The Mitochondrial Genome of Linichthys laticeps (Cypriniformes: Cyprinidae): Characterization and Phylogeny
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling, DNA Extraction, and High-Throughput Sequencing
2.2. Sequence Assembly, Annotation, and Analysis
2.3. Phylogenetic Analysis
3. Results and Discussion
3.1. Genome Organization and Base Composition
3.2. Protein-Coding Genes and Codon Usage
3.3. Evolutionary Rates and Patterns
3.4. Ribosomal and Transfer RNA Genes
3.5. Control Region
3.6. Phylogenetic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.H.; Zhang, Y.P. Genetics and evolution of mitochondrial DNA in fish. Acta Hydrobiol. Sin. 2000, 24, 384–391. [Google Scholar]
- Brown, K.H. Fish mitochondrial genomics: Sequence, inheritance and functional variation. J. Fish Biol. 2008, 72, 355–374. [Google Scholar] [CrossRef]
- Zhang, R.; Deng, L.; Lv, X.; Tang, Q. Complete mitochondrial genomes of two catfishes (Siluriformes, Bagridae) and their phylogenetic implications. Zookeys 2022, 1115, 103–116. [Google Scholar] [CrossRef]
- Zhang, R.; Tang, Q.; Deng, L. The complete mitochondrial genome of Microphysogobio elongatus (Teleostei, Cyprinidae) and its phylogenetic implications. Zookeys 2021, 1061, 57–73. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Sato, Y.; Sado, T.; Miya, M.; Iwasaki, W. MitoFish, MitoAnnotator, and MiFish Pipeline: Updates in ten years. Mol. Biol. Evol. 2023, 40, msad035. [Google Scholar] [CrossRef] [PubMed]
- Fricke, R.; Eschmeyer, W.N.; Van der Laan, R. Eschmeyer’s Catalog of Fishes: Genera, Species, References. Electronic Version. Available online: https://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (accessed on 25 August 2023).
