Comparative Analysis and Phylogenetic Insights of Cas14-Homology Proteins in Bacteria and Archaea
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Cas14 Mining
2.2. Domain and Phylogenetic Analysis
2.3. CRISPR Array/Repeats Prediction
3. Results
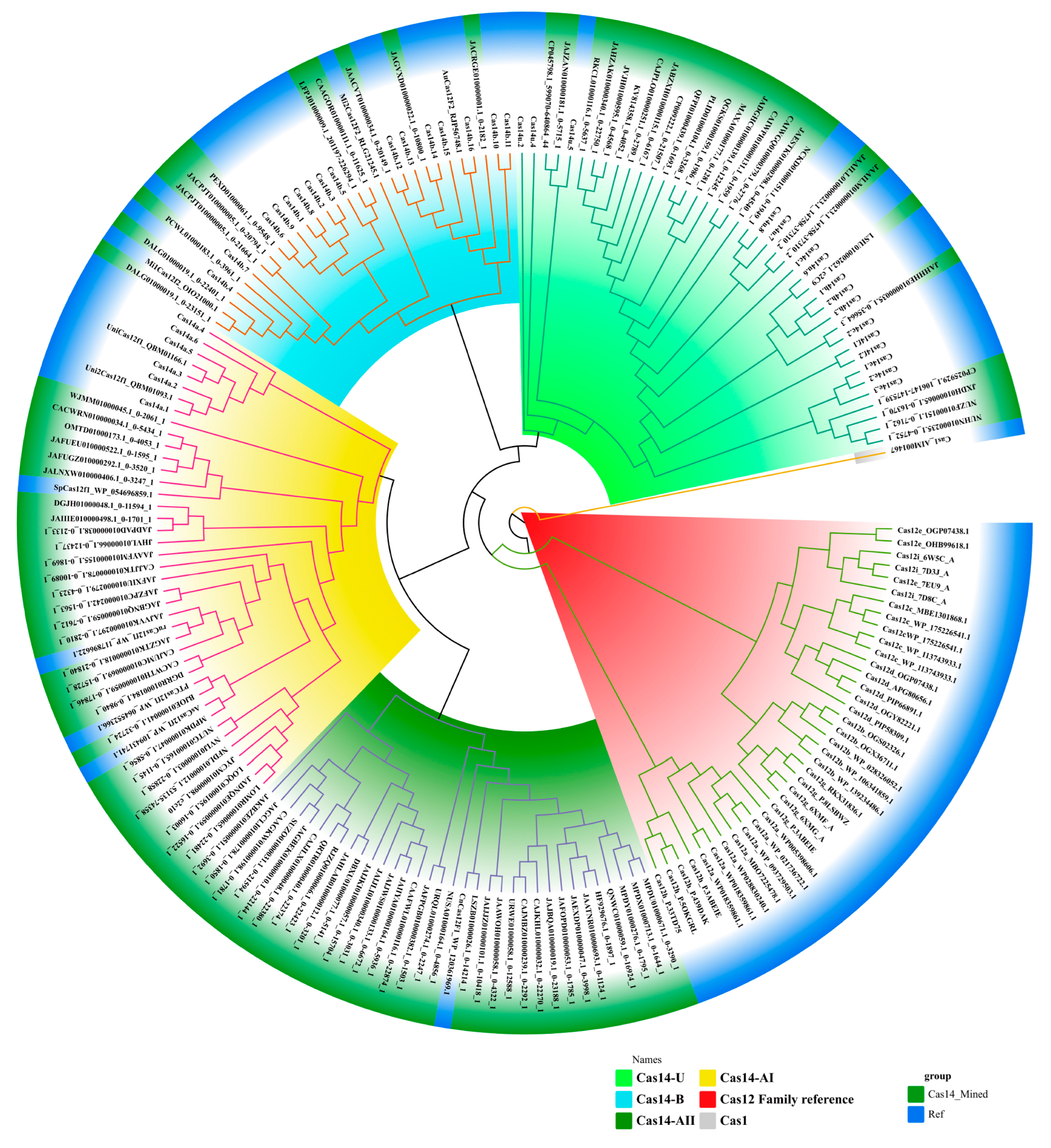
3.1. Classification of Cas14-Homology Proteins
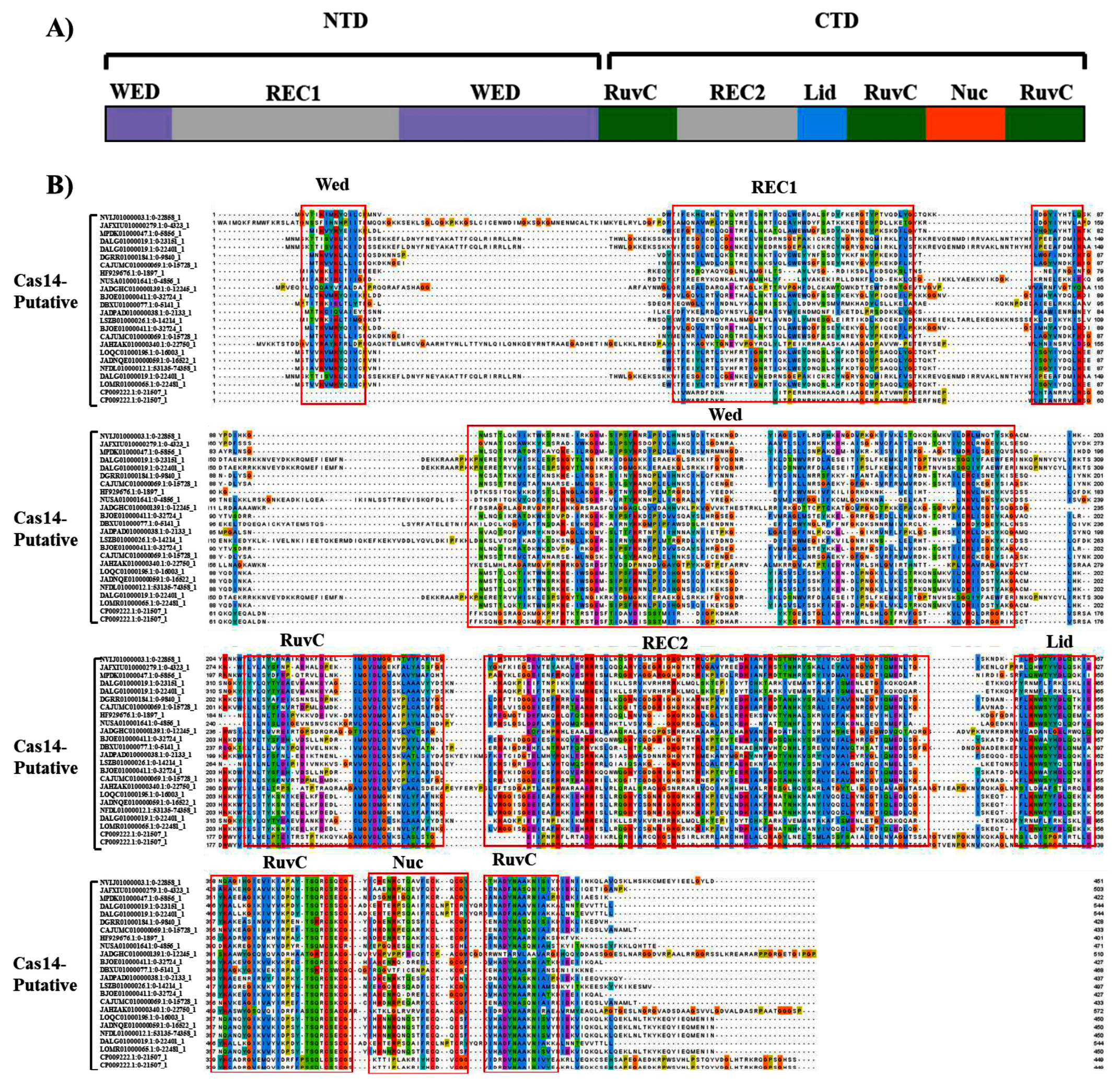
3.2. Domain Organization of Cas14-Homology Proteins
3.3. Putatively Functional Cas14 Proteins
3.4. Distribution of Ca14-Homology Proteins in Bacteria and Archaea
3.5. Distribution of Putatively Functional Cas14 Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mali, P.; Esvelt, K.M.; Church, G.M. Cas9 as a Versatile Tool for Engineering Biology. Nat. Methods 2013, 10, 957–963. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.L.E. CRISPR Rewrites the Future of Medicine. Cris. J. 2022, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Sousa, A.A.; Walton, R.T.; Tak, Y.E.; Hsu, J.Y.; Clement, K.; Welch, M.M.; Horng, J.E.; Malagon-Lopez, J.; Scarfò, I.; et al. Engineered CRISPR–Cas12a Variants with Increased Activities and Improved Targeting Ranges for Gene, Epigenetic and Base Editing. Nat. Biotechnol. 2019, 37, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Tu, M.; Lin, L.; Cheng, Y.; He, X.; Sun, H.; Xie, H.; Fu, J.; Liu, C.; Li, J.; Chen, D.; et al. A New Lease of Life’: FnCpf1 Possesses DNA Cleavage Activity for Genome Editing in Human Cells. Nucleic Acids Res. 2017, 45, 11295–11304. [Google Scholar] [CrossRef]
- Liu, W.; Li, L.; Jiang, J.; Wu, M.; Lin, P. Applications and Challenges of CRISPR-Cas Gene-Editing to Disease Treatment in Clinics. Precis. Clin. Med. 2021, 4, 179–191. [Google Scholar] [CrossRef]
- Liu, J.J.; Orlova, N.; Oakes, B.L.; Ma, E.; Spinner, H.B.; Baney, K.L.M.; Chuck, J.; Tan, D.; Knott, G.J.; Harrington, L.B.; et al. CasX Enzymes Comprise a Distinct Family of RNA-Guided Genome Editors. Nature 2019, 566, 218–223. [Google Scholar] [CrossRef]
