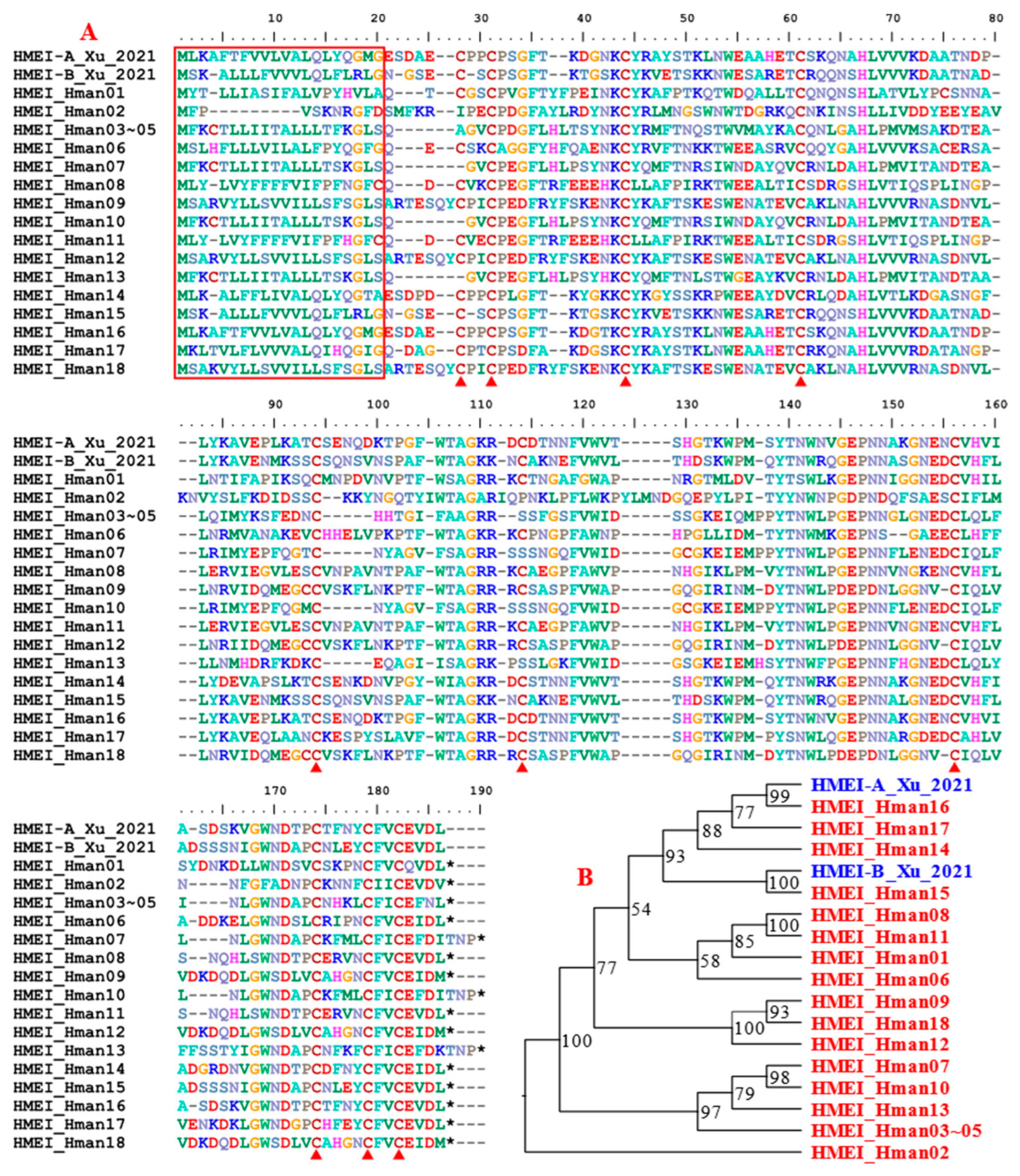
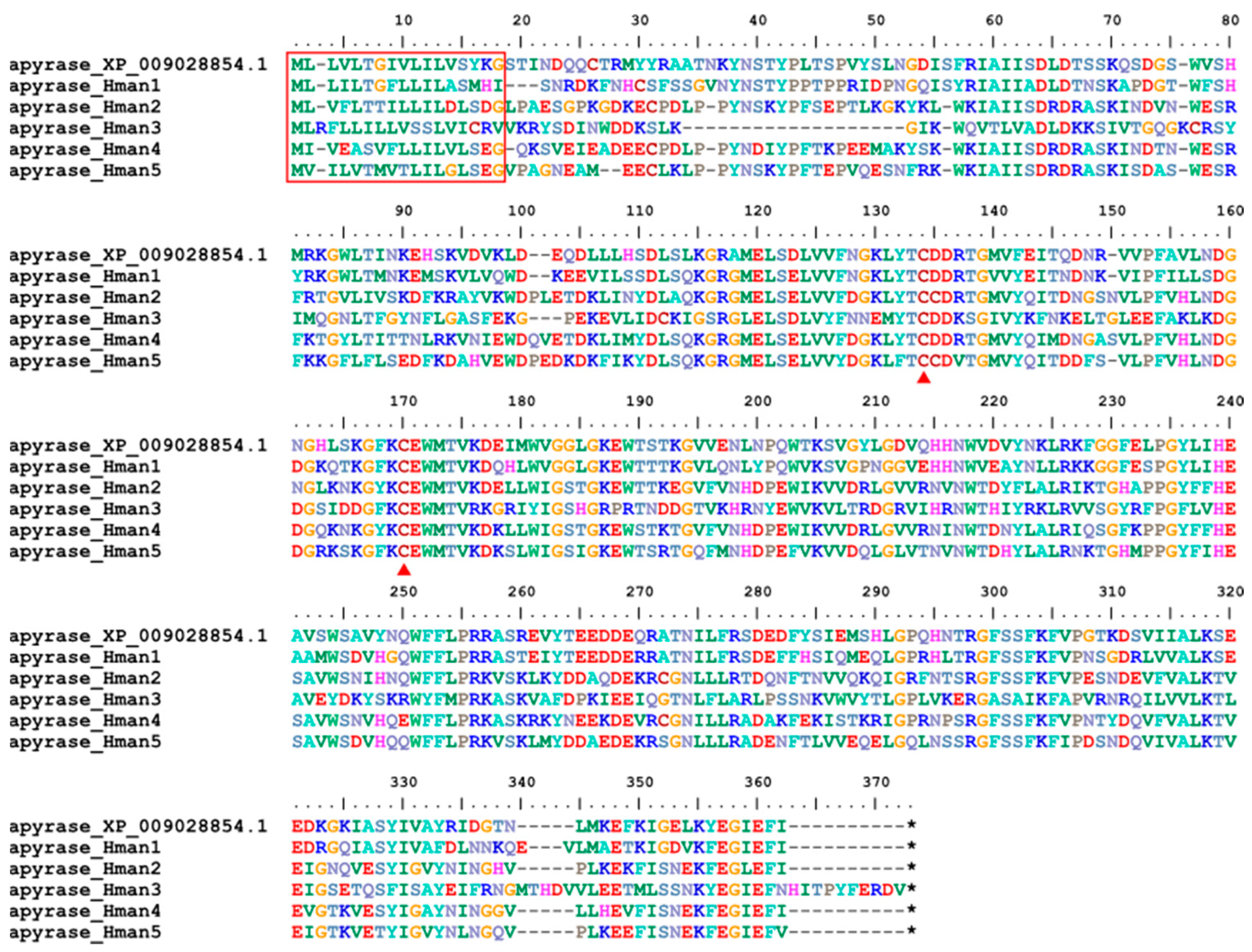
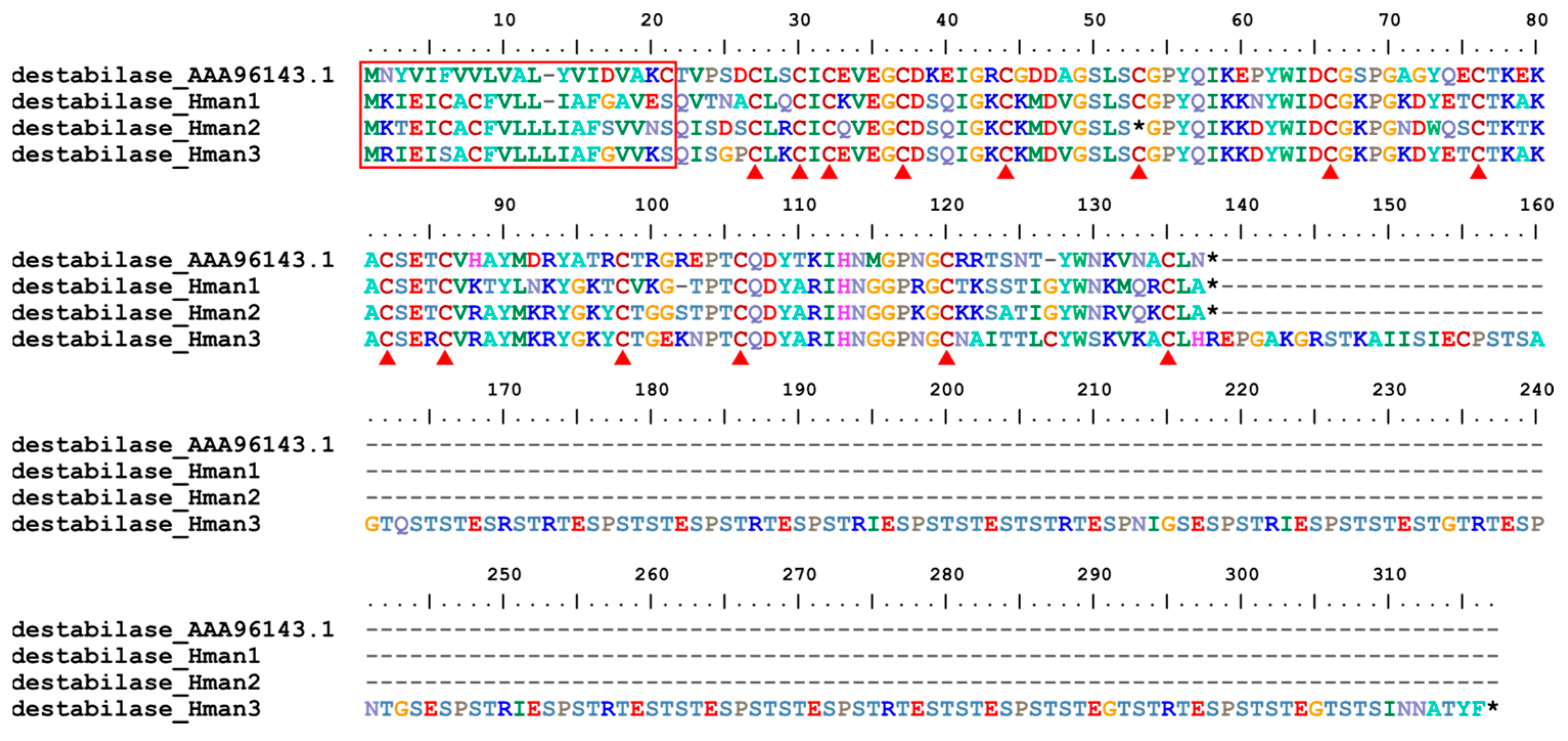
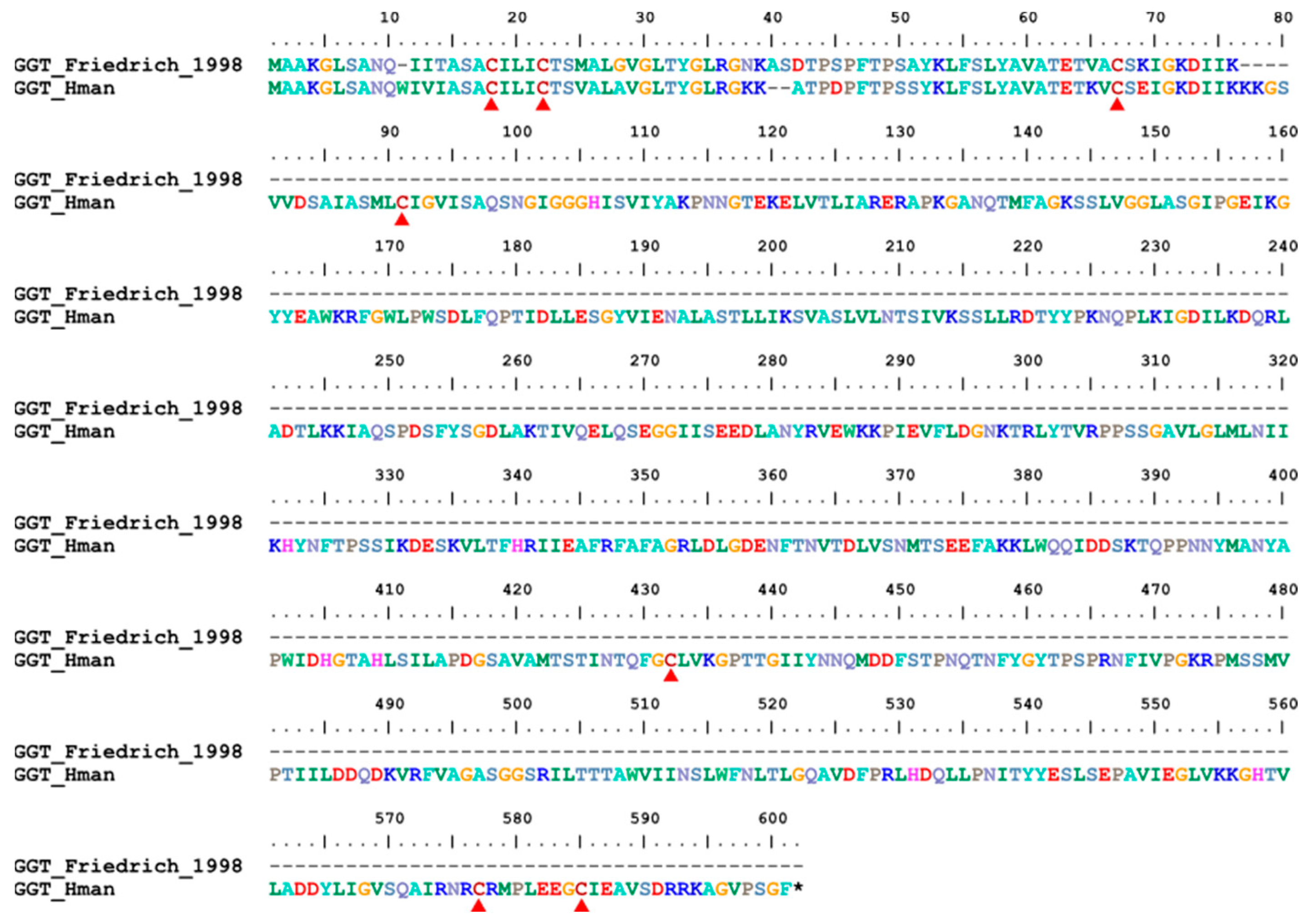
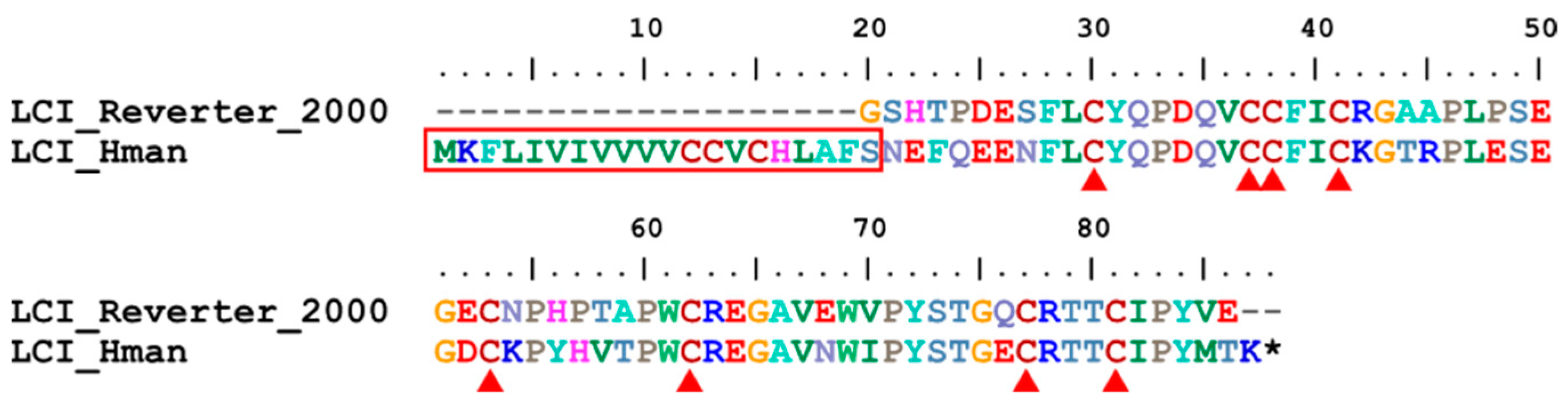
3.4. Genetic Variation of Antithrombotic Proteins
3.5. Anticoagulation of Recombinant Hirudins
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- DeLoughery, T.G. Hemostasis and Thrombosis; Springer Nature Switzerland AG: Cham, Switzerland, 2019. [Google Scholar]
- WHO. The Top 10 Causes of Death; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Mackman, N.; Bergmeier, W.; Stouffer, G.A.; Weitz, J.I. Therapeutic strategies for thrombosis: New targets and approaches. Nat. Rev. Drug Discov. 2020, 19, 333–352. [Google Scholar] [CrossRef] [PubMed]
- Elantably, D.; Mourad, A.; Elantably, A.; Effat, M. Warfarin induced leukocytoclastic vasculitis: An extraordinary side effect. J. Thromb. Thrombolysis 2020, 49, 149–152. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.J.; Wang, Y.N.; Song, Q.H.; Qiu, K.; Liu, M. Use of anticoagulant therapy and cerebral microbleeds: A systematic review and meta-analysis. J. Neurol. 2021, 268, 1666–1679. [Google Scholar] [CrossRef] [PubMed]
- Al-Husein, B.A.; Al-Azzam, S.I.; Alzoubi, K.H.; Khabour, O.F.; Nusair, M.B.; Alzayadeen, S. Investigating the effect of demographics, clinical characteristics, and polymorphism of MDR-1, CYP1A2, CYP3A4, and CYP3A5 on clopidogrel resistance. J. Cardiovasc. Pharmacol. 2018, 72, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Giahchi, F.; Mohammadi, M. Reteplase versus streptokinase in management of ST-segment elevation myocardial infarction; a letter to the editor. Adv. J. Emerg. Med. 2019, 3, e34. [Google Scholar] [PubMed]
- Sawyer, R.T. Leech Biology and Behaviour; Oxford University Press: Oxford, UK, 1986. [Google Scholar]
- Kvist, S.; Manzano-Marín, A.; de Carle, D.; Trontelj, P.; Siddall, M.E. Draft genome of the European medicinal leech Hirudo medicinalis (Annelida, Clitellata, Hirudiniformes) with emphasis on anticoagulants. Sci. Rep. 2020, 10, 9885. [Google Scholar] [CrossRef]
- Sket, B.; Trontelj, P. Global diversity of leeches (Hirudinea) in freshwater. Hydrobiologia 2008, 595, 129–137. [Google Scholar] [CrossRef]
- Sig, A.K.; Guney, M.; Guclu, A.U.; Ozmen, E. Medicinal leech therapy—An overall perspective. Integr. Med. Res. 2017, 6, 337–343. [Google Scholar] [CrossRef]
- Ma, C.J.; Li, X.; Chen, H. Research progress in the use of leeches for medical purposes. Tradit. Med. Res. 2021, 6, 56–69. [Google Scholar] [CrossRef]
- Shakouri, A.; Adljouy, N.; Balkani, S.; Mohamadi, M.; Hamishehkar, H.; Abdolalizadeh, J.; Shakouri, S.K. Effectiveness of topical gel of medical leech (Hirudo medicinalis) saliva extract on patients with knee osteoarthritis: A randomized clinical trial. Complement. Ther. Clin. Pract. 2018, 31, 352–359. [Google Scholar] [CrossRef]
- Hohmann, C.D.; Stange, R.; Steckhan, N.; Robens, S.; Ostermann, T.; Paetow, A.; Michalsen, A. The effectiveness of leech therapy in chronic low back pain. Dtsch. Arztebl. Int. 2018, 115, 785–792. [Google Scholar] [CrossRef]
- Hamidizadeh, N.; Azizi, A.; Zarshenas, M.M.; Ranjbar, S. Leech therapy in treatment of cutaneous leishmaniasis: A case report. J. Integr. Med. 2017, 15, 407–410. [Google Scholar] [CrossRef]
- Michalsen, A.; Roth, M.; Dobos, G. Medicinal Leech Therapy; Georg Thieme Verlag: Stuttgart, Germany, 2007. [Google Scholar]
- Elyassi, A.R.; Terres, J.; Rowshan, H.H. Medicinal leech therapy on head and neck patients: A review of literature and proposed protocol. Oral. Surg. Oral. Med. Oral. Pathol. Oral. Radiol. 2013, 116, e167–e172. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Ren, J.X.; Wang, J.J.; Ding, L.S.; Zhao, J.J.; Liu, S.Y.; Gao, H.M. Chinese medicinal leech: Ethnopharmacology, phytochemistry, and pharmacological activities. Evid. Based Complement. Altern. Med. 2016, 2016, 7895935. [Google Scholar] [CrossRef] [PubMed]
- Song, J.X.; Lyu, Y.; Wang, M.M.; Zhang, J.; Gao, L.; Tong, X.L. Treatment of human urinary kallidinogenase combined with maixuekang capsule promotes good functional outcome in ischemic stroke. Front. Physiol. 2018, 9, 84. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.M.A.; Jameel, S.S.; Zaman, F.; Jilani, S.; Sultana, A.; Khan, S.A. A systematic overview of the medicinal importance of sanguivorous leeches. Altern. Med. Rev. 2011, 16, 59–65. [Google Scholar]
