High Mitochondrial Haplotype Diversity Found in Three Pre-Hispanic Groups from Colombia
Abstract
1. Introduction
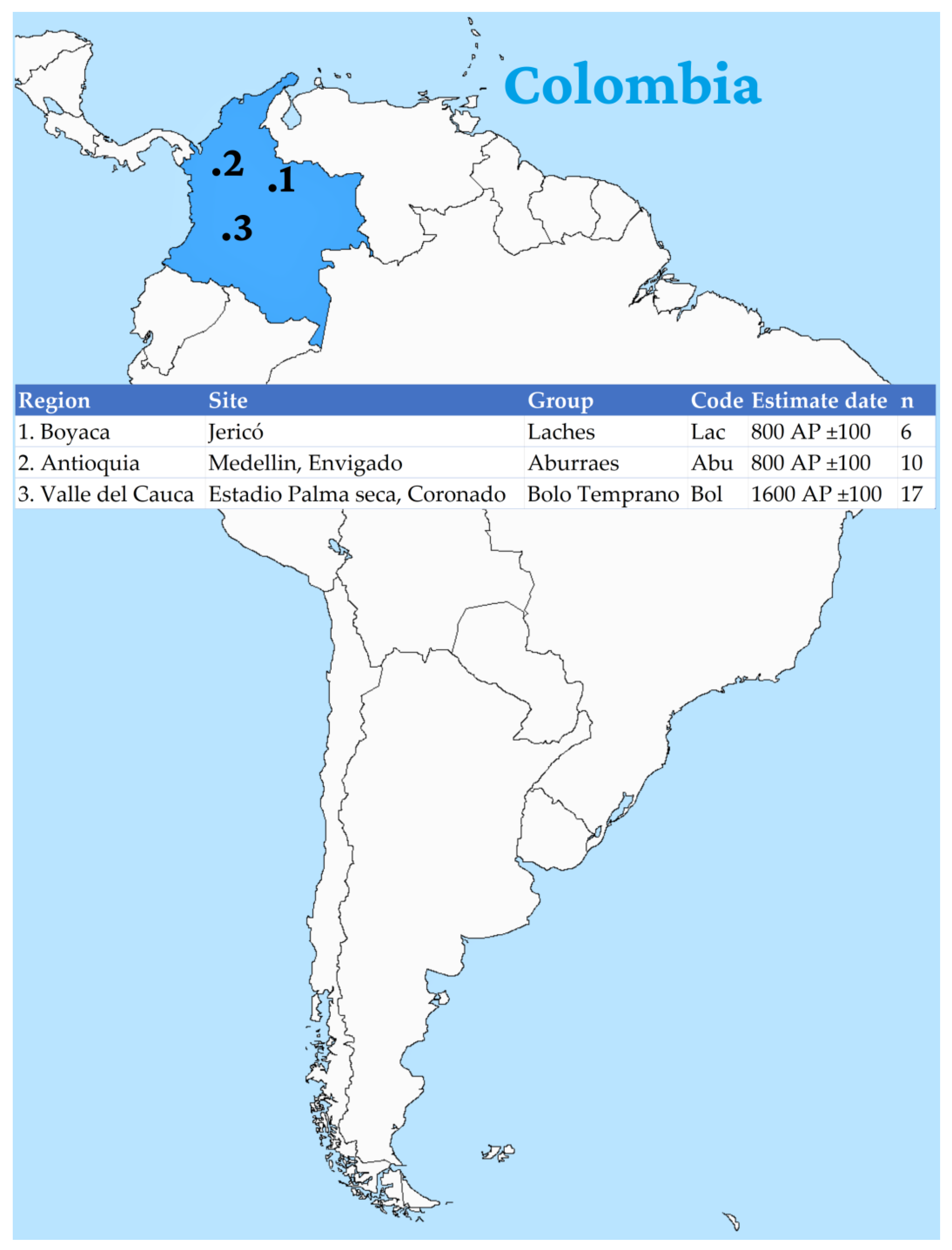
2. Materials and Methods
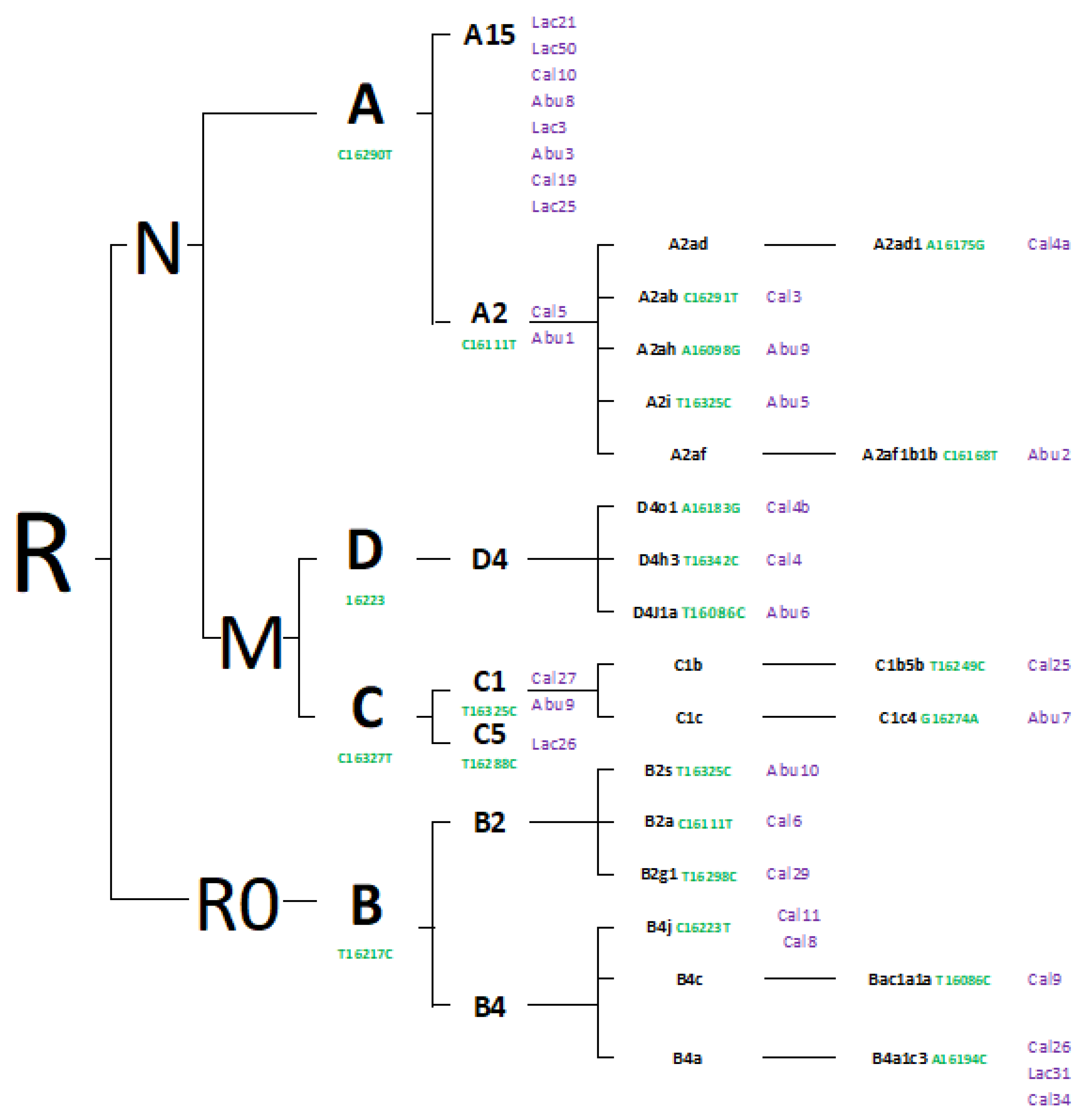
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reichel-Dolmatoff, G. Colombia Indígena-Periodo Prehispánico. In Manual de Historia de Colombia Tomo i; Instituto Colombiano de Cultura: Bogota, Colombia, 1978; pp. 33–115. [Google Scholar]
- Langebaek, C.H. Los Muiscas. La Historia Milenaria de un Pueblo Chibcha; Debate: Bogotá, Colombia, 2019. [Google Scholar]
- López, H.S.; Barbosa, M.V. Early prehispanic settlement in the Magdalena Valley in Tolima, Colombia. Balance and perspectives. Quat. Int. 2019, 505, 55–68. [Google Scholar] [CrossRef]
- Lanning, E. Pleistocene man in South America. World Archaeol. 1970, 2, 90–111. [Google Scholar] [CrossRef] [PubMed]
- Etter, A.; McAlpine, C.; Possingham, H. Historical patterns and drivers of landscape change in Colombia since 1500: A regionalized spatial approach. Ann. Assoc. Am. Geogr. 2008, 98, 2–23. [Google Scholar] [CrossRef]
- Van der Hammen, T. The quaternary of Colombia: Introduction to a research project and a series of publications. Palaeogeogr. Palaeoclim. Palaeoecol. 1973, 14, 1–7. [Google Scholar] [CrossRef]
- Dickau, R.; Aceituno, F.J.; Loaiza, N.; López, C.; Cano, M.; Herrera, L.; Restrepo, C.; Ranere, A.J. Radiocarbon chronology of terminal Pleistocene to middle Holocene human occupation in the Middle Cauca Valley, Colombia. Quat. Int. 2015, 363, 43–54. [Google Scholar] [CrossRef]
- Cooke, R. Human Settlement of Central America and Northernmost South America (14,000–8000BP). Quat. Int. 1998, 49–50, 177–190. [Google Scholar] [CrossRef]
- Rodriguez-Cuenca, J.V. Los Chibchas, Pobladores Antiguos de los Andes Orientales: Adaptaciones Bioculturales; Fundación de Investigaciones Arqueológicas Nacionales: Bogotá, Colombia, 1999. [Google Scholar]
- Hoopes, J.W.; Fonseca, O. Goldwork and Chibchan identity: Endogenous change and diffuse unity in the Isthmo-Colombian area. In Gold and Power in Ancient Costa Rica, Panama, and Colombia; Dumbarton Oaks: Washington, DC, USA, 2003; pp. 49–89. [Google Scholar]
- Yang, N.N.; Mazières, S.; Bravi, C.; Ray, N.; Wang, S.; Burley, M.-W.; Bedoya, G.; Rojas, W.; Parra, M.V.; Molina, J.A.; et al. Contrasting patterns of nuclear and mtDNA diversity in Native American populations. Ann. Hum. Genet. 2010, 74, 525–538. [Google Scholar] [CrossRef]
- Wang, S.; Lewis, C.M.; Jakobsson, M.; Ramachandran, S.; Ray, N.; Bedoya, G.; Rojas, W.; Parra, M.V.; A Molina, J.; Gallo, C.; et al. Genetic variation and population structure in Native Americans. PLoS Genet. 2007, 3, e185. [Google Scholar] [CrossRef]