- Xu, X.; Chemparathy, A.; Zeng, L.; Kempton, H.R.; Shang, S.; Nakamura, M.; Qi, L.S. Engineered Miniature CRISPR-Cas System for Mammalian Genome Regulation and Editing. Mol. Cell 2021, 81, 4333–4345. [Google Scholar] [CrossRef]
- Bigelyte, G.; Young, J.K.; Karvelis, T.; Budre, K.; Zedaveinyte, R.; Djukanovic, V.; Van Ginkel, E.; Paulraj, S.; Gasior, S.; Jones, S.; et al. Miniature Type V-F CRISPR-Cas Nucleases Enable Targeted DNA Modification in Cells. Nat. Commun. 2021, 10, 957–963. [Google Scholar] [CrossRef] [PubMed]
- Freije, C.A.; Myhrvold, C.; Boehm, C.K.; Lin, A.E.; Welch, N.L.; Carter, A.; Metsky, H.C.; Luo, C.Y.; Abudayyeh, O.O.; Gootenberg, J.S.; et al. Programmable Inhibition and Detection of RNA Viruses Using Cas13. Mol. Cell 2019, 76, 826–837. [Google Scholar] [CrossRef]
- Pausch, P.; Soczek, K.M.; Herbst, D.A.; Tsuchida, C.A.; Al-Shayeb, B.; Banfield, J.F.; Nogales, E.; Doudna, J.A. DNA Interference States of the Hypercompact CRISPR–CasΦ Effector. Nat. Struct. Mol. Biol. 2021, 28, 652–661. [Google Scholar] [CrossRef]
- Doudna, J.A. The Promise and Challenge of Therapeutic Genome Editing. Nature 2020, 578, 229–236. [Google Scholar] [CrossRef]
- Harrington, L.B.; Harrington, L.B.; Burstein, D.; Chen, J.S.; Paez-espino, D.; Ma, E.; Witte, I.P.; Cofsky, J.C.; Kyrpides, N.C.; Banfield, J.F.; et al. Programmed DNA Destruction by Miniature CRISPR-Cas14 Enzymes. Science 2018, 362, 839–842. [Google Scholar] [CrossRef] [PubMed]
- Karvelis, T.; Bigelyte, G.; Young, J.K.; Hou, Z.; Zedaveinyte, R.; Budre, K.; Paulraj, S.; Djukanovic, V.; Gasior, S.; Silanskas, A.; et al. PAM Recognition by Miniature CRISPR-Cas12f Nucleases Triggers Programmable Double-Stranded DNA Target Cleavage. Nucleic Acids Res. 2020, 48, 5016–5023. [Google Scholar] [CrossRef]