- Fritsma, G.A. Monitoring the direct thrombin inhibitors. Clin. Lab. Sci. 2013, 26, 54–57. [Google Scholar] [CrossRef]
- Müller, C.; Lukas, P.; Böhmert, M.; Hildebrandt, J.P. Hirudin or hirudin-like factor—That is the question: Insights from the analyses of natural and synthetic HLF variants. FEBS Lett. 2020, 594, 841–850. [Google Scholar] [CrossRef]
- Mousa, R.; Hidmi, T.; Pomyalov, S.; Lansky, S.; Khouri, L.; Shalev, D.E.; Shoham, G.; Metanis, N. Diselenide crosslinks for enhanced and simplified oxidative protein folding. Commun. Chem. 2021, 4, 30. [Google Scholar] [CrossRef]
- Montinari, M.R.; Minelli, S. From ancient leech to direct thrombin inhibitors and beyond: New from old. Biomed. Pharmacother. 2022, 149, 112878. [Google Scholar] [CrossRef]
- Chen, J.R.; Xie, X.F.; Zhang, H.Q.; Li, G.M.; Yin, Y.P.; Cao, X.Y.; Gao, Y.Q.; Li, Y.N.; Zhang, Y.; Peng, F.; et al. Pharmacological activities and mechanisms of hirudin and its derivatives—A review. Front. Pharmacol. 2021, 12, 660757. [Google Scholar]
- Cheng, R.M.; Tang, X.P.; Long, A.L.; Mwangi, J.; Lai, R.; Sun, R.P.; Long, C.B.; Zhang, Z.Q. Purification and characterization of a novel anti-coagulant from the leech Hirudinaria manillensis. Zool. Res. 2019, 40, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Zavalova, L.; Lukyanov, S.; Baskova, I.; Snezhkov, E.; Akopov, S.; Berezhnoy, S.; Bogdanova, E.; Barsova, E.; Sverdlov, E.D. Genes from the medicinal leech (Hirudo medicinalis) coding for unusual enzymes that specifically cleave endo-epsilon (γ-Glu)-Lys isopeptide bonds and help to dissolve blood clots. Mol. Gen. Genet. 1996, 253, 20–25. [Google Scholar] [CrossRef]
- Reverter, D.; Fernández-Catalán, C.; Baumgartner, R.; Pfänder, R.; Huber, R.; Bode, W.; Vendrell, J.; Holak, T.A.; Avilés, F.X. Structure of a novel leech carboxypeptidase inhibitor determined free in solution and in complex with human carboxypeptidase A2. Nat. Struct. Biol. 2000, 7, 322–328. [Google Scholar]
- Jin, P.; Kang, Z.; Zhang, N.; Du, G.C.; Chen, J. High-yield novel leech hyaluronidase to expedite the preparation of specific hyaluronan oligomers. Sci. Rep. 2014, 4, 4471. [Google Scholar] [CrossRef] [PubMed]
- Gronwald, W.; Bomke, J.; Maurer, T.; Domogalla, B.; Huber, F.; Schumann, F.; Kremer, W.; Fink, F.; Rysiok, T.; Frech, M.; et al. Structure of the leech protein saratin and characterization of its binding to collagen. J. Mol. Biol. 2008, 381, 913–927. [Google Scholar] [CrossRef]
- Kvist, S.; Min, G.S.; Siddall, M.E. Diversity and selective pressures of anticoagulants in three medicinal leeches (Hirudinida: Hirudinidae, Macrobdellidae). Ecol. Evol. 2013, 3, 918–933. [Google Scholar] [CrossRef] [PubMed]
- Iwama, R.E.; Tessler, M.; Kvist, S. Leech anticoagulants are ancestral and likely to be multifunctional. Zool. J. Linn. Soc. 2022, 196, 137–148. [Google Scholar] [CrossRef]
- Babenko, V.V.; Podgorny, O.V.; Manuvera, V.A.; Kasianov, A.S.; Manolov, A.I.; Grafskaia, E.N.; Shirokov, D.A.; Kurdyumov, A.S.; Vinogradov, D.V.; Nikitina, A.S.; et al. Draft genome sequences of Hirudo medicinalis and salivary transcriptome of three closely related medicinal leeches. BMC Genom. 2020, 21, 331. [Google Scholar]
- Zheng, F.S.; Zhang, M.; Yang, X.W.; Wu, F.L.; Wang, G.; Feng, X.X.; Ombati, R.; Zuo, R.L.; Yang, C.J.; Liu, J.; et al. Prostaglandin E1 is an efficient molecular tool for forest leech blood sucking. Front. Vet. Sci. 2021, 7, 615915. [Google Scholar] [CrossRef]
- Zheng, J.H.; Wang, X.B.; Feng, T.; Rehman, S.U.; Yan, X.Y.; Shan, H.Q.; Ma, X.C.; Zhou, W.G.; Xu, W.H.; Lu, L.Y.; et al. Molecular mechanisms underlying hematophagia revealed by comparative analyses of leech genomes. Gigascience 2023, 12, giad023. [Google Scholar] [CrossRef] [PubMed]
- Müller, C.; Haase, M.; Lemke, S.; Hildebrandt, J.P. Hirudins and hirudin-like factors in Hirudinidae: Implications for function and phylogenetic relationships. Parasitol. Res. 2017, 116, 313–325. [Google Scholar] [CrossRef] [PubMed]
