Finding Predictors of Leg Defects in Pigs Using CNV-GWAS
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample for Analysis
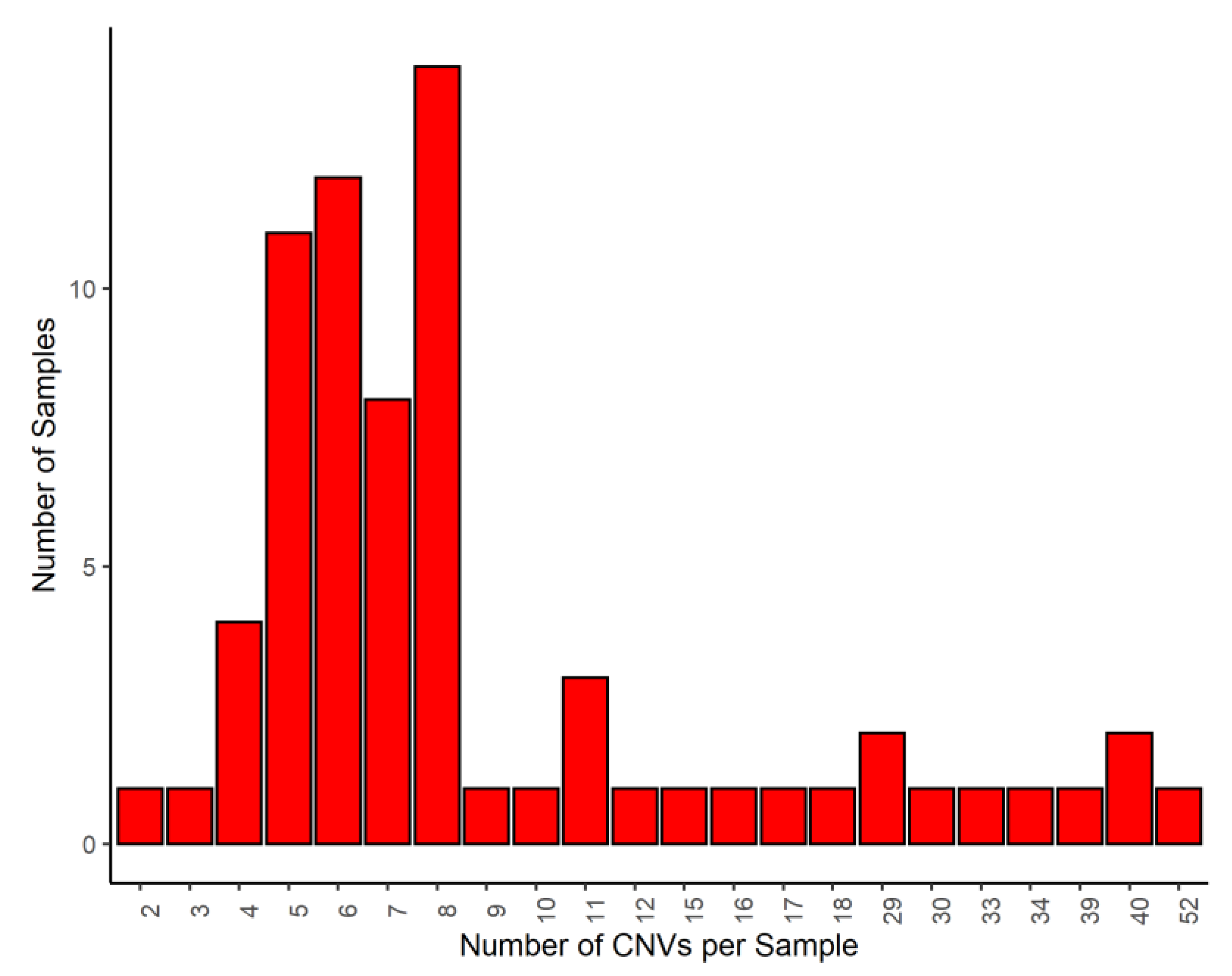
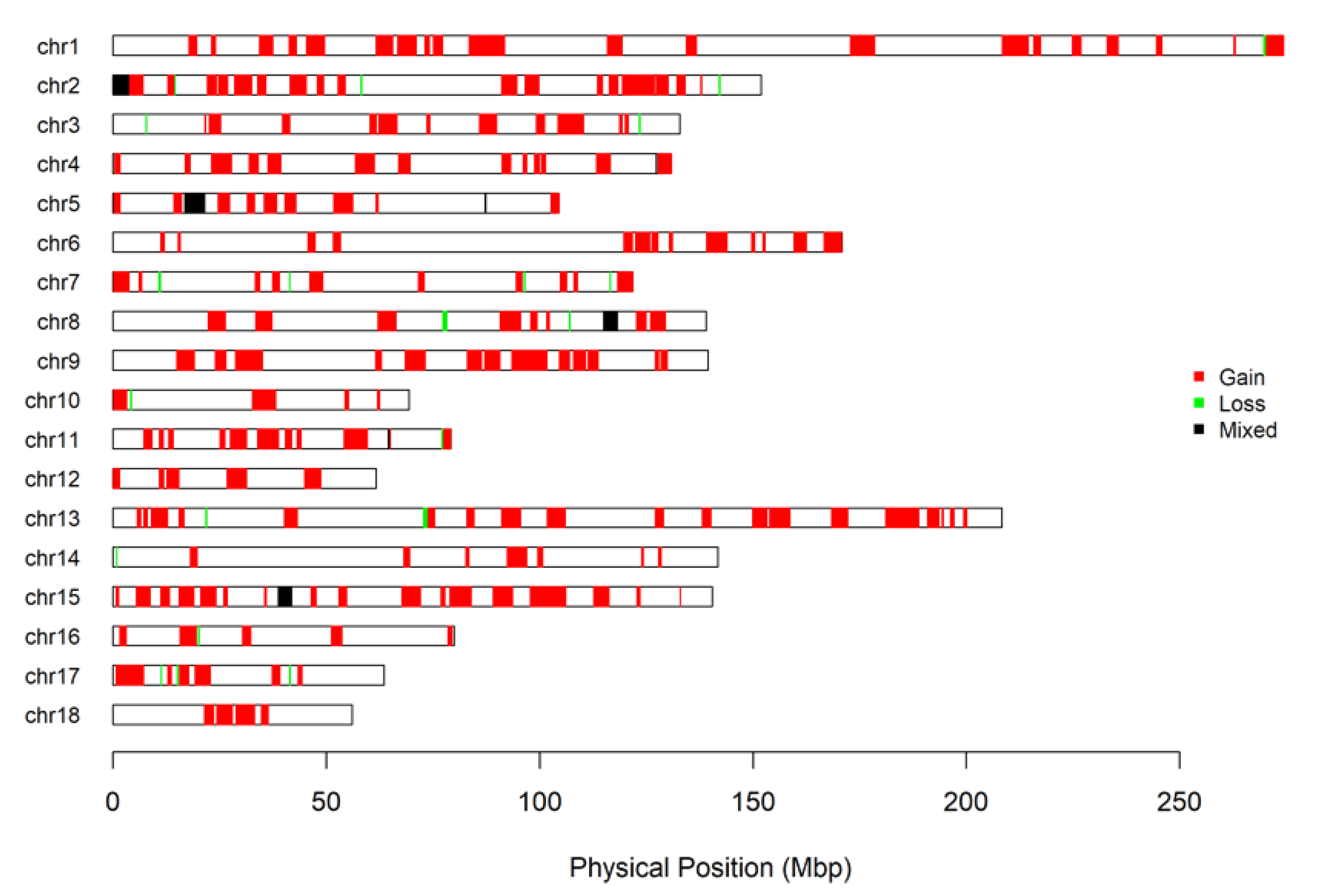
2.2. Calling CNV
2.3. Annotation of Quantitative Trait Loci Located in CNVs Regions
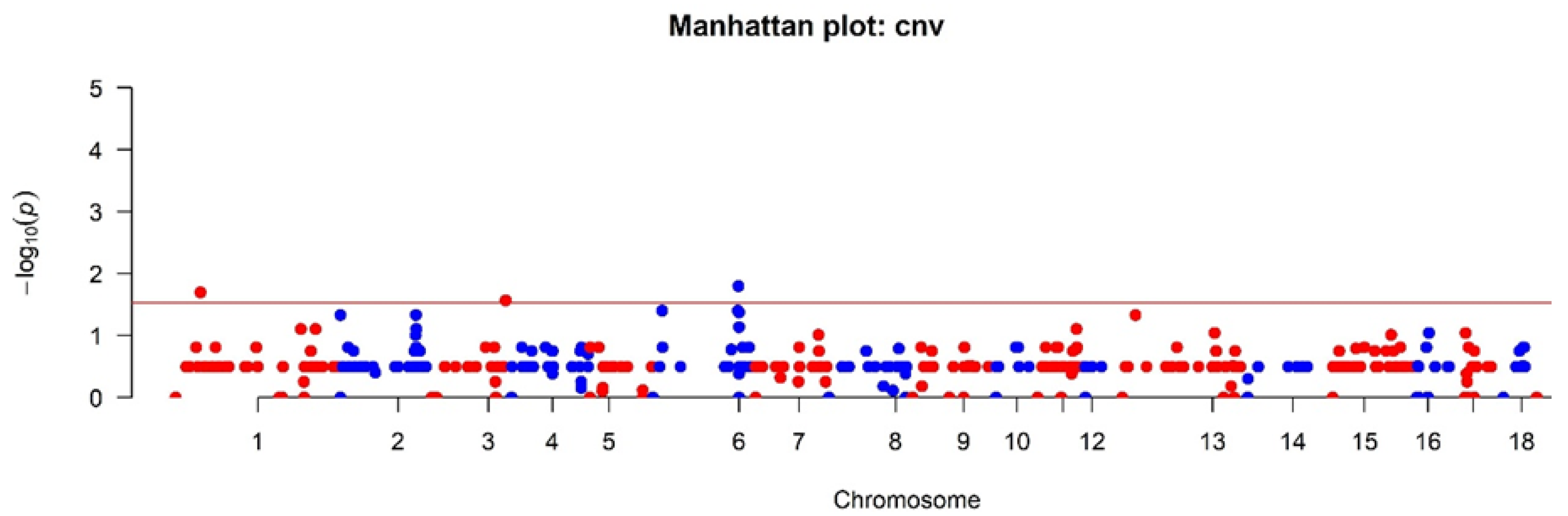
2.4. Association Analysis of CNV with Phenotype
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- The International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature 2008, 455, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Shao, X.; Lv, N.; Liao, J.; Long, J.; Xue, R.; Ai, N.; Xu, D.; Fan, X. Copy number variation is highly correlated with differential gene expression: A pan-cancer study. BMC Med. Genet. 2019, 20, 175. [Google Scholar] [CrossRef] [PubMed]
- Shlien, A.; Malkin, D. Copy number variations and cancer. Genome Med. 2009, 1, 62. [Google Scholar] [CrossRef] [PubMed]
- Hilger, A.C.; Dworschak, G.C.; Reutter, H.M. Lessons Learned from CNV Analysis of Major Birth Defects. Int. J. Mol. Sci. 2020, 21, 8247. [Google Scholar] [CrossRef]
- Wright, D.; Boije, H.; Meadows, J.R.; Bed’hom, B.; Gourichon, D.; Vieaud, A.; Tixier-Boichard, M.; Rubin, C.J.; Imsland, F.; Hallböök, F.; et al. Copy number variation in intron 1 of SOX5 causes the Pea-comb phenotype in chickens. PLoS Genet. 2009, 5, e1000512. [Google Scholar] [CrossRef] [PubMed]
- Sundström, E.; Imsland, F.; Mikko, S.; Wade, C.; Sigurdsson, S.; Pielberg, G.R.; Golovko, A.; Curik, I.; Seltenhammer, M.H.; Sölkner, J.; et al. Copy number expansion of the STX17 duplication in melanoma tissue from Grey horses. BMC Genom. 2012, 13, 365. [Google Scholar] [CrossRef] [PubMed]
- Salmon Hillbertz, N.H.; Isaksson, M.; Karlsson, E.K.; Hellmén, E.; Pielberg, G.R.; Savolainen, P.; Wade, C.M.; von Euler, H.; Gustafson, U.; Hedhammar, A.; et al. Duplication of FGF3, FGF4, FGF19 and ORAOV1 causes hair ridge and predisposition to dermoid sinus in Ridgeback dogs. Nat. Genet. 2007, 39, 1318–1320. [Google Scholar] [CrossRef] [PubMed]