- Kim, D.Y.; Chung, Y.; Lee, Y.; Jeong, D.; Park, K.H.; Chin, H.J.; Lee, J.M.; Park, S.; Ko, S.; Ko, J.H.; et al. Hypercompact Adenine Base Editors Based on Transposase B Guided by Engineered RNA. Nat. Chem. Biol. 2022, 18, 1005–1013. [Google Scholar] [CrossRef]
- Okano, K.; Sato, Y.; Hizume, T.; Honda, K. Genome Editing by Miniature CRISPR/Cas12f1 Enzyme in Escherichia coli. J. Biosci. Bioeng. 2021, 132, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Y.; Pan, D.; Yu, H.; Zhang, Y.; Chen, W.; Li, F.; Wu, Z.; Ji, Q. Guide RNA Engineering Enables Efficient CRISPR Editing with a Miniature Syntrophomonas palmitatica Cas12f1 Nuclease. Cell Rep. 2022, 40, 111418. [Google Scholar] [CrossRef]
- Takeda, S.N.; Nakagawa, R.; Okazaki, S.; Hirano, H.; Kobayashi, K.; Kusakizako, T.; Nishizawa, T.; Yamashita, K.; Nishimasu, H.; Nureki, O. Structure of the Miniature Type V-F CRISPR-Cas Effector Enzyme. Mol. Cell 2021, 81, 558–570. [Google Scholar] [CrossRef] [PubMed]
- Naeem, M.; Alkhnbashi, O.S. Current Bioinformatics Tools to Optimize CRISPR/Cas9 Experiments to Reduce Off-Target Effects. Int. J. Mol. Sci. 2023, 24, 6261. [Google Scholar] [CrossRef]
- Sharrar, A.; Arake de Tacca, L.; Collingwood, T.; Meacham, Z.; Rabuka, D.; Staples-Ager, J.; Schelle, M. Discovery and Characterization of Novel Type V Cas12f Nucleases with Diverse Protospacer Adjacent Motif Preferences. Cris. J. 2023, 6, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Koonin, E.V. Annotation and Classification of CRISPR-Cas Systems. Methods Mol. Biol. 2015, 1311, 47–75. [Google Scholar] [CrossRef] [PubMed]
- York, A. Metagenomics: Mining for CRISPR-Cas. Nat. Rev. Microbiol. 2017, 15, 133. [Google Scholar] [CrossRef]
- Cornwell, W.; Nakagawa, S. Phylogenetic Comparative Methods. Curr. Biol. 2017, 27, R333–R336. [Google Scholar] [CrossRef]