- Tubtimon, J.; Jeratthitikul, E.; Sutcharit, C.; Kongim, B.; Panha, S. Systematics of the freshwater leech genus Hirudinaria Whitman, 1886 (Arhynchobdellida, Hirudinidae) from northeastern Thailand. Zookeys 2014, 452, 15–33. [Google Scholar]
- Jeratthitikul, E.; Jiranuntskul, P.; Nakano, T.; Sutcharit, C.; Panha, S. A new species of buffalo leech in the genus Hirudinaria Whitman, 1886 (Arhynchobdellida, Hirudinidae) from Thailand. Zookeys 2020, 933, 1–14. [Google Scholar] [CrossRef]
- Electricwala, A.; Hartwell, R.; Scawen, M.D.; Atkinson, T. The complete amino acid sequence of a hirudin variant from the leech Hirudinaria manillensis. J. Protein Chem. 1993, 12, 365–370. [Google Scholar] [CrossRef]
- Liu, F.; Guo, Q.S.; Shi, H.Z.; Cheng, B.X.; Lu, Y.X.; Gou, L.; Wang, J.; Shen, W.B.; Yan, S.M.; Wu, M.J. Genetic variation in Whitmania pigra, Hirudo nipponica and Poecilobdella manillensis, three endemic and endangered species in China using SSR and TRAP markers. Gene 2016, 579, 172–182. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, B.; Gong, Y.; Yu, X.; Lv, J.Y. Anticoagulant active substances extraction and anti-thrombin activity analysis of several species of leeches. Acta Sci. Nat. Univ. Sun 2012, 51, 92–96. [Google Scholar]
- Guan, D.L.; Yang, J.; Liu, Y.K.; Li, Y.; Mi, D.; Ma, L.B.; Wang, Z.Z.; Xu, S.Q.; Qiu, Q. Draft genome of the Asian buffalo leech Hirudinaria manillensis. Front. Genet. 2020, 10, 1321. [Google Scholar] [CrossRef]
- Hu, J.; Fan, J.; Sun, Z.; Liu, S. NextPolish: A fast and efficient genome polishing tool for long-read assembly. Bioinformatics 2020, 36, 2253–2255. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Zhang, H.W.; Song, L.; Wang, X.T.; Cheng, H.Y.; Wang, C.F.; Meyer, C.A.; Liu, T.; Tang, M.; Aluru, S.; Yue, F.; et al. Fast alignment and preprocessing of chromatin profiles with Chromap. Nat. Commun. 2021, 12, 6566. [Google Scholar] [CrossRef] [PubMed]
- Durand, N.C.; Shamim, M.S.; Machol, I.; Rao, S.S.P.; Huntley, M.H.; Lander, E.S.; Aiden, E.L. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 2016, 3, 95–98. [Google Scholar] [CrossRef]
- Zhou, C.; McCarthy, S.A.; Durbin, R. YaHS: Yet another Hi-C scaffolding tool. Bioinformatics 2023, 39, btac808. [Google Scholar] [CrossRef] [PubMed]
- Durand, N.C.; Robinson, J.T.; Shamim, M.S.; Machol, I.; Mesirov, J.P.; Lander, E.S.; Aiden, E.L. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 2016, 3, 99–101. [Google Scholar] [CrossRef]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; dePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef] [PubMed]
- Seppey, M.; Manni, M.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness. Methods Mol. Biol. 2019, 1962, 227–245. [Google Scholar]
- Rhie, A.; Walenz, B.P.; Koren, S.; Phillippy, A.M. Merqury: Reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 2020, 21, 245. [Google Scholar] [CrossRef]
- Flynn, J.M.; Hubley, R.; Goubert, C.; Rosen, J.; Clark, A.G.; Feschotte, C.; Smit, A.F. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. USA 2020, 117, 9451–9457. [Google Scholar] [CrossRef]
- Bao, W.D.; Kojima, K.K.; Kohany, O. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 2015, 6, 11. [Google Scholar] [CrossRef]
- Majoros, W.H.; Pertea, M.; Salzberg, S.L. TigrScan and GlimmerHMM: Two open source ab initio eukaryotic gene-finders. Bioinformatics 2004, 20, 2878–2879. [Google Scholar] [CrossRef]
- Korf, I. Gene finding in novel genomes. BMC Bioinform. 2004, 5, 59. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Delcher, A.L.; Mount, S.M.; Wortman, J.R.; Smith, R.K.J.; Hannick, L.I.; Maiti, R.; Ronning, C.M.; Rusch, D.B.; Town, C.D.; et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003, 31, 5654–5666. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Hoff, K.J.; Lange, S.; Lomsadze, A.; Borodovsky, M.; Stanke, M. BRAKER1: Unsupervised RNA-seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 2016, 32, 767–769. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Salzberg, S.L.; Zhu, W.; Pertea, M.; Allen, J.E.; Orvis, J.; White, O.; Buell, C.R.; Wortman, J.R. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 2008, 9, R7. [Google Scholar] [CrossRef]
- Pertea, G.; Pertea, M. GFF utilities: GffRead and GffCompare. F1000Research 2020, 9, 304. [Google Scholar] [CrossRef]