- Fontanesi, L.; Beretti, F.; Riggio, V.; Gómez González, E.; Dall’Olio, S.; Davoli, R.; Russo, V.; Portolano, B. Copy number variation and missense mutations of the agouti signaling protein (ASIP) gene in goat breeds with different coat colors. Cytogenet. Genome Res. 2009, 126, 333–347. [Google Scholar] [CrossRef]
- Sun, G.; Liang, X.; Qin, K.; Qin, Y.; Shi, X.; Cong, P.; Mo, D.; Liu, X.; Chen, Y.; He, Z. Functional Analysis of KIT Gene Structural Mutations Causing the Porcine Dominant White Phenotype Using Genome Edited Mouse Models. Front. Genet. 2020, 11, 138. [Google Scholar] [CrossRef]
- Chen, C.; Qiao, R.; Wei, R.; Guo, Y.; Ai, H.; Ma, J.; Ren, J.; Huang, L. A comprehensive survey of copy number variation in 18 diverse pig populations and identification of candidate copy number variable genes associated with complex traits. BMC Genom. 2012, 13, 733. [Google Scholar] [CrossRef]
- Johansson, A.; Pielberg, G.; Andersson, L.; Edfors-Lilja, I. Polymorphism at the porcine Dominant white/KIT locus influence coat colour and peripheral blood cell measures. Anim. Genet. 2005, 36, 288–296. [Google Scholar] [CrossRef]
- Rubin, C.J.; Megens, H.J.; Martinez Barrio, A.; Maqbool, K.; Sayyab, S.; Schwochow, D.; Wang, C.; Carlborg, Ö.; Jern, P.; Jørgensen, C.B.; et al. Strong signatures of selection in the domestic pig genome. Proc. Natl. Acad. Sci. USA 2012, 109, 19529–19536. [Google Scholar] [CrossRef]
- Fowler, K.E.; Pong-Wong, R.; Bauer, J.; Clemente, E.J.; Reitter, C.P.; Affara, N.A.; Waite, S.; Walling, G.A.; Griffin, D.K. Genome wide analysis reveals single nucleotide polymorphisms associated with fatness and putative novel copy number variants in three pig breeds. BMC Genom. 2013, 14, 784. [Google Scholar] [CrossRef]
- Zappaterra, M.; Gioiosa, S.; Chillemi, G.; Zambonelli, P.; Davoli, R. Muscle transcriptome analysis identifies genes involved in ciliogenesis and the molecular cascade associated with intramuscular fat content in Large White heavy pigs. PLoS ONE 2020, 15, e0233372. [Google Scholar] [CrossRef]