- Monsalve, M.V.; Cardenas, F.; Guhl, F.; Delaney, A.D.; Devine, D.V. Phylogenetic analysis of mtDNA lineages in South American mummies. Ann. Hum. Genet. 1996, 60, 293–303. [Google Scholar] [CrossRef]
- Fernández, N.J. La Arqueología Molecular Aplicada a la Solución de Problemas Prehistóricos: Análisis de ADN Mitocondrial en Momias y Restos Óseos Prehispánicos; Universidad Nacional de Colombia: Bogotá, Colombia, 1999. [Google Scholar]
- Silva, A.; Briceño, I.; Burgos, J.; Torres, D.; Villegas, V.; Gómez, A.; Bernal, J.E.; Rodríguez, J.V. Mitochondrial DNA analysis on pre-Columbian bone remains of the Herrera period. Biomédica 2008, 28, 569–577. [Google Scholar] [CrossRef][Green Version]
- Gutiérrez, J. Determinación de la Estructura Genetica en un Grupo Poblacional Muisca Mediante el Análisis de Polimorfismos en el ADN Mitochondrial; Universidad Pontificia Javeriana: Bogotá, Colombia, 2007. [Google Scholar]
- Sánchez Collazos, M.C. Secuenciación de la Región Control del ADN Mitocondrial a Partir de Fragmentos Óseos Prehispánicos Hallados en el Sector de Candelaria La Nueva en Bogotá. Master′s Thesis, Pontificia Universidad Javeriana, Bogotá, Colombia, 1 September 2007. [Google Scholar]
- Casas-Vargas, A.; Gómez, A.; Briceño, I.; Díaz-Matallana, M.; Bernal, J.E.; Rodríguez, J.V. High genetic diversity on a sample of pre-Columbian bone remains from Guane territories in northwestern Colombia. Am. J. Phys. Anthr. 2011, 146, 637–649. [Google Scholar] [CrossRef] [PubMed]
- Noguera-Santamaría, M.C.; Anderson, C.E.; Uricoechea, D.; Durán, C.; Briceño-Balcázar, I.; Bernal-Villegas, J. Mitochondrial DNA analysis suggests a Chibchan migration into Colombia. Univ. Sci. 2015, 20, 261–278. [Google Scholar] [CrossRef]
- Hummel, S. Authenticity of results. In Ancient DNA Typing; Hummel, S., Ed.; Springer: Berlin/Heidelberg, Germany, 2003; pp. 131–157. [Google Scholar]
- Cooper, A.; Poinar, H.N. Ancient DNA: Do It Right or Not at All. Science 2000, 289, 1139. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, M.T.P.; Bandelt, H.-J.; Hofreiter, M.; Barnes, I. Assessing ancient DNA studies. Trends Ecol. Evol. 2005, 20, 541–544. [Google Scholar] [CrossRef]
- Andrews, R.M.; Kubacka, I.; Chinnery, P.F.; Lightowlers, R.N.; Turnbull, D.M.; Howell, N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 1999, 23, 147. [Google Scholar] [CrossRef]
- Fehren-Schmitz, L.; Reindel, M.; Cagigao, E.T.; Hummel, S.; Herrmann, B. Pre-Columbian population dynamics in coastal southern Peru: A diachronic investigation of mtDNA patterns in the Palpa region by ancient DNA analysis. Am. J. Phys. Anthr. 2009, 141, 208–221. [Google Scholar] [CrossRef]
- Pääbo, S.; Poinar, H.; Serre, D.; Jaenicke-Després, V.; Hebler, J.; Rohland, N.; Kuch, M.; Krause, J.; Vigilant, L.; Hofreiter, M. Genetic analyses from ancient DNA. Annu. Rev. Genet. 2004, 38, 645–679. [Google Scholar] [CrossRef]