- Sharma, K.; Singh, A.K.; Maddipatla, D.K.; Deshwal, G.K.; Rao, P.S.; Sharma, H. Eggnog: Process optimization and characterization of a dairy-based beverage. J. Dairy. Res. 2023, 90, 205–212. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Lomsadze, A.; Borodovsky, M. Identification of protein coding regions in RNA transcripts. Nucleic Acids Res. 2015, 43, e78. [Google Scholar] [CrossRef] [PubMed]
- Slater, G.S.C.; Birney, E. Automated generation of heuristics for biological sequence comparison. BMC Bioinform. 2005, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Dainat, J. AGAT: Another Gff Analysis Toolkit to Handle Annotations in Any GTF/GFF Format, v0.7.0; Zenodo: Meyrin, Switzerland, 2021. [Google Scholar] [CrossRef]
- Shen, W.; Le, S.; Li, Y.; Hu, F.Q. SeqKit: A cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS ONE 2016, 11, e0163962. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European molecular biology open software suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Hu, Z.L.; Zhang, N.; Gu, F.; Li, Y.; Deng, X.J.; Chen, G.P. Expression, purification and characterization of recombinant targeting bifunctional hirudin in Pichia pastoris. Afr. J. Biotechnol. 2009, 8, 5582–5588. [Google Scholar]
- Chinese Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China; Medicine Science and Technology Press: Beijing, China, 2020.
- Hong, S.J.; Kang, K.W. Purification of granulin-like polypeptide from the blood-sucking leech, Hirudo nipponia. Protein Expr. Purif. 1999, 16, 340–346. [Google Scholar] [CrossRef]
- Tang, X.P.; Chen, M.R.; Duan, Z.L.; Mwangi, J.; Li, P.P.; Lai, R. Isolation and characterization of poecistasin, an anti-thrombotic antistasin-type serine protease inhibitor from leech Poecilobdella manillensis. Toxins 2018, 10, 429. [Google Scholar] [CrossRef]
- Xu, K.H.; Zhou, M.; Wu, F.L.; Tang, X.P.; Lu, Q.M.; Lai, R.; Long, C.B. Identification and characterization of a novel elastase inhibitor from Hirudinaria manillensis. Chin. J. Nat. Med. 2021, 19, 540–544. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, T.; Kröger, B.; Koerwer, W.; Strube, K.H.; Meyer, T.; Bialojan, S. An isopeptide bond splitting enzyme from Hirudo medicinalis similar to γ-glutamyl transpeptidase. Eur. J. Biochem. 1998, 256, 297–302. [Google Scholar] [CrossRef]
- Markwardt, F. Untersuchungen über hirudin. Naturwissenschaften 1955, 42, 537–538. [Google Scholar] [CrossRef]
- Vitali, J.; Martin, P.D.; Malkowski, M.G.; Robertson, W.D.; Lazar, J.B.; Winant, R.C.; Johnson, P.H.; Edwards, B.F. The structure of a complex of bovine α-thrombin and recombinant hirudin at 2.8-A resolution. J. Biol. Chem. 1992, 267, 17670–17678. [Google Scholar] [CrossRef] [PubMed]
- Dodt, J.; Müller, H.P.; Seemüller, U.; Chang, J.Y. The complete amino acid sequence of hirudin, a thrombin specific inhibitor: Application of colour carboxymethylation. FEBS Lett. 1984, 165, 180–184. [Google Scholar] [CrossRef]
- Harvey, R.P.; Degryse, E.; Stefani, L.; Schamber, F.; Cazenave, J.P.; Courtney, M.; Tolstoshev, P.; Lecocq, J.P. Cloning and expression of a cDNA coding for the anticoagulant hirudin from the bloodsucking leech, Hirudo medicinalis. Proc. Natl. Acad. Sci. USA 1986, 83, 1084–1088. [Google Scholar] [CrossRef] [PubMed]
- Müller, C.; Mescke, K.; Liebig, S.; Mahfoud, H.; Lemke, S.; Hildebrandt, J.P. More than just one: Multiplicity of hirudins and hirudin-like factors in the medicinal leech, Hirudo medicinalis. Mol. Genet. Genom. 2016, 291, 227–240. [Google Scholar] [CrossRef] [PubMed]
- Scacheri, E.; Nitti, G.; Valsasina, B.; Orsini, G.; Visco, C.; Ferrera, M.; Sawyer, R.T.; Sarmientos, P. Novel hirudin variants from the leech Hirudinaria manillensis. Amino acid sequence, cDNA cloning and genomic organization. Eur. J. Biochem. 1993, 214, 295–304. [Google Scholar] [CrossRef]
- Strube, K.H.; Kröger, B.; Bialojan, S.; Otte, M.; Dodt, J. Isolation, sequence analysis, and cloning of haemadin. An anticoagulant peptide from the Indian leech. J. Biol. Chem. 1993, 268, 8590–8595. [Google Scholar] [CrossRef]
- Tong, L.; Dai, S.X.; Kong, D.J.; Yang, P.P.; Tong, X.; Tong, X.R.; Bi, X.X.; Su, Y.; Zhao, Y.Q.; Liu, Z.C. The genome of medicinal leech (Whitmania pigra) and comparative genomic study for exploration of bioactive ingredients. BMC Genom. 2022, 23, 76. [Google Scholar] [CrossRef]