- Wang, L.; Xu, L.; Liu, X.; Zhang, T.; Li, N.; El Hay, H.; Zhang, Y.; Yan, H.; Zhao, K.; Liu, G.E.; et al. Copy number variation-based genome wide association study reveals additional variants contributing to meat quality in Swine. Sci. Rep. 2015, 5, 12535. [Google Scholar] [CrossRef]
- Zhou, Y.; Utsunomiya, Y.T.; Xu, L.; Hay, E.H.A.; Bickhart, D.M.; Sonstegard, T.S.; Van Tassell, C.P.; Garcia, J.F.; Liu, G.E. Comparative analyses across cattle genders and breeds reveal the pitfalls caused by false positive and lineage-differential copy number variations. Sci. Rep. 2016, 6, 29219. [Google Scholar] [CrossRef]
- Zhou, Y.; Connor, E.E.; Wiggans, G.R.; Lu, Y.; Tempelman, R.J.; Schroeder, S.G.; Chen, H.; Liu, G.E. Genome-wide copy number variant analysis reveals variants associated with 10 diverse production traits in Holstein cattle. BMC Genom. 2018, 19, 314. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hadley, D.; Liu, R.; Glessner, J.; Grant, S.F.; Hakonarson, H.; Bucan, M. PennCNV: An integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007, 17, 1665–1674. [Google Scholar] [CrossRef]
- Colella, S.; Yau, C.; Taylor, J.M.; Mirza, G.; Butler, H.; Clouston, P.; Bassett, A.S.; Seller, A.; Holmes, C.C.; Ragoussis, J. QuantiSNP: An Objective Bayes Hidden-Markov Model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007, 35, 2013–2025. [Google Scholar] [CrossRef]
- Pique-Regi, R.; Cáceres, A.; González, J.R. R-Gada: A fast and flexible pipeline for copy number analysis in association studies. BMC Bioinform. 2010, 11, 380. [Google Scholar] [CrossRef]
- Qiu, Y.; Ding, R.; Zhuang, Z.; Wu, J.; Yang, M.; Zhou, S.; Ye, Y.; Geng, Q.; Xu, Z.; Huang, S.; et al. Genome-wide detection of CNV regions and their potential association with growth and fatness traits in Duroc pigs. BMC Genom. 2021, 22, 332. [Google Scholar] [CrossRef]