- Parson, W.; Gusmão, L.; Hares, D.; Irwin, J.; Mayr, W.; Morling, N.; Pokorak, E.; Prinz, M.; Salas, A.; Schneider, P.; et al. DNA Commission of the International Society for Forensic Genetics: Revised and extended guidelines for mitochondrial DNA typing. Forensic Sci. Int. Genet. 2014, 13, 134–142. [Google Scholar] [CrossRef]
- Behar, D.M.; van Oven, M.; Rosset, S.; Metspalu, M.; Loogväli, E.-L.; Silva, N.M.; Kivisild, T.; Torroni, A.; Villems, R. A “Copernican” reassessment of the human mitochondrial DNA tree from its root. Am. J. Hum. Genet. 2012, 90, 675–684. [Google Scholar] [CrossRef][Green Version]
- Fan, L.; Yao, Y.-G. An update to MitoTool: Using a new scoring system for faster mtDNA haplogroup determination. Mitochondrion 2013, 13, 360–363. [Google Scholar] [CrossRef]
- Kloss-Brandstätter, A.; Pacher, D.; Schönherr, S.; Weissensteiner, H.; Binna, R.; Specht, G.; Kronenberg, F. HaploGrep: A fast and reliable algorithm for automatic classification of mitochondrial DNA haplogroups. Hum. Mutat. 2010, 32, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Bandelt, H.J.; Forster, P.; Rohl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Lott, M.T.; Leipzig, J.N.; Derbeneva, O.; Xie, H.M.; Chalkia, D.; Sarmady, M.; Procaccio, V.; Wallace, D.C. mtDNA variation and analysis using Mitomap and Mitomaster. Curr. Protoc. Bioinform. 2013, 44, 1.23.1–1.23.26. [Google Scholar] [CrossRef] [PubMed]
- van Oven, M.; Kayser, M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 2009, 30, E386–E394. [Google Scholar] [CrossRef] [PubMed]
- Röck, A.W.; Dür, A.; van Oven, M.; Parson, W. Concept for estimating mitochondrial DNA haplogroups using a maximum likelihood approach (EMMA). Forensic Sci. Int. Genet. 2013, 7, 601–609. [Google Scholar] [CrossRef]
- Parson, W.; Dür, A. EMPOP—A forensic mtDNA database. Forensic Sci. Int. Genet. 2007, 1, 88–92. [Google Scholar] [CrossRef]
- Derenko, M.; Malyarchuk, B.; Grzybowski, T.; Denisova, G.; Dambueva, I.; Perkova, M.; Dorzhu, C.; Luzina, F.; Lee, H.K.; Vanecek, T.; et al. Phylogeographic analysis of mitochondrial DNA in Northern Asian populations. Am. J. Hum. Genet. 2007, 81, 1025–1041. [Google Scholar] [CrossRef]
- Tanaka, M.; Cabrera, V.M.; González, A.M.; Larruga, J.M.; Takeyasu, T.; Fuku, N.; Guo, L.-J.; Hirose, R.; Fujita, Y.; Kurata, M.; et al. Mitochondrial genome variation in eastern Asia and the peopling of Japan. Genome Res. 2004, 14, 1832–1850. [Google Scholar] [CrossRef]
- Behar, D.M.; Rosset, S.; Blue-Smith, J.; Balanovsky, O.; Tzur, S.; Comas, D.; Mitchell, R.J.; Quintana-Murci, L.; Tyler-Smith, C.; Wells, R.S.; et al. The genographic project public participation mitochondrial DNA database. PLoS Genet. 2007, 3, e104. [Google Scholar] [CrossRef]