- Yang, L.; Mayden, R.L. Phylogenetic relationships, subdivision, and biogeography of the cyprinid tribe Labeonini (sensu Rainboth, 1991) (Teleostei: Cypriniformes), with comments on the implications of lips and associated structures in the labeonin classification. Mol. Phylogenet. Evol. 2010, 54, 254–265. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.P.; Yang, J.X.; Chen, X.Y.; Wang, W.Y. Phylogenetic relationships of the Chinese Labeoninae (Teleostei, Cypriniformes) derived from two nuclear and three mitochondrial genes. Zool. Scr. 2010, 39, 559–571. [Google Scholar] [CrossRef]
- Yang, L.; Arunachalam, M.; Sado, T.; Levin, B.A.; Golubtsov, A.S.; Freyhof, J.; Friel, J.P.; Chen, W.J.; Hirt, M.V.; Manickam, R.; et al. Molecular phylogeny of the cyprinid tribe Labeonini (Teleostei: Cypriniformes). Mol. Phylogenet. Evol. 2012, 65, 362–379. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.Y.; Zhou, Q.; Xiang, H.M.; Wang, J.X.; Lan, X.Y.; Luo, Q.H.; Jiang, W.S. Complete mitochondrial genome of Rectoris luxiensis (Teleostei, Cyprinidae): Characterisation and phylogenetic implications. Biodivers. Data J. 2023, 11, e96066. [Google Scholar] [CrossRef]
- Zhang, E.; Fang, F. Linichthys: A new genus of Chinese cyprinid fishes (Teleostei: Cypriniformes). Copeia 2005, 1, 61–67. [Google Scholar] [CrossRef]
- Lin, R.D.; Zhang, C.G. Description of a new species of the Barbine genus Barbodes from China (Cypriniformes: Cyprinides). Acta Zootaxonomica Sin. 1986, 11, 108–110. [Google Scholar]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; dePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Juhling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Putz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Nguyen, M.A.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, S.K.; Liu, T.; Wang, X.; Zeng, S.; Min, W.W.; Zhou, L. Characterization of the complete mitochondrial genome of an endangered fish Semilabeo obscurus (Cyprinidae; Labeoninae; Semilabeo). Conserv. Genet. Resour. 2019, 11, 147–150. [Google Scholar] [CrossRef]