- Ding, R.; Zhuang, Z.; Qiu, Y.; Wang, X.; Wu, J.; Zhou, S.; Ruan, D.; Xu, C.; Hong, L.; Gu, T.; et al. A composite strategy of genome-wide association study and copy number variation analysis for carcass traits in a Duroc pig population. BMC Genom. 2022, 23, 590. [Google Scholar] [CrossRef]
- Getmantseva, L.; Kolosova, M.; Bakoev, F.; Zimina, A.; Bakoev, S. Genomic Regions and Candidate Genes Linked to Capped Hock in Pig. Life 2021, 11, 510. [Google Scholar] [CrossRef]
- Da Silva, V.; Ramos, M.; Groenen, M.; Crooijmans, R.; Johansson, A.; Regitano, L.; Coutinho, L.; Zimmer, R.; Waldron, L.; Geistlinger, L. CNVRanger: Association analysis of CNVs with gene expression and quantitative phenotypes. Bioinformatics 2020, 36, 972–973. [Google Scholar] [CrossRef]
- Fonseca, P.A.S.; Suárez-Vega, A.; Marras, G.; Cánovas, Á. GALLO: An R package for genomic annotation and integration of multiple data sources in livestock for positional candidate loci. GigaScience 2020, 9, giaa149. [Google Scholar] [CrossRef]
- Gley, K.; Murani, E.; Haack, F.; Trakooljul, N.; Zebunke, M.; Puppe, B.; Wimmers, K.; Ponsuksili, S. Haplotypes of coping behavior associated QTL regions reveal distinct transcript profiles in amygdala and hippocampus. Behav. Brain Res. 2019, 372, 112038. [Google Scholar] [CrossRef]
- Etterlin, P.E.; Ytrehus, B.; Lundeheim, N.; Heldmer, E.; Österberg, J.; Ekman, S. Effects of free-range and confined housing on joint health in a herd of fattening pigs. BMC Vet. Res. 2014, 10, 208. [Google Scholar] [CrossRef]
- Slišković, I.; Eich, H.; Müller-McNicoll, M. Exploring the multifunctionality of SR proteins. Biochem. Soc. Trans. 2022, 50, 187–198. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612, Erratum in Nucleic Acids Res. 2021, 49, 10800. [Google Scholar] [CrossRef]
- Kahindi, R.K.; Regassa, A.; Htoo, J.K.; Nyachoti, C.M. Growth performance and expression of genes encoding enzymes involved in methionine and cysteine metabolism in piglets fed increasing sulphur amino acid to lysine ratio during enterotoxigenic Escherichia coli challenge. Can. J. Anim. Sci. 2018, 98, 333–340. [Google Scholar] [CrossRef]