- Raff, J.A.; Rzhetskaya, M.; Tackney, J.; Hayes, M.G. Mitochondrial diversity of Iñupiat people from the Alaskan North Slope provides evidence for the origins of the Paleo- and Neo-Eskimo peoples. Am. J. Phys. Anthr. 2015, 157, 603–614. [Google Scholar] [CrossRef]
- Torroni, A.; Schurr, T.G.; Yang, C.C.; Szathmary, E.J.; Williams, R.C.; Schanfield, M.S.; A Troup, G.; Knowler, W.C.; Lawrence, D.N.; Weiss, K.M. Native American mitochondrial DNA analysis indicates that the Amerind and the Nadene populations were founded by two independent migrations. Genetics 1992, 130, 153–162. Available online: https://www.ncbi.nlm.nih.gov/pubmed/1346260 (accessed on 18 January 2022). [CrossRef] [PubMed]
- Reich, D.; Patterson, N.; Campbell, D.; Tandon, A.; Mazieres, S.; Ray, N.; Parra, M.V.; Rojas, W.; Duque, C.; Mesa, N.; et al. Reconstructing native American population history. Nature 2012, 488, 370. [Google Scholar] [CrossRef] [PubMed]
- Tamm, E.; Kivisild, T.; Reidla, M.; Metspalu, M.; Smith, D.G.; Mulligan, C.J.; Bravi, C.M.; Rickards, O.; Martinez-Labarga, C.; Khusnutdinova, E.K.; et al. Beringian standstill and spread of Native American founders. PLoS ONE 2007, 2, e829. [Google Scholar] [CrossRef]
- Achilli, A.; Perego, U.A.; Lancioni, H.; Olivieri, A.; Gandini, F.; Kashani, B.H.; Battaglia, V.; Grugni, V.; Angerhofer, N.; Rogers, M.P.; et al. Reconciling migration models to the Americas with the variation of North American native mitogenomes. Proc. Natl. Acad. Sci. USA 2013, 110, 14308–14313. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, M.T.P.; Jenkins, D.L.; Götherstrom, A.; Naveran, N.; Sanchez, J.J.; Hofreiter, M.; Thomsen, P.F.; Binladen, J.; Higham, T.F.G.; Yohe, R.M.; et al. DNA from pre-Clovis human coprolites in Oregon, North America. Science 2008, 320, 786–789. [Google Scholar] [CrossRef] [PubMed]
- Raff, J.A.; Bolnick, D.A.; Tackney, J.; O’Rourke, D.H. Ancient DNA perspectives on American colonization and population history. Am. J. Phys. Anthr. 2011, 146, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Achilli, A.; Perego, U.A.; Bravi, C.M.; Coble, M.D.; Kong, Q.-P.; Woodward, S.R.; Salas, A.; Torroni, A.; Bandelt, H.-J. The phylogeny of the four Pan-American MtDNA Haplogroups: Implications for evolutionary and disease studies. PLoS ONE 2008, 3, e1764. [Google Scholar] [CrossRef]
- Perego, U.A.; Lancioni, H.; Tribaldos, M.; Angerhofer, N.; Ekins, J.E.; Olivieri, A.; Woodward, S.R.; Pascale, J.M.; Cooke, R.; Motta, J.; et al. Decrypting the mitochondrial gene pool of modern Panamanians. PLoS ONE 2012, 7, e38337. [Google Scholar] [CrossRef]