- Jacques, F.; Bolivar, P.; Pietras, K.; Hammarlund, E.U. Roadmap to the Study of Gene and Protein Phylogeny and evolution—A Practical Guide. PLoS ONE 2023, 18, e0279597. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Makarova, K.S. Origins and Evolution of CRISPR-Cas Systems. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180087. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary Classification of CRISPR–Cas Systems: A Burst of Class 2 and Derived Variants. Nat. Rev. Microbiol. 2020, 18, 67–83. [Google Scholar] [CrossRef]
- Makarova, K.S.; Koonin, E.V. Evolution and Classification of CRISPR-Cas Systems and Cas Protein Families. In CRISPR-Cas Systems: RNA-Mediated Adaptive Immunity in Bacteria and Archaea; Springer: Berlin/Heidelberg, Germany, 2013; ISBN 9783642346576. [Google Scholar]
- Lange, S.J.; Alkhnbashi, O.S.; Rose, D.; Will, S.; Backofen, R. CRISPRmap: An Automated Classification of Repeat Conservation in Prokaryotic Adaptive Immune Systems. Nucleic Acids Res. 2013, 41, 8034–8044. [Google Scholar] [CrossRef] [PubMed]
- Biswas, A.; Staals, R.H.J.; Morales, S.E.; Fineran, P.C.; Brown, C.M. CRISPRDetect: A Flexible Algorithm to Define CRISPR Arrays. BMC Genom. 2016, 17, 356. [Google Scholar] [CrossRef]
- Crawley, A.B.; Henriksen, J.R.; Barrangou, R. CRISPRdisco: An Automated Pipeline for the Discovery and Analysis of CRISPR-Cas Systems. Cris. J. 2018, 1, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Abby, S.S.; Néron, B.; Ménager, H.; Touchon, M.; Rocha, E.P.C. MacSyFinder: A Program to Mine Genomes for Molecular Systems with an Application to CRISPR-Cas Systems. PLoS ONE 2014, 9, e110726. [Google Scholar] [CrossRef]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an Update of CRISRFinder, Includes a Portable Version, Enhanced Performance and Integrates Search for Cas Proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef]
- Chai, G.; Yu, M.; Jiang, L.; Duan, Y.; Huang, J. HMMCAS: A Web Tool for the Identification and Domain Annotations of CAS Proteins. IEEE/ACM Trans. Comput. Biol. Bioinforma. 2019, 16, 1313–1315. [Google Scholar] [CrossRef]