- Chen, W.; Miao, K.; Wang, J.; Wang, H.; Sun, W.; Yuan, S.; Luo, S.; Hu, C.; Chang, Q. Five new mitogenomes sequences of Calidridine sandpipers (Aves: Charadriiformes) and comparative mitogenomics of genus Calidris. Peer J. 2022, 10, e13268. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, X. Characterization and phylogenetic analysis of the complete mitogenome of a rare cavefish, Sinocyclocheilus multipunctatus (Cypriniformes: Cyprinidae). Genes Genom. 2018, 40, 1033–1040. [Google Scholar] [CrossRef]
- Zhang, R.; Zhu, T.; Luo, Q. The complete mitochondrial genome of the freshwater fish Onychostoma ovale (Cypriniformes, Cyprinidae): Genome characterization and phylogenetic analysis. Genes 2023, 14, 1227. [Google Scholar] [CrossRef]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef]
- Yang, Z.; Bielawski, J.P. Statistical methods for detecting molecular adaptation. Trends. Ecol. Evol. 2000, 15, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Wolstenholme, D.R. Animal mitochondrial DNA: Structure and evolution. Int. Rev. Cytol. 1992, 141, 173–216. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.-X.; Hewitt, G.M. Insect mitochondrial control region: A review of its structure, evolution and usefulness in evolutionary studies. Biochem. Syst. Ecol. 1997, 25, 99–120. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, K.; Peng, Y.; Zhou, J.; Liu, Y.; Liu, B. Novel gene rearrangement in the mitochondrial genome of three Garra and insights into the phylogenetic relationships of Labeoninae. Front. Genet. 2022, 13, 922634. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.P.; Chen, X.Y.; Yang, J.X. Molecular phylogeny and systematic revision of Bangana sensu lato (Teleostei, Cyprinidae). J. Zool. Syst. Evol. Res. 2019, 57, 884–891. [Google Scholar] [CrossRef]
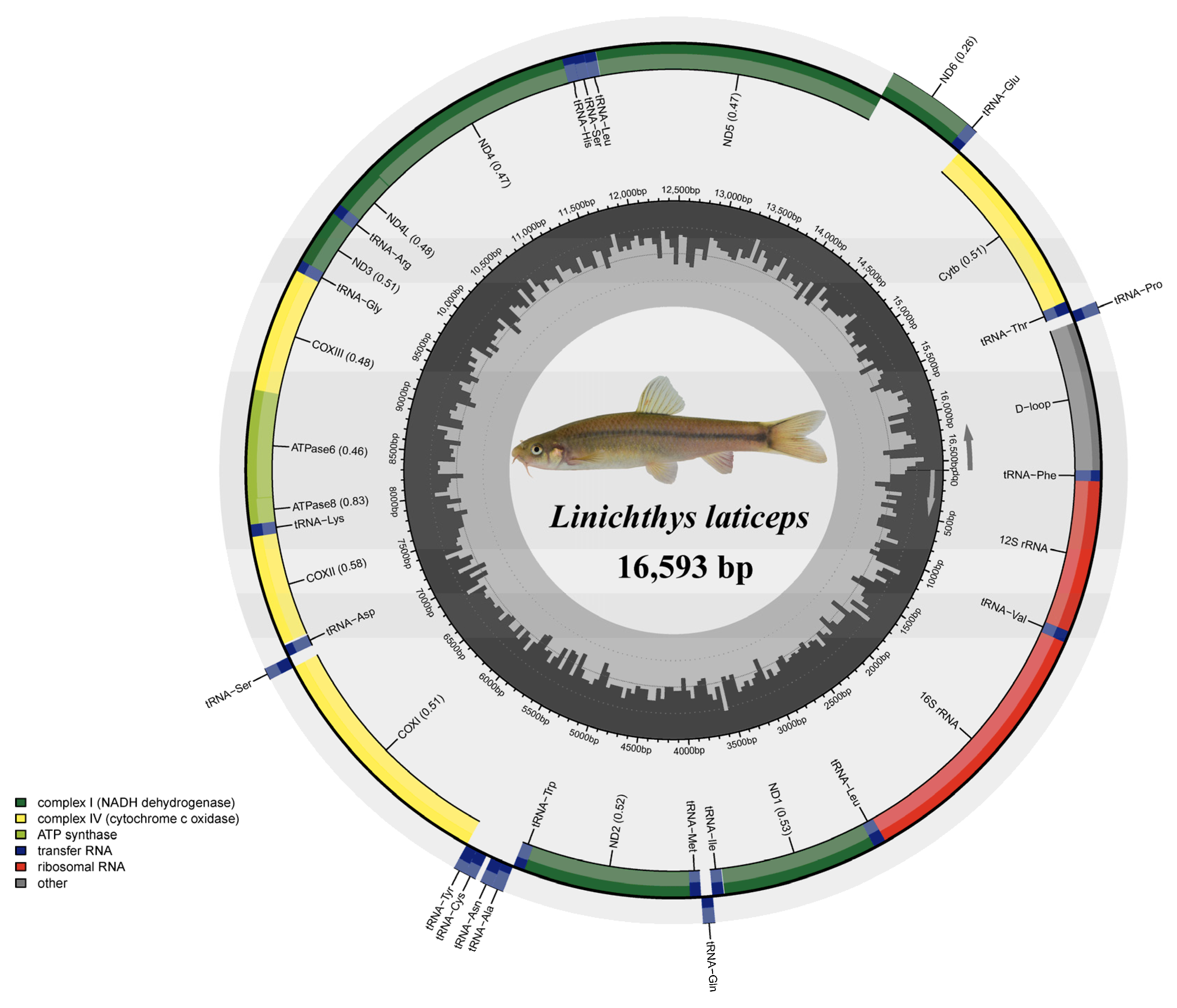