- Taboada-Echalar, P.; Álvarez-Iglesias, V.; Heinz, T.; Vidal-Bralo, L.; Gómez-Carballa, A.; Catelli, L.; Pardo-Seco, J.; Pastoriza, A.; Carracedo, Á.; Torres-Balanza, A.; et al. The genetic legacy of the pre-Colonial period in contemporary Bolivians. PLoS ONE 2013, 8, e58980. [Google Scholar] [CrossRef]
- Alves-Silva, J.; Santos, M.d.S.; Guimarães, P.E.; Ferreira, A.C.; Bandelt, H.-J.; Pena, S.D.; Prado, V.F. The ancestry of Brazilian mtDNA lineages. Am. J. Hum. Genet. 2000, 67, 444–461. [Google Scholar] [CrossRef]
- Motti, J.M.B.; Hagelberg, E.; Lindo, J.; Malhi, R.S.; Bravi, C.M.; A Guichón, R. Primer genoma mitocondrial en restos humanos de la Costa de Santa Cruz, Argentina. Magallania 2015, 43, 119–131. [Google Scholar] [CrossRef]
- Herrnstadt, C.; Elson, J.L.; Fahy, E.; Preston, G.; Turnbull, D.M.; Anderson, C.; Ghosh, S.S.; Olefsky, J.M.; Beal, M.F.; Davis, R.E.; et al. Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am. J. Hum. Genet. 2002, 70, 1152–1171. [Google Scholar] [CrossRef] [PubMed]
- Kemp, B.M.; González-Oliver, A.; Malhi, R.S.; Monroe, C.; Schroeder, K.B.; McDonough, J.; Rhett, G.; Resendéz, A.; Peñaloza-Espinosa, R.I.; Buentello-Malo, L.; et al. Evaluating the farming/language dispersal hypothesis with genetic variation exhibited by populations in the Southwest and Mesoamerica. Proc. Natl. Acad. Sci. USA 2010, 107, 6759–6764. [Google Scholar] [CrossRef] [PubMed]
- Allard, M.W.; Polanskey, D.; Wilson, M.R.; Monson, K.L.; Budowle, B. Evaluation of variation in control region sequences for Hispanic individuals in the SWGDAM mtDNA data set. J. Forensic Sci. 2006, 51, 566–573. [Google Scholar] [CrossRef]
- Malhi, R.S.; Schultz, B.A.; Smith, D.G. Distribution of mitochondrial DNA lineages among Native American tribes of Northeastern North America. Hum. Biol. 2001, 73, 17–55. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Bellis, C.; Zlojutro, M.; E Melton, P.; Blangero, J.; E Curran, J. Large scale mitochondrial sequencing in Mexican Americans suggests a reappraisal of Native American origins. BMC Evol. Biol. 2011, 11, 293. [Google Scholar] [CrossRef]
- Soares, P.; Rito, T.; Trejaut, J.; Mormina, M.; Hill, C.; Tinkler-Hundal, E.; Braid, M.; Clarke, D.J.; Loo, J.-H.; Thomson, N.; et al. Ancient voyaging and Polynesian origins. Am. J. Hum. Genet. 2011, 88, 239–247. [Google Scholar] [CrossRef]
- Prieto, L.; Zimmermann, B.; Goios, A.; Rodriguez, M.A.; Paneto, G.G.; Alves, C.; Alonso, A.; Fridman, C.; Cardoso, S.; Lima, G.; et al. The GHEP–EMPOP collaboration on mtDNA population data—A new resource for forensic casework. Forensic. Sci. Int. Genet. 2011, 5, 146–151. [Google Scholar] [CrossRef]
- Fagundes, N.J.; Kanitz, R.; Eckert, R.; Valls, A.C.; Bogo, M.R.; Salzano, F.M.; Smith, D.G.; Silva, W.A.; Zago, M.A.; Ribeiro-Dos-Santos, A.K.; et al. Mitochondrial population genomics supports a single pre-Clovis origin with a coastal route for the peopling of the Americas. Am. J. Hum. Genet. 2008, 82, 583–592. [Google Scholar] [CrossRef]