- Mitrofanov, A.; Alkhnbashi, O.S.; Shmakov, S.A.; Makarova, K.S.; Koonin, E.V.; Backofen, R. CRISPRidentify: Identification of CRISPR Arrays Using Machine Learning Approach. Nucleic Acids Res. 2021, 49, e20. [Google Scholar] [CrossRef]
- Kong, X.; Zhang, H.; Li, G.; Wang, Z.; Kong, X.; Wang, L.; Xue, M.; Zhang, W.; Wang, Y.; Lin, J.; et al. Engineered CRISPR-OsCas12f1 and RhCas12f1 with Robust Activities and Expanded Target Range for Genome Editing. Nat. Commun. 2023, 14, 2046. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in Homology Search: HMMER3 and Convergent Evolution of Coiled-Coil Regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef]
- Russel, J.; Pinilla-Redondo, R.; Mayo-Muñoz, D.; Shah, S.A.; Sørensen, S.J. CRISPRCasTyper: Automated Identification, Annotation, and Classification of CRISPR-Cas Loci. Cris. J. 2020, 3, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Wiedenheft, B.; Zhou, K.; Jinek, M.; Coyle, S.M.; Ma, W.; Doudna, J.A. Structural Basis for DNase Activity of a Conserved Protein Implicated in CRISPR-Mediated Genome Defense. Structure 2009, 17, 904–912. [Google Scholar] [CrossRef]
- Xiao, R.; Li, Z.; Wang, S.; Han, R.; Chang, L. Structural Basis for Substrate Recognition and Cleavage by the Dimerization-Dependent CRISPR-Cas12f Nuclease. Nucleic Acids Res. 2021, 49, 4120–4128. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, G.T.; Dhingra, Y.; Sashital, D.G. Miniature CRISPR-Cas12 Endonucleases—Programmed DNA Targeting in a Smaller Package. Curr. Opin. Struct. Biol. 2022, 77, 102466. [Google Scholar] [CrossRef]
- Pausch, P.; Al-Shayeb, B.; Bisom-Rapp, E.; Tsuchida, C.A.; Li, Z.; Cress, B.F.; Knott, G.J.; Jacobsen, S.E.; Banfield, J.F.; Doudna, J.A. Crispr-Casf from Huge Phages Is a Hypercompact Genome Editor. Science 2020, 369, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Xin, C.; Yin, J.; Yuan, S.; Ou, L.; Liu, M.; Zhang, W.; Hu, J. Comprehensive Assessment of Miniature CRISPR-Cas12f Nucleases for Gene Disruption. Nat. Commun. 2022, 13, 5623. [Google Scholar] [CrossRef]