- Tanaka, Y.; Suzuki, G.; Matsuwaki, T.; Hosokawa, M.; Serrano, G.; Beach, T.G.; Yamanouchi, K.; Hasegawa, M.; Nishihara, M. Progranulin regulates lysosomal function and biogenesis through acidification of lysosomes. Hum. Mol. Genet. 2017, 26, 969–988. [Google Scholar] [CrossRef]
- Mittl, P.R.E.; Di Marco, S.; Fendrich, G.; Pohlig, G.; Heim, J.; Sommerhoff, C.; Fritz, H.; Priestle, J.P.; Grütter, M.G. A new structural class of serine protease inhibitors revealed by the structure of the hirustasin-kallikrein complex. Structure 1997, 5, 253–264. [Google Scholar] [CrossRef]
- Dunwiddie, C.; Thornberry, N.A.; Bull, H.G.; Sardana, M.; Friedman, P.A.; Jacobs, J.W.; Simpson, E. Antistasin, a leech-derived inhibitor of factor Xa. Kinetic analysis of enzyme inhibition and identification of the reactive site. J. Biol. Chem. 1989, 264, 16694–16699. [Google Scholar] [CrossRef]
- Nutt, E.; Gasic, T.; Rodkey, J.; Gasic, G.J.; Jacobs, J.W.; Friedman, P.A.; Simpson, E. The amino acid sequence of antistasin. A potent inhibitor of factor Xa reveals a repeated internal structure. J. Biol. Chem. 1988, 263, 10162–10167. [Google Scholar] [CrossRef]
- Blankenship, D.T.; Brankamp, R.G.; Manley, G.D.; Cardin, A.D. Amino acid sequence of ghilanten: Anticoagulant-antimetastatic principle of the south American leech, Haementeria ghilianii. Biochem. Bioph Res. Co. 1990, 166, 1384–1389. [Google Scholar] [CrossRef]
- Han, J.H.; Law, S.W.; Keller, P.M.; Kniskern, P.J.; Silberklang, M.; Tung, J.S.; Gasic, T.B.; Gasic, G.J.; Friedman, P.A.; Ellis, R.W. Cloning and expression of cDNA encoding antistasin, a leech-derived protein having anti-coagulant and anti-metastatic properties. Gene 1989, 75, 47–57. [Google Scholar] [CrossRef]
- Faria, F.; Kelen, E.M.; Sampaio, C.A.; Bon, C.; Duval, N.; Chudzinski-Tavassi, A.M. A new factor Xa inhibitor (lefaxin) from the Haementeria depressa leech. Thromb. Haemost. 1999, 82, 1469–1473. [Google Scholar] [CrossRef] [PubMed]
- Chopin, V.; Salzet, M.; Baert, J.l.; Vandenbulcke, F.; Sautiére, P.E.; Kerckaert, J.P.; Malecha, J. Therostasin, a novel clotting factor Xa inhibitor from the rhynchobdellid leech, Theromyzon tessulatum. J. Biol. Chem. 2000, 275, 32701–32707. [Google Scholar] [CrossRef] [PubMed]
- Söllner, C.; Mentele, R.; Eckerskorn, C.; Fritz, H.; Sommerhoff, C.P. Isolation and characterisation of hirustasin, an antistasin-type serine-proteinase inhibitor from the medical leech Hirudo medicinalis. Eur. J. Biochem. 1994, 219, 937–943. [Google Scholar] [CrossRef]
- Jung, H.I.; Kim, S.I.; Ha, K.S.; Joe, C.O.; Kang, K.W. Isolation and characterization of guamerin, a new human leukocyte elastase inhibitor from Hirudo nipponia. J. Biol. Chem. 1995, 270, 13879–13884. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.R.; Kang, K.W. Amino acid sequence of piguamerin, an antistasin-type protease inhibitor from the blood sucking leech Hirudo nipponia. Eur. J. Biochem. 1998, 254, 692–697. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.R.; Hong, S.J.; Ha, K.S.; Joe, C.O.; Kang, K.W. A cysteine-rich serine protease inhibitor (guamerin II) from the non-blood sucking leech Whitmania edentula: Biochemical characterization and amino acid sequence analysis. J. Enzym. Inhib. 1996, 10, 81–91. [Google Scholar] [CrossRef]
- Moser, M.; Auerswald, E.; Mentele, R.; Eckerskorn, C.; Fritz, H.; Fink, E. Bdellastasin, a serine protease inhibitor of the antistasin family from the medical leech (Hirudo medicinalis)—Primary structure, expression in yeast, and characterisation of native and recombinant inhibitor. Eur. J. Biochem. 1998, 253, 212–220. [Google Scholar] [CrossRef]
- Seemüller, U.; Eulitz, M.; Fritz, H.; Strobl, A. Structure of the elastase-cathepsin G inhibitor of the leech Hirudo medicinalis. Hoppe Seylers Z. Physiol. Chem. 1980, 361, 1841–1846. [Google Scholar]
- Renesto, P.; Ferrer-Lopez, P.; Chignard, M. Eglin C and heparin inhibition of platelet activation induced by cathepsin G or human neutrophils. Ann. N. Y Acad. Sci. 1991, 624, 321–324. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.Y.; Knecht, R.; Maschler, R.; Seemüller, U. Elastase-cathepsin G inhibitors eglin b and eglin c differ by a single Tyr—His substitution. A micro-method for the identification of amino-acid substitution. Biol. Chem. Hoppe Seyler 1985, 366, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Fritz, H.; Oppitz, K.H.; Gebhardt, M.; Oppitz, I.; Werle, E.; Marx, R. On the presence of a trypsin-plasmin inhibitor in hirudin. Hoppe Seylers Z. Physiol. Chem. 1969, 350, 91–92. [Google Scholar]