- González-Oliver, A.; Márquez-Morfín, L.; Jiménez, J.C.; Torre-Blanco, A. Founding Amerindian mitochondrial DNA lineages in ancient Maya from Xcaret, Quintana Roo. Am. J. Phys. Anthr. 2001, 116, 230–235. [Google Scholar] [CrossRef]
- Starikovskaya, E.B.; Sukernik, R.I.; Derbeneva, O.A.; Volodko, N.V.; Ruiz-Pesini, E.; Torroni, A.; Brown, M.D.; Lott, M.T.; Hosseini, S.H.; Huoponen, K.; et al. Mitochondrial DNA diversity in indigenous populations of the southern extent of Siberia, and the origins of Native American haplogroups. Ann. Hum. Genet. 2005, 69, 67–89. [Google Scholar] [CrossRef] [PubMed]
- Whittington, S.L.; Reed, D.M. Bones of the Maya: Studies of Ancient Skeletons; University of Alabama Press: Tuscaloosa, AL, USA, 2006. [Google Scholar]
- Lalueza-Fox, C.; Gilbert, M.; Martínez-Fuentes, A.; Calafell, F.; Bertranpetit, J. Mitochondrial DNA from pre-Columbian Ciboneys from Cuba and the prehistoric colonization of the Caribbean. Am. J. Phys. Anthr. 2003, 121, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Lalueza-Fox, C.; Calderon, F.L.; Calafell, F.; Morera, B.; Bertranpetit, J. MtDNA from extinct Tainos and the peopling of the Caribbean. Ann. Hum. Genet. 2001, 65, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Perego, U.A.; Achilli, A.; Angerhofer, N.; Accetturo, M.; Pala, M.; Olivieri, A.; Kashani, B.H.; Ritchie, K.H.; Scozzari, R.; Kong, Q.-P.; et al. Distinctive Paleo-Indian migration routes from Beringia marked by two rare mtDNA haplogroups. Curr. Biol. 2009, 19, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zillges, H. The Genetic History of the Otomi in the Central Mexican Valley. Ph.D. Thesis, University of Pennsylvania, Philadelphia, PA, USA, 2013. [Google Scholar]
- Derenko, M.; Malyarchuk, B.; Grzybowski, T.; Denisova, G.; Rogalla, U.; Perkova, M.; Dambueva, I.; Zakharov, I. Origin and post-glacial dispersal of mitochondrial DNA Haplogroups C and D in northern Asia. PLoS ONE 2010, 5, e15214. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekar, A.; Kumar, S.; Sreenath, J.; Sarkar, B.N.; Urade, B.P.; Mallick, S.; Bandopadhyay, S.S.; Barua, P.; Barik, S.S.; Basu, D.; et al. Updating phylogeny of mitochondrial DNA Macrohaplogroup M in India: Dispersal of Modern human in South Asian corridor. PLoS ONE 2009, 4, e7447. [Google Scholar] [CrossRef] [PubMed]
- Mesa, N.R.; Mondragón, M.C.; Soto, I.D.; Parra, M.V.; Duque, C.; Ortíz-Barrientos, D.; García, L.F.; Velez, I.D.; Bravo, M.L.; Múnera, J.G.; et al. Autosomal, mtDNA, and Y-chromosome diversity in Amerinds: Pre- and post-Columbian patterns of gene flow in South America. Am. J. Hum. Genet. 2000, 67, 1277–1286. [Google Scholar] [CrossRef] [PubMed]