- Tong, B.; Dong, H.; Cui, Y.; Jiang, P.; Jin, Z.; Zhang, D. The Versatile Type V CRISPR Effectors and Their Application Prospects. Front. Cell Dev. Biol. 2021, 8, 622103. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Song, L.; Yuan, B.; Zhang, C.; Cao, J.; Chen, J.; Qiu, J.; Tai, Y.; Chen, J.; Qiu, Z.; et al. TadA Reprogramming to Generate Potent Miniature Base Editors with High Precision. Nat. Commun. 2023, 14, 413. [Google Scholar] [CrossRef] [PubMed]

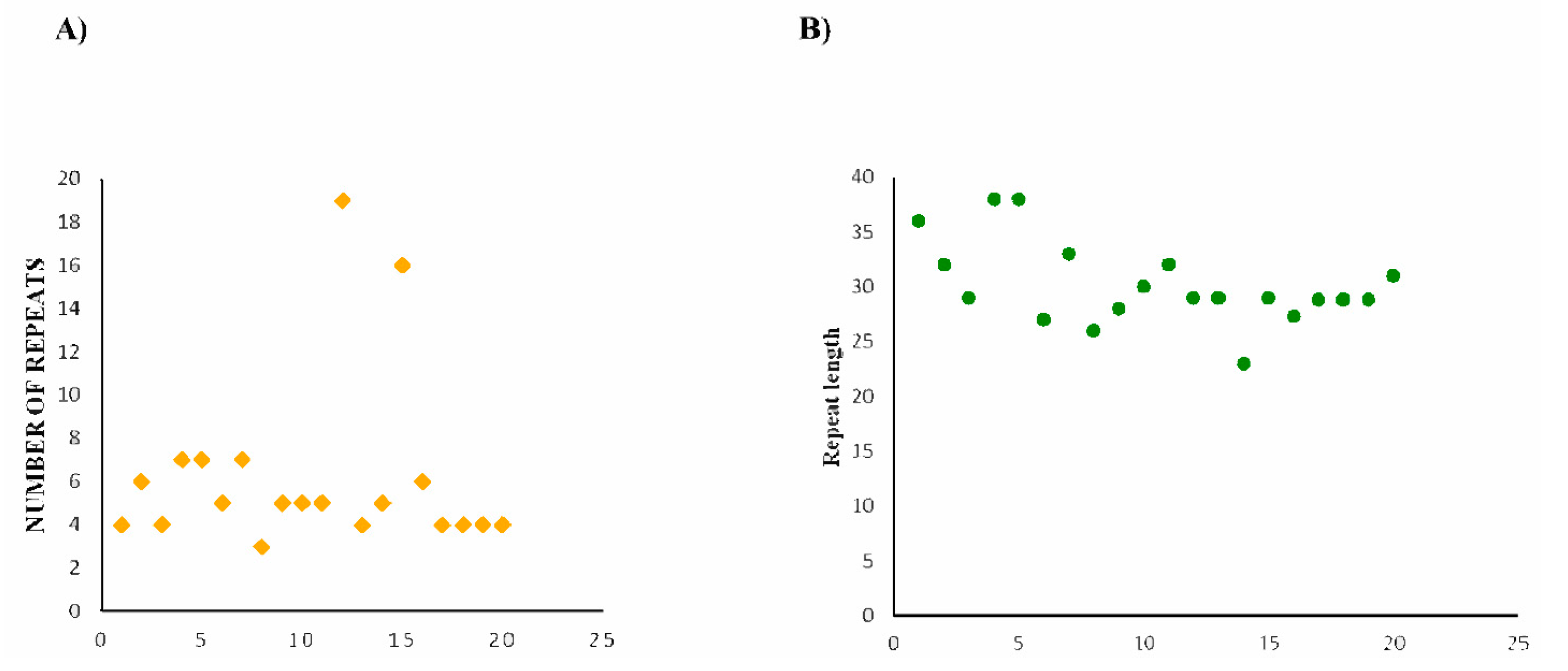
| Sequence ID | Consensus Repeat | Repeat Subtype |
|---|---|---|
| BJOE01000041.1:0-32724_1 | CTCCAAACAGAATCATGCTTCTATGACTGTTCCGAG | V-F1 |
| CAJUMC010000069.1:0-15728_7 | CTTACACCATATACCTACGCATAGTTCGAGTC | V-F1 |
| CP009222.1:0-21507_8 | GTTCTTCCCACGCACACGAAGAAGATCCC | V-F2 |
| DALG01000019.1:0-22401_10 | AGTTGCATCTCTCATCTCGTTAATTCGTGCGCTGAAAC | V-F1 |
| DALG01000019.1:0-23151_11 | AGTTGCATCTCTCATCTCGTTAATTCGTGCGCTGAAAC | V-F1 |
| DBXU01000077.1:0-5141_12 | GCTGTGACTCATAGCAAAAAAGAAGGT | V-F1 |
| DGRR01000184.1:0-9840_14 | GATTATATCTGCTTGTATGGGTATACTGCGAGA | V-F1 |
| HF929676.1:0-1897_15 | TACACACTACATAGTCATTATATAAC | V-F1 |
| JADGHC010000139.1:0-12245_16 | GGGACTTCCCCGAGCGCGAGGACGACGG | V-F2 |
| JADPAD010000038.1:0-2133_17 | GTTTAAGAATAACAATAGTTGTATTTAAAT | V-F1 |
| JAFXIU010000279.1:0-4323_19 | GTTGCAACACGCGCATAAGGATGACTTGAAGG | V-F1 |
| MPDK01000047.1:0-5856_23 | GTTCACACTCCACAAGCTAGCTCGCAAAC | V-F1 |
| NUSA01000164.1:0-4856_24 | GTTTTGAATAAACTATGTAGAATGTGAAT | V-F1 |
| NVIJ01000003.1:0-22858_25 | ATTTAAATACATCTTATGTTAGT | V-F1 |
| LSZB01000026.1:0-14214_33 | ATTTACATTTCACATAGTTAAACTAAAAC | V-F1 |
| JAHZAK010000340.1:0-22750-1_36 | GTGTTCCCCGTATGTGCGGGGGTGAGC | V-F2 |