- Hood, R.L.; Allen, C.E. Cellularity of porcine adipose tissue: Effects of growth and adiposity. J. Lipid Res. 1977, 18, 275–284. [Google Scholar] [CrossRef]
- Le, T.H.; Christensen, O.F.; Nielsen, B.; Sahana, G. Genome-wide association study for conformation traits in three Danish pig breeds. Genet. Sel. Evol. 2017, 49, 12. [Google Scholar] [CrossRef]
- Phoomak, C.; Silsirivanit, A.; Park, D.; Sawanyawisuth, K.; Vaeteewoottacharn, K.; Wongkham, C.; Lam, E.W.; Pairojkul, C.; Lebrilla, C.B.; Wongkham, S. O-GlcNAcylation mediates metastasis of cholangiocarcinoma through FOXO3 and MAN1A1. Oncogene 2018, 37, 5648–5665. [Google Scholar] [CrossRef]
- Hamester, F.; Legler, K.; Wichert, B.; Kelle, N.; Eylmann, K.; Rossberg, M.; Ding, Y.; Kürti, S.; Schmalfeldt, B.; Milde-Langosch, K.; et al. Prognostic relevance of the Golgi mannosidase MAN1A1 in ovarian cancer: Impact of N-glycosylation on tumour cell aggregation. Br. J. Cancer 2019, 121, 944–953. [Google Scholar] [CrossRef]
- Ivanov, D.O.; Novikova, V.P.; Pokhlebkina, A.A. Congenital disorders of glycosylation. Pediatrician 2018, 9, 5–15. [Google Scholar] [CrossRef]
- Csaki, L.S.; Reue, K. Lipins: Multifunctional lipid metabolism proteins. Annu. Rev. Nutr. 2010, 30, 257–272. [Google Scholar] [CrossRef]
- Donkor, J.; Sariahmetoglu, M.; Dewald, J.; Brindley, D.N.; Reue, K. Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J. Biol. Chem. 2007, 282, 3450–3457. [Google Scholar] [CrossRef]
- Shi, H.; Luo, J.; Zhu, J.; Li, J.; Sun, Y.; Lin, X.; Zhang, L.; Yao, D.; Shi, H. PPAR γ Regulates Genes Involved in Triacylglycerol Synthesis and Secretion in Mammary Gland Epithelial Cells of Dairy Goats. PPAR Res. 2013, 2013, 310948. [Google Scholar] [CrossRef]
- Bionaz, M.; Loor, J.J. ACSL1, AGPAT6, FABP3, LPIN1, and SLC27A6 are the most abundant isoforms in bovine mammary tissue and their expression is affected by stage of lactation. J. Nutr. 2008, 138, 1019–1024. [Google Scholar] [CrossRef]