- Melton, P.E.; Briceño, I.; Gómez, A.; Devor, E.; Bernal, J.; Crawford, M. Biological relationship between central and South American Chibchan speaking populations: Evidence from mtDNA. Am. J. Phys. Anthr. 2007, 133, 753–770. [Google Scholar] [CrossRef]
- Keyeux, G.; Rodas, M.C.; Bernal Villegas, J. Haplogrupos fundadores del DNA mitocondrial en poblaciones colombianas: Aporte a los estudios en América. In Ordóñez V, Coordinator. Geografía Humana de Colombia: Variación Biológica y Cultural en Colombia. Colección V Centenario, 1st ed.; Instituto Colombiano de Cultura Hispánica: Bogotá, Colombia, 2001. [Google Scholar]
- Ossa, H.; Aquino, J.; Pereira, R.; Ibarra, A.; Ossa, R.H.; Pérez, L.A.; Granda, J.D.; Lattig, M.C.; Groot, H.; de Carvalho, E.F.; et al. Outlining the ancestry landscape of Colombian admixed populations. PLoS ONE 2016, 11, e0164414. [Google Scholar] [CrossRef]
- Hoffecker, J.F.; Elias, S.A.; O’Rourke, D.H.; Scott, G.R.; Bigelow, N.H. Beringia and the global dispersal of modern humans. Evol. Anthropol. Issues News Rev. 2016, 25, 64–78. [Google Scholar] [CrossRef]
- Dixon, E.J. Late Pleistocene colonization of North America from Northeast Asia: New insights from large-scale paleogeographic reconstructions. In Mobility and Ancient Society in Asia and the Americas; Springer: Cham, Switzerland, 2015; pp. 169–184. [Google Scholar] [CrossRef]
- Relethford, J.H. Biological anthropology, population genetics, and race. In The Oxford Handbook of Philosophy and Race; Oxford Handbooks; Oxford Academic: Oxford, UK, 2017. [Google Scholar] [CrossRef]
- Kivisild, T. Maternal ancestry and population history from whole mitochondrial genomes. Investig. Genet. 2015, 6, 3. [Google Scholar] [CrossRef] [PubMed]
- Wallace Douglas, C. Mitochondrial DNA variation in human radiation and disease. Cell 2015, 163, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Llamas, B.; Fehren-Schmitz, L.; Valverde, G.; Soubrier, J.; Mallick, S.; Rohland, N.; Nordenfelt, S.; Valdiosera, C.; Richards, S.M.; Rohrlach, A.; et al. Ancient mitochondrial DNA provides high-resolution time scale of the peopling of the Americas. Sci. Adv. 2016, 2, e1501385. [Google Scholar] [CrossRef] [PubMed]
- Lindo, J.; Achilli, A.; Perego, U.A.; Archer, D.; Valdiosera, C.; Petzelt, B.; Mitchell, J.; Worl, R.; Dixon, E.J.; Fifield, T.E.; et al. Ancient individuals from the North American Northwest Coast reveal 10,000 years of regional genetic continuity. Proc. Natl. Acad. Sci. USA 2017, 114, 4093–4098. [Google Scholar] [CrossRef]
- Langebaek, C.H. Arqueología Colombiana: Ciencia, Pasado y Exclusion; Colciencias, Instituto Colombiano para el Desarrollo de la Ciencia y la Tecnología Francisco José de Caldas: Bogota, Colombia, 2003; Volume 3. [Google Scholar]