- Fink, E.; Rehm, H.; Gippner, C.; Bode, W.; Eulitz, M.; Machleidt, W.; Fritz, H. The primary structure of bdellin B-3 from the leech Hirudo medicinalis. Bdellin B-3 is a compact proteinase inhibitor of a “non-classical” Kazal type. It is present in the leech in a high molecular mass form. Biol. Chem. Hoppe Seyler 1986, 367, 1235–1242. [Google Scholar] [CrossRef]
- Laskowski, M.J.; Kato, I. Protein inhibitors of proteinases. Annu. Rev. Biochem. 1980, 49, 593–626. [Google Scholar] [CrossRef]
- Campos, I.T.N.; Silva, M.M.; Azzolini, S.S.; Souza, A.F.; Sampaio, C.A.M.; Fritz, H.; Tanaka, A.S. Evaluation of phage display system and leech-derived tryptase inhibitor as a tool for understanding the serine proteinase specificities. Arch. Biochem. Biophys. 2004, 425, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Sommerhoff, C.P.; Söllner, C.; Mentele, R.; Piechottka, G.P.; Auerswald, E.A.; Fritz, H. A Kazal-type inhibitor of human mast cell tryptase: Isolation from the medical leech Hirudo medicinalis, characterization, and sequence analysis. Biol. Chem. Hoppe Seyler 1994, 375, 685–694. [Google Scholar] [CrossRef] [PubMed]
- Pohlig, G.; Fendrich, G.; Knecht, R.; Eder, B.; Piechottka, G.; Sommerhoff, C.P.; Heim, J. Purification, characterization and biological evaluation of recombinant leech-derived tryptase inhibitor (rLDTI) expressed at high level in the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 1996, 241, 619–626. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, A.S.; Silva, M.M.; Torquato, R.J.; Noguti, M.A.; Sampaio, C.A.; Fritz, H.; Auerswald, E.A. Functional phage display of leech-derived tryptase inhibitor (LDTI): Construction of a library and selection of thrombin inhibitors. FEBS Lett. 1999, 458, 11–16. [Google Scholar] [CrossRef]
- Zhou, Y.L.; Xu, Z.D.; Liu, Z.Q. Impact of neutrophil extracellular traps on thrombosis formation: New findings and future perspective. Front. Cell Infect. Microbiol. 2022, 12, 910908. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.P.; Alshafie, T.A.; Cruz, C.P.; Fan, C.Y.; Brown, A.T.; Wang, Y.; Eidt, J.F.; Moursi, M.M. Saratin, an inhibitor of collagen-platelet interaction, decreases venous anastomotic intimal hyperplasia in a canine dialysis access model. Vasc. Endovasc. Surg. 2003, 37, 259–269. [Google Scholar] [CrossRef] [PubMed]
- White, T.C.; Bernyl, M.A.; Robinson, D.K.; Yin, H.; DeGrado, W.F.; Hanson, S.R.; McCarthy, O.J.T. The leech product saratin is a potent inhibitor of platelet integrin α2β1 and von Willebrand factor binding to collagen. FEBS J. 2007, 274, 1481–1491. [Google Scholar] [CrossRef]
- Cruz, C.P.; Eidt, J.; Drouilhet, J.; Brown, A.T.; Wang, Y.; Barnes, C.S.; Moursi, M.M. Saratin, an inhibitor of von Willebrand factor-dependent platelet adhesion, decreases platelet aggregation and intimal hyperplasia in a rat carotid endarterectomy model. J. Vasc. Surg. 2001, 34, 724–729. [Google Scholar] [CrossRef]
- Cheng, B.X.; Kuang, S.T.; Shao, G.Y.; Tian, Q.Q.; Gao, T.Y.; Che, X.F.; Ao, H.W.; Zhang, K.; Liu, F. Molecular cloning and functional analysis of HnSaratin from Hirudo nipponia. Gene 2023, 869, 147401. [Google Scholar] [CrossRef]
- Barnes, C.S.; Krafft, B.; Frech, M.; Hofmann, U.R.; Papendieck, A.; Dahlems, U.; Gellissen, G.; Hoylaerts, M.F. Production and characterization of saratin, an inhibitor of von Willebrand factor-dependent platelet adhesion to collagen. Semin. Thromb. Hemost. 2001, 27, 337–348. [Google Scholar] [CrossRef]
- Huizinga, E.G.; Schouten, A.; Connoly, T.M.; Kroon, J.; Sixma, J.J.; Gros, P. The structure of leech anti-platelet protein, an inhibitor of haemostasis. Acta Crystallogr. D Biol. Crystallogr. 2001, 57, 1071–1078. [Google Scholar] [CrossRef]
- Pérez, L.A.A.; Tabachnick, W.J. Apyrase activity and adenosine diphosphate induced platelet aggregation inhibition by the salivary gland proteins of Culicoides variipennis, the North American vector of bluetongue viruses. Vet. Parasitol. 1996, 61, 327–338. [Google Scholar] [CrossRef]