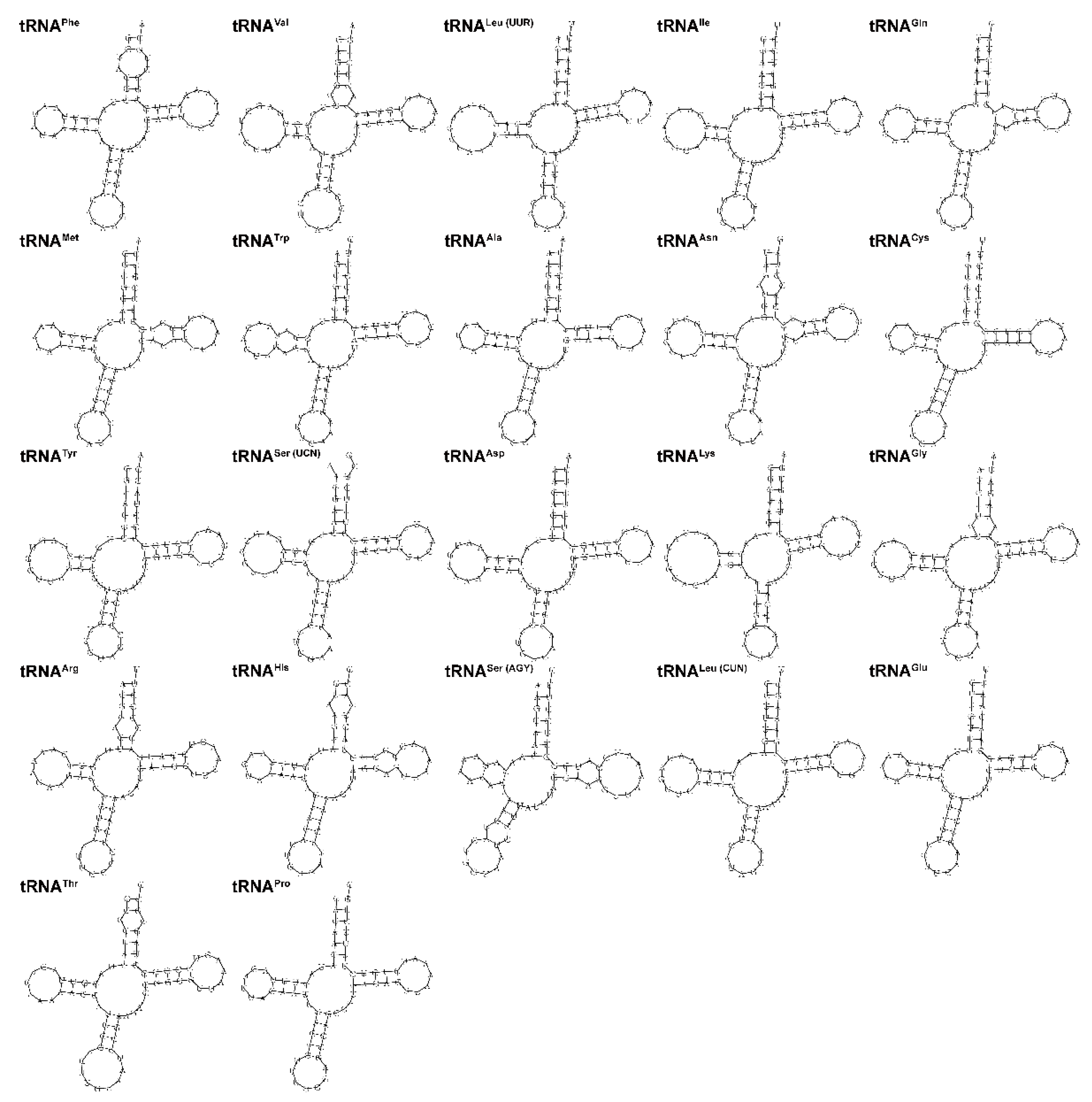

| Gene | Location | Size (bp) | Codon | Anticodon | Strand * | Intergenetic Nucleotides | ||
|---|---|---|---|---|---|---|---|---|
| From | To | Start | Stop | |||||
| tRNAPhe | 1 | 69 | 69 | GAA | H | 0 | ||
| 12S rRNA | 70 | 1023 | 954 | H | 0 | |||
| tRNAVal | 1024 | 1095 | 72 | TAC | H | 0 | ||
| 16S rRNA | 1096 | 2778 | 1683 | H | 0 | |||
| tRNALeu(UUR) | 2779 | 2854 | 76 | TAA | H | 0 | ||
| ND1 | 2856 | 3830 | 975 | ATG | TAA | H | 1 | |
| tRNAIle | 3836 | 3907 | 72 | H | 5 | |||
| tRNAGln | 3906 | 3976 | 71 | GAT | L | −2 | ||
| tRNAMet | 3978 | 4046 | 69 | TTG | H | 1 | ||
| ND2 | 4047 | 5091 | 1045 | ATG | T-- | CAT | H | 0 |
| tRNATrp | 5092 | 5162 | 71 | H | 0 | |||
| tRNAAla | 5165 | 5233 | 69 | TCA | L | 2 | ||
| tRNAAsn | 5235 | 5307 | 73 | TGC | L | 1 | ||
| tRNACys | 5341 | 5407 | 67 | GTT | L | 33 | ||
| tRNATyr | 5409 | 5479 | 71 | GCA | L | 1 | ||
| COI | 5481 | 7031 | 1551 | GTG | TAA | GTA | H | 1 |
| tRNASer(UCN) | 7032 | 7102 | 71 | L | 0 | |||
| tRNAAsp | 7106 | 7177 | 72 | TGA | H | 3 | ||
| COII | 7191 | 7881 | 691 | ATG | T-- | GTC | H | 13 |
| tRNALys | 7882 | 7957 | 76 | H | 0 | |||
| ATPase8 | 7959 | 8123 | 165 | ATG | TAA | TTT | H | 1 |
| ATPase6 | 8117 | 8799 | 683 | ATG | TA- | H | −7 | |
| COIII | 8800 | 9584 | 785 | ATG | TA- | H | 0 | |
| tRNAGly | 9585 | 9656 | 72 | H | 0 | |||
| ND3 | 9657 | 10,005 | 349 | ATG | T-- | TCC | H | 0 |
| tRNAArg | 10,006 | 10,075 | 70 | H | 0 | |||
| ND4L | 10,076 | 10,372 | 297 | ATG | TAA | TCG | H | 0 |
| ND4 | 10,366 | 11,746 | 1381 | ATG | T-- | H | −7 | |
| tRNAHis | 11,747 | 11,815 | 69 | H | 0 | |||
| tRNASer(AGY) | 11,816 | 11,884 | 69 | GTG | H | 0 | ||
| tRNALeu(CUN) | 11,886 | 11,958 | 73 | GCT | H | 1 | ||
| ND5 | 11,962 | 13,785 | 1824 | ATG | TAA | TAG | H | 3 |