- Rey, D.; Areces, C.; Enríquez-De-Salamanca, M.; Parga-Lozano, C.; Abd-El-Fatah, S.; Fernández, M.; Arnaiz-Villena, A. Los primeros pobladores de América y sus relaciones con poblaciones del Océano Pacífico según los genes HLA. Inmunología 2012, 31, 83–91. [Google Scholar] [CrossRef]
- Walbert, D. Disease and catastrophe. In North Carolina History: A Digital Textbook; NCpedia: Kinston, NC, USA, 2016. [Google Scholar]
- Taylor, R. Diseases That Changed History. In White Coat Tales; Springer: Cham, Switzerland, 2016. [Google Scholar]
- O’Fallon, B.D.; Fehren-Schmitz, L. Native Americans experienced a strong population bottleneck coincident with European contact. Proc. Natl. Acad. Sci. USA 2011, 108, 20444–20448. [Google Scholar] [CrossRef]
- Balter, M. Genes confirm Europeans’ blow to Native Americans. Science 2011, 334, 1335. [Google Scholar] [CrossRef]
- Jobling, M.; Hurles, M.; Tyler-Smith, C. Human Evolutionary Genetics, 2nd ed.; Taylor & Francis Group: New York, NY, USA, 2013. [Google Scholar]
- O’Rourke, D.H. Human migrations: The two roads taken. Curr. Biol. 2009, 19, R203–R205. [Google Scholar] [CrossRef][Green Version]
- Stoneking, M.; Krause, J. Learning about human population history from ancient and modern genomes. Nat. Rev. Genet. 2011, 12, 603–614. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uricoechea Patiño, D.; Collins, A.; García, O.J.R.; Santos Vecino, G.; Cuenca, J.V.R.; Bernal, J.E.; Benavides Benítez, E.; Vergara Muñoz, S.; Briceño Balcázar, I. High Mitochondrial Haplotype Diversity Found in Three Pre-Hispanic Groups from Colombia. Genes 2023, 14, 1853. https://doi.org/10.3390/genes14101853
Uricoechea Patiño D, Collins A, García OJR, Santos Vecino G, Cuenca JVR, Bernal JE, Benavides Benítez E, Vergara Muñoz S, Briceño Balcázar I. High Mitochondrial Haplotype Diversity Found in Three Pre-Hispanic Groups from Colombia. Genes. 2023; 14(10):1853. https://doi.org/10.3390/genes14101853
Chicago/Turabian StyleUricoechea Patiño, Daniel, Andrew Collins, Oscar Julián Romero García, Gustavo Santos Vecino, José Vicente Rodríguez Cuenca, Jaime E. Bernal, Escilda Benavides Benítez, Saray Vergara Muñoz, and Ignacio Briceño Balcázar. 2023. "High Mitochondrial Haplotype Diversity Found in Three Pre-Hispanic Groups from Colombia" Genes 14, no. 10: 1853. https://doi.org/10.3390/genes14101853
APA StyleUricoechea Patiño, D., Collins, A., García, O. J. R., Santos Vecino, G., Cuenca, J. V. R., Bernal, J. E., Benavides Benítez, E., Vergara Muñoz, S., & Briceño Balcázar, I. (2023). High Mitochondrial Haplotype Diversity Found in Three Pre-Hispanic Groups from Colombia. Genes, 14(10), 1853. https://doi.org/10.3390/genes14101853