- Rigbi, M.; Orevi, M.; Eldor, A. Platelet aggregation and coagulation inhibitors in leech saliva and their roles in leech therapy. Semin. Thromb. Hemost. 1996, 22, 273–278. [Google Scholar] [CrossRef]
- Agoncillo, A. Meta-analysis on the safety and efficacy of lumbrokinase in peripheral arterial disease. Eur. Heart J. Acute Ca 2021, 10, zuab020.219. [Google Scholar] [CrossRef]
- Mihara, H.; Sumi, H.; Yoneta, T.; Mizumoto, H.; Ikeda, R.; Seiki, M.; Maruyama, M. A novel fibrinolytic enzyme extracted from the earthworm, Lumbricus rubellus. Jpn. J. Physiol. 1991, 41, 461–472. [Google Scholar] [CrossRef]
- Iannucci, N.B.; Camperi, S.A.; Cascone, O. Purification of lumbrokinase from Eisenia fetida using aqueous two-phase systems and anion-exchange chromatography. Sep. Purif. Technol. 2008, 64, 131–134. [Google Scholar] [CrossRef]
- Bobrovsky, P.; Manuvera, V.; Baskova, I.; Nemirova, S.; Medvedev, A.; Lazarev, V. Recombinant destabilase from Hirudo medicinalis is able to dissolve human blood clots In vitro. Curr. Issues Mol. Biol. 2021, 43, 2068–2081. [Google Scholar] [CrossRef]
- Baskova, I.P.; Nikonov, G.I. Destabilase: An enzyme of medicinal leech salivary gland secretion hydrolyzes the isopeptide bonds in stabilized fibrin. Biokhimiia 1985, 50, 424–431. [Google Scholar]
- Kurdyumov, A.S.; Manuvera, V.A.; Baskova, I.P.; Lazarev, V.N. A comparison of the enzymatic properties of three recombinant isoforms of thrombolytic and antibacterial protein--Destabilase-Lysozyme from medicinal leech. BMC Biochem. 2015, 16, 27. [Google Scholar] [CrossRef]
- Castellano, I.; Merlino, A. γ-glutamyl transpeptidases: Structure and function. In γ-Glutamyl Transpeptidases; Springer: Basel, Switzerland, 2013. [Google Scholar]
- Reverter, D.; Vendrell, J.; Canals, F.; Horstmann, J.; Avilés, F.X.; Fritz, H.; Sommerhoff, C.P. A carboxypeptidase inhibitor from the medical leech Hirudo medicinalis. Isolation, sequence analysis, cDNA cloning, recombinant expression, and characterization. J. Biol. Chem. 1998, 273, 32927–32933. [Google Scholar] [CrossRef]
- Arolas, J.L.; Castillo, V.; Bronsoms, S.; Aviles, F.X.; Ventura, S. Designing out disulfide bonds of leech carboxypeptidase inhibitor: Implications for its folding, stability and function. J. Mol. Biol. 2009, 392, 529–546. [Google Scholar] [CrossRef]
- Jung, H. Hyaluronidase: An overview of its properties, applications, and side effects. Arch. Plast. Surg. 2020, 47, 297–300. [Google Scholar] [CrossRef]
- Linker, A.; Hoffman, P.; Meyer, K. The hyaluronidase of the leech: An endoglucuronidase. Nature 1957, 180, 810–811. [Google Scholar] [CrossRef]
- Das, B.K. An overview on hirudotherapy/leech therapy. Ind. Res. J. Pharm. Sci. 2014, 1, 33–45. [Google Scholar]
- Hovingh, P.; Linker, A. Hyaluronidase activity in leeches (Hirudinea). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1999, 124, 319–326. [Google Scholar] [CrossRef]
- Feyertag, F.; Alvarez-Ponce, D. Disulfide bonds enable accelerated protein evolution. Mol. Biol. Evol. 2017, 34, 1833–1837. [Google Scholar] [CrossRef]
- Andrade, M.A.; Perez-Iratxeta, C.; Ponting, C.P. Protein repeats: Structures, functions, and evolution. J. Struct. Biol. 2001, 134, 117–131. [Google Scholar] [CrossRef]
- Salzet, M. Anticoagulants and inhibitors of platelet aggregation derived from leeches. FEBS Lett. 2001, 492, 187–192. [Google Scholar] [CrossRef]
- Electricwala, A.; Sawyer, R.T.; Jones, C.P.; Atkinson, T. Isolation of thrombin inhibitor from the leech Hirudinaria manillensis. Blood Coagul. Fibrinolysis 1991, 2, 83–89. [Google Scholar] [CrossRef]
- Markwardt, F. Hirudin as alternative anticoagulant—A historical review. Semin. Thromb. Hemost. 2002, 28, 405–414. [Google Scholar] [CrossRef]






| Annotation Approach | Gene Number | Total CDS Length (Mb) | N50 of CDS (bp) | No. of Hirudins |
|---|---|---|---|---|
| GlimmerHMM | 56,061 | 31,047,090 | 873 | 2 |
| SNAP | 45,826 | 26,341,403 | 762 | 2 |
| PASA | 37,111 | 33,145,509 | 1122 | 0 |
| Stringtie | 32,919 | 40,238,799 | 1554 | 0 |
| BRAKER | 25,331 | 36,676,815 | 2151 | 3 |
| EVidenceModeler | 21,828 | 26,863,541 | 1731 | 0 |
| Annotation Approach | Gene Number | Percentage |