| LOQC01000195.1:0-16003-1_48 | ATTTCAATACATCTATTGTTATGTTTTAAC | V-F1 |
| JADNQE010000059.1:0-16522-1_52 | ATTTCAATACATCTATTGTTATGTTTTAAC | V-F1 |
| LOMR01000065.1:0-22481-1_53 | ATTTCAATACATCTATTGTTATGTTTTAAC | V-F1 |
| NFDL01000012.1:53135-74358-1_58 | AGGAAAAACATAATAATAGATGTATTGAAAT | V-F1 |
| BJOE01000041.1:0-32724_1 | CTCCAAACAGAATCATGCTTCTATGACTGTTCCGAG | V-F1 |
| CAJUMC010000069.1:0-15728_7 | CTTACACCATATACCTACGCATAGTTCGAGTC | V-F1 |
| CP009222.1:0-21507_8 | GTTCTTCCCACGCACACGAAGAAGATCCC | V-F2 |
| DALG01000019.1:0-22401_10 | AGTTGCATCTCTCATCTCGTTAATTCGTGCGCTGAAAC | V-F1 |
| Group | All | Putative | ||
|---|---|---|---|---|
| Bacteria | Nitrospirae | Nitrospirae | 0 | 0 |
| FCB group | Fibrobacteres | 0 | 0 | |
| Bacteroidetes | 14 | 1 | ||
| Chlorobi | 0 | 0 | ||
| Gemmatimonadetes | 0 | 0 | ||
| PVC group | Verrucomicrobia | 0 | 0 | |
| Planctomycetes | 0 | 0 | ||
| Chlamydiae | 0 | 0 | ||
| Terrabacteria group | Deinococcus-Thermus | 0 | 0 | |
| Firmicutes | 37 | 16 | ||
| Armatimonadetes | 0 | 0 | ||
| Chloroflexi | 0 | 0 | ||
| Actinobacteria | 16 | 3 | ||
| Candidatus Melainabacteria | 0 | 0 | ||
| Cyanobacteria | 1 | 1 | ||
| Candidatus Eremiobacteraeota | 0 | 0 | ||
| Proteobacteria | Gammaproteobacteria | 0 | 0 | |
| Alphaproteobacteria | 2 | 0 | ||
| Betaproteobacteria | 1 | 0 | ||
| unclassified Proteobacteria | 0 | 0 | ||
| environmental samples | 0 | 0 | ||
| delta/epsilon subdivisions | 0 | 0 | ||
| Zetaproteobacteria | 0 | 0 | ||
| Oligoflexia | 0 | 0 | ||
| Acidithiobacillia | 0 | 0 | ||
| Candidatus Lambdaproteobacteria | 0 | 0 | ||
| Candidatus Muproteobacteria | 0 | 0 | ||
| Hydrogenophilalia | 0 | 0 | ||
| Aquificae | Aquificae | 0 | 0 | |
| Thermotogae | Thermotogae | 0 | 0 | |
| Deferribacteres | Deferribacteres | 0 | 0 | |
| Chrysiogenetes | Chrysiogenetes | 0 | 0 | |
| Thermodesulfobacteria | Thermodesulfobacteria | 0 | 0 | |
| Spirochaetes | Spirochaetes | 0 | 0 | |