- Zhou, F.; Fan, X.; Miao, Y. LPIN1 promotes triglycerides synthesis and is transcriptionally regulated by PPARG in buffalo mammary epithelial cells. Sci. Rep. 2022, 12, 2390. [Google Scholar] [CrossRef]
- Hood, S.E.; Kofler, X.V.; Chen, Q.; Scott, J.; Ortega, J.; Lehmann, M. Nuclear translocation ability of Lipin differentially affects gene expression and survival in fed and fasting Drosophila. J. Lipid Res. 2020, 61, 1720–1732. [Google Scholar] [CrossRef] [PubMed]
- Reue, K. The lipin family: Mutations and metabolism. Curr. Opin. Lipidol. 2009, 20, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Balboa, M.A.; de Pablo, N.; Meana, C.; Balsinde, J. The role of lipins in innate immunity and inflammation. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 1328–1337. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Getmantseva, L.; Kolosova, M.; Fede, K.; Korobeinikova, A.; Kolosov, A.; Romanets, E.; Bakoev, F.; Romanets, T.; Yudin, V.; Keskinov, A.; et al. Finding Predictors of Leg Defects in Pigs Using CNV-GWAS. Genes 2023, 14, 2054. https://doi.org/10.3390/genes14112054
Getmantseva L, Kolosova M, Fede K, Korobeinikova A, Kolosov A, Romanets E, Bakoev F, Romanets T, Yudin V, Keskinov A, et al. Finding Predictors of Leg Defects in Pigs Using CNV-GWAS. Genes. 2023; 14(11):2054. https://doi.org/10.3390/genes14112054
Chicago/Turabian StyleGetmantseva, Lyubov, Maria Kolosova, Kseniia Fede, Anna Korobeinikova, Anatoly Kolosov, Elena Romanets, Faridun Bakoev, Timofey Romanets, Vladimir Yudin, Anton Keskinov, and et al. 2023. "Finding Predictors of Leg Defects in Pigs Using CNV-GWAS" Genes 14, no. 11: 2054. https://doi.org/10.3390/genes14112054
APA StyleGetmantseva, L., Kolosova, M., Fede, K., Korobeinikova, A., Kolosov, A., Romanets, E., Bakoev, F., Romanets, T., Yudin, V., Keskinov, A., & Bakoev, S. (2023). Finding Predictors of Leg Defects in Pigs Using CNV-GWAS. Genes, 14(11), 2054. https://doi.org/10.3390/genes14112054