|---|---|---|
| NR | 17,411 | 68.73% |
| TrEmbl | 17,421 | 68.77% |
| EggNOG | 15,952 | 62.97% |
| Pfam | 15,675 | 61.88% |
| integration | 18,373 | 72.53% |
| No. | Protein | Leech Species | Accession/Reference | Function |
|---|---|---|---|---|
| 1 | hirudin | Hirudo medicinalis | ALA22933.1 | coagulation inhibitor |
| 2 | granulin | Hirudo nipponia | [75] | coagulation inhibitor |
| 3 | antistasin | Haementeria officinalis | AAA29192.1 | coagulation inhibitor |
| 4 | lefaxin | Haementeria depressa | P86681.1 | coagulation inhibitor |
| 5 | therostasin | Theromyzon tessulatum | AAF73958.1 | coagulation inhibitor |
| 6 | hirustasin | H. medicinalis | P80302.1 | coagulation inhibitor |
| 7 | guamerin | H. nipponia | P46443.1 | coagulation inhibitor |
| 8 | piguamerin | H. nipponia | P81499.1 | coagulation inhibitor |
| 9 | bdellastasin | H. medicinalis | P82107.1 | coagulation inhibitor |
| 10 | poecistasin | H. manillensis | [76] | coagulation inhibitor |
| 11 | eglin | H. medicinails | PDB: 4H4F | coagulation inhibitor |
| 12 | bdellin | H. nipponia | AAK58688.1 | coagulation inhibitor |
| 13 | LDTI | H. medicinails | P80424.1 | coagulation inhibitor |
| 14 | HMEI | H. manillensis | [77] | coagulation inhibitor |
| 15 | saratin | H. officinalis | PDB: 2K13 | platelet aggregation inhibitor |
| 16 | apyrase | Helobdella robusta | XP_009028854.1 | platelet aggregation inhibitor |
| 17 | lumbrokinase | L. rubellus | AAN28692.1 | platelet aggregation inhibitor |
| 18 | destabilase | H. medicinails | AAA96143.1 | fibrinolysis enhancer |
| 19 | GGT | H. medicinails | [78] | fibrinolysis enhancer |
| 20 | LCI | H. medicinails | [28] | fibrinolysis enhancer |
| 21 | hyaluronidase | H. nipponia | AHV78514.1 | tissue penetration enhancer |
| No. | Gene Family | Guan et al. [42] | Zheng et al. [35] | BRAKER Prediction | BRAKER-Plus Prediction |
|---|---|---|---|---|---|
| 1 | hirudin | 1 | 3 | 3 | 5 |
| 2 | progranulin | 1 | 1 | 1 | 1 |
| 3 | antistasin | 0 | 2 | 2 | 2 |
| 4 | lefaxin | 3 | 3 | 3 | 3 |
| 5 | therostasin | 0 | 1 | 0 | 1 |
| 6 | hirustasin/hirustasin-like | 1/3 | 1/5 | 1/11 | 1/12 |
| 7 | guamerin | 0 | 0 | 1 | 1 |
| 8 | piguamerin | 0 | 0 | 1 | 1 |
| 9 | bdellastasin | 0 | 1 | 1 | 1 |
| 10 | poecistasin | 0 | 1 | 0 | 2 |
| 11 | eglin | 0 | 3 | 3 | 4 |
| 12 | bdellin | 0 | 0 | 0 | 1 |
| 13 | LDTI | 1 | 1 | 1 | 1 |
| 14 | HMEI | 4 | 9 | 15 | 18 |
| 15 | saratin | 1 | 1 | 2 | 2 |
| 16 | apyrase | 5 | 5 | 5 | 5 |
| 17 | lumbrokinase | 3 | 3 | 3 | 3 |
| 18 | destabilase | 0 | 1 | 3 | 3 |
| 19 | GGT | 1 | 1 | 1 | 1 |
| 20 | LCI | 1 | 0 | 1 | 1 |
| 21 | hyaluronidase | 3 | 3 | 3 | 3 |
| — | total | 28 | 46 | 61 | 72 |
| Hirudin | Target Species | Protein | Accession | Identity |
|---|---|---|---|---|
| hirudin_Hman1 | H. manillensis | HM1 | Q07558.1 | 76.83% |
| hirudin_Hman2 | H. manillensis | HLF8 | APA20852.1 | 57.63% |
| hirudin_Hman3 | H. manillensis | HLF7 | APA20868.1 | 62.96% |
| hirudin_Hman4 | H. manillensis | HLF6 | APA20866.1 | 73.68% |
| hirudin_Hman5 | H. manillensis | HM1 | Q07558.1 | 68.33% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Zhao, F.; Huang, Z.; Hu, Q.; Meng, R.; Lin, Y.; Qi, J.; Lin, G. Revisiting the Asian Buffalo Leech (Hirudinaria manillensis) Genome: Focus on Antithrombotic Genes and Their Corresponding Proteins. Genes 2023, 14, 2068. https://doi.org/10.3390/genes14112068
Liu Z, Zhao F, Huang Z, Hu Q, Meng R, Lin Y, Qi J, Lin G. Revisiting the Asian Buffalo Leech (Hirudinaria manillensis) Genome: Focus on Antithrombotic Genes and Their Corresponding Proteins. Genes. 2023; 14(11):2068. https://doi.org/10.3390/genes14112068
Chicago/Turabian StyleLiu, Zichao, Fang Zhao, Zuhao Huang, Qingmei Hu, Renyuan Meng, Yiquan Lin, Jianxia Qi, and Gonghua Lin. 2023. "Revisiting the Asian Buffalo Leech (Hirudinaria manillensis) Genome: Focus on Antithrombotic Genes and Their Corresponding Proteins" Genes 14, no. 11: 2068. https://doi.org/10.3390/genes14112068
APA StyleLiu, Z., Zhao, F., Huang, Z., Hu, Q., Meng, R., Lin, Y., Qi, J., & Lin, G. (2023). Revisiting the Asian Buffalo Leech (Hirudinaria manillensis) Genome: Focus on Antithrombotic Genes and Their Corresponding Proteins. Genes, 14(11), 2068. https://doi.org/10.3390/genes14112068