| Fusobacteria | Fusobacteria | 0 | 0 | |
| Acidobacteria | Acidobacteria | 0 | 0 | |
| Dictyoglomi | Dictyoglomi | 0 | 0 | |
| Calditrichaeota | Calditrichaeota | 0 | 0 | |
| Nitrospinae/Tectomicrobia group | Nitrospinae/Tectomicrobia group | 0 | 0 | |
| Krumholzibacteriota | Krumholzibacteriota | 0 | 0 | |
| Caldiserica/Crysericota group | Caldiserica/Crysericota group | 0 | 0 | |
| Coprothermobacteria | Coprothermobacteria | 0 | 0 | |
| Elusimicrobia | Elusimicrobia | 0 | 0 | |
| Synergistetes | Synergistetes | 0 | 0 | |
| unclassified Bacteria | unclassified Bacteria | 0 | 0 | |
| environmental samples | environmental samples | 0 | 0 | |
| Archaea | Asgard group | 0 | 0 | |
| Candidatus Thermoplasmatota | 0 | 0 | ||
| DPANN group | 9 | 3 | ||
| Euyarchaeota | 1 | 0 | ||
| TACK group | 0 | 0 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ullah, N.; Yang, N.; Guan, Z.; Xiang, K.; Wang, Y.; Diaby, M.; Chen, C.; Gao, B.; Song, C. Comparative Analysis and Phylogenetic Insights of Cas14-Homology Proteins in Bacteria and Archaea. Genes 2023, 14, 1911. https://doi.org/10.3390/genes14101911
Ullah N, Yang N, Guan Z, Xiang K, Wang Y, Diaby M, Chen C, Gao B, Song C. Comparative Analysis and Phylogenetic Insights of Cas14-Homology Proteins in Bacteria and Archaea. Genes. 2023; 14(10):1911. https://doi.org/10.3390/genes14101911
Chicago/Turabian StyleUllah, Numan, Naisu Yang, Zhongxia Guan, Kuilin Xiang, Yali Wang, Mohamed Diaby, Cai Chen, Bo Gao, and Chengyi Song. 2023. "Comparative Analysis and Phylogenetic Insights of Cas14-Homology Proteins in Bacteria and Archaea" Genes 14, no. 10: 1911. https://doi.org/10.3390/genes14101911
APA StyleUllah, N., Yang, N., Guan, Z., Xiang, K., Wang, Y., Diaby, M., Chen, C., Gao, B., & Song, C. (2023). Comparative Analysis and Phylogenetic Insights of Cas14-Homology Proteins in Bacteria and Archaea. Genes, 14(10), 1911. https://doi.org/10.3390/genes14101911