| ND6 | 13,782 | 14,303 | 522 | ATG | TAA | L | −4 | |
| tRNAGlu | 14,304 | 14,372 | 69 | L | 0 | |||
| Cyt b | 14,378 | 15,518 | 1141 | ATG | T-- | H | 5 | |
| tRNAThr | 15,519 | 15,590 | 72 | H | 0 | |||
| tRNAPro | 15,590 | 15,659 | 70 | L | −1 | |||
| control region | 15,660 | 16,593 | 934 | H | 0 | |||
| Location | Size (bp) | A | T | G | C | A + T | G + C | A + T Skew | G + C Skew |
|---|---|---|---|---|---|---|---|---|---|
| Genome | 16,593 | 31.4 | 25.7 | 16.0 | 26.9 | 57.1 | 42.9 | 0.100 | −0.254 |
| PCGs | 11,400 | 29.3 | 27.7 | 15.5 | 27.5 | 57.0 | 43.0 | 0.028 | −0.279 |
| 1st codon position | 3800 | 27.0 | 22.0 | 25.7 | 25.3 | 49.0 | 51.0 | 0.102 | 0.008 |
| 2nd codon position | 3800 | 18.5 | 40.7 | 13.6 | 27.2 | 59.2 | 40.8 | −0.375 | −0.333 |
| 3rd codon position | 3800 | 42.3 | 20.4 | 7.3 | 30.0 | 62.7 | 37.3 | 0.349 | −0.609 |
| rRNA | 2637 | 35.1 | 19.9 | 20.5 | 24.5 | 55.0 | 45.0 | 0.276 | −0.089 |
| tRNA | 1563 | 29.0 | 26.5 | 23.2 | 21.3 | 55.5 | 44.5 | 0.045 | 0.043 |
| control region | 934 | 33.4 | 33.3 | 13.5 | 19.8 | 66.7 | 33.3 | 0.001 | −0.189 |
| Amino Acid | Codon | Count | RSCU | Amino Acid | Codon | Count | RSCU |
|---|---|---|---|---|---|---|---|
| Phe | UUU | 108 | 0.94 | Tyr | UAU | 59 | 1.04 |
| Phe | UUC | 123 | 1.06 | Tyr | UAC | 54 | 0.96 |
| Leu | UUA | 145 | 1.39 | stop codon | UAA | 6 | 4 |
| Leu | UUG | 16 | 0.15 | stop codon | UAG | 0 | 0 |
| Leu | CUU | 85 | 0.82 | His | CAU | 32 | 0.59 |
| Leu | CUC | 102 | 0.98 | His | CAC | 76 | 1.41 |
| Leu | CUA | 235 | 2.26 | Gln | CAA | 88 | 1.74 |
| Leu | CUG | 42 | 0.4 | Gln | CAG | 13 | 0.26 |
| Ile | AUU | 178 | 1.21 | Asn | AAU | 52 | 0.85 |
| Ile | AUC | 116 | 0.79 | Asn | AAC | 70 | 1.15 |
| Met | AUA | 144 | 1.62 | Lys | AAA | 72 | 1.89 |
| Met | AUG | 34 | 0.38 | Lys | AAG | 4 | 0.11 |
| Val | GUU | 54 | 0.99 | Asp | GAU | 21 | 0.55 |
| Val | GUC | 35 | 0.64 | Asp | GAC | 55 | 1.45 |
| Val | GUA | 109 | 1.99 | Glu | GAA | 80 | 1.57 |
| Val | GUG | 21 | 0.38 | Glu | GAG | 22 | 0.43 |
| Ser | UCU | 33 | 0.86 | Cys | UGU | 6 | 0.48 |
| Ser | UCC | 55 | 1.44 | Cys | UGC | 19 | 1.52 |
| Ser | UCA | 83 | 2.17 | Trp | UGA | 106 | 1.78 |
| Ser | UCG | 9 | 0.24 | Trp | UGG | 13 | 0.22 |
| Pro | CCU | 19 | 0.36 | Arg | CGU | 7 | 0.37 |
| Pro | CCC | 74 | 1.38 | Arg | CGC | 16 | 0.84 |
| Pro | CCA | 106 | 1.98 | Arg | CGA | 45 | 2.37 |
| Pro | CCG | 15 | 0.28 | Arg | CGG | 8 | 0.42 |
| Thr | ACU | 39 | 0.51 | Ser | AGU | 11 | 0.29 |
| Thr | ACC | 112 | 1.46 | Ser | AGC | 38 | 1 |
| Thr | ACA | 137 | 1.79 | stop codon | AGA | 0 | 0 |
| Thr | ACG | 19 | 0.25 | stop codon | AGG | 0 | 0 |
| Ala | GCU | 49 | 0.59 | Gly | GGU | 24 | 0.39 |
| Ala | GCC | 140 | 1.69 | Gly | GGC | 54 | 0.87 |
| Ala | GCA | 136 | 1.64 | Gly | GGA | 116 | 1.88 |
| Ala | GCG | 7 | 0.08 | Gly | GGG | 53 | 0.86 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, R.; Zhu, T.; Li, H.; Deng, L. The Mitochondrial Genome of Linichthys laticeps (Cypriniformes: Cyprinidae): Characterization and Phylogeny. Genes 2023, 14, 1938. https://doi.org/10.3390/genes14101938
Zhang R, Zhu T, Li H, Deng L. The Mitochondrial Genome of Linichthys laticeps (Cypriniformes: Cyprinidae): Characterization and Phylogeny. Genes. 2023; 14(10):1938. https://doi.org/10.3390/genes14101938
Chicago/Turabian StyleZhang, Renyi, Tingting Zhu, Hongmei Li, and Lei Deng. 2023. "The Mitochondrial Genome of Linichthys laticeps (Cypriniformes: Cyprinidae): Characterization and Phylogeny" Genes 14, no. 10: 1938. https://doi.org/10.3390/genes14101938
APA StyleZhang, R., Zhu, T., Li, H., & Deng, L. (2023). The Mitochondrial Genome of Linichthys laticeps (Cypriniformes: Cyprinidae): Characterization and Phylogeny. Genes, 14(10), 1938. https://doi.org/10.3390/genes14101938







