The Satellite DNA Catalogues of Two Serrasalmidae (Teleostei, Characiformes): Conservation of General satDNA Features over 30 Million Years
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling, Cytogenetic Analysis, DNA Extraction and WGS
2.2. Bioinformatic Analyses—Satellitome Characterization and Comparison
2.3. Primers Design, Probes Manufacturing and Fluorescence In Situ Hybridization (FISH)
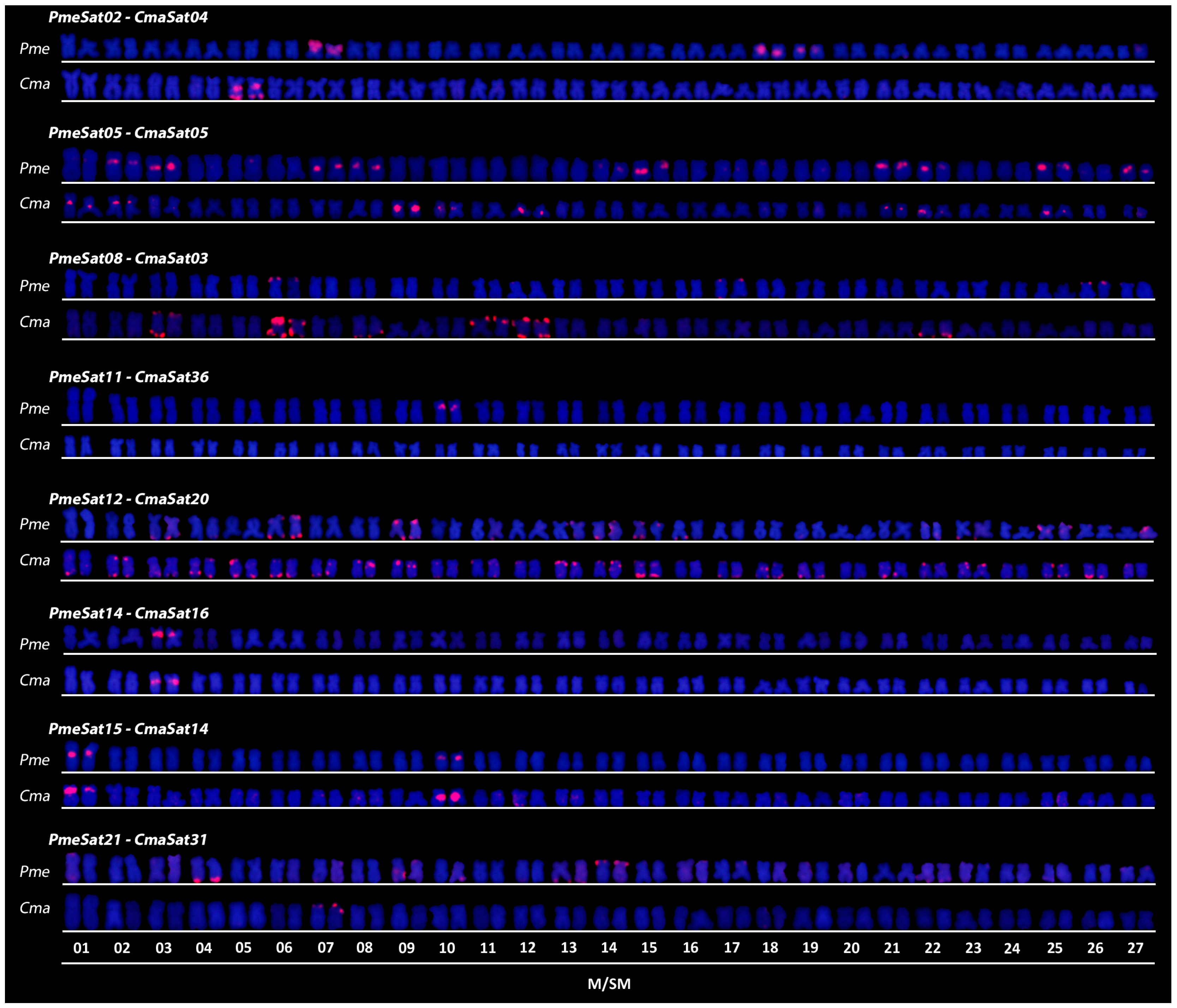
3. Results
3.1. Satellitome Description of P. mesopotamicus and C. macropomum Reveals Several Shared satDNAs
3.2. FISH Reveals Maintenance of Chromosomal Clustering Sites of satDNAs in P. mesopotamicus and C. macropomum
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Charlesworth, B.; Sniegowski, P.; Stephan, L.W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 1994, 371, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Plohl, M.; Meštrović, N.; Mravinac, B. Satellite DNA evolution. In Repetitive DNA; Karger Publishers: Basel, Switzerland, 2012; Volume 7, pp. 126–152. [Google Scholar]
- Garrido-Ramos, M.A. Satellite DNA: An evolving topic. Genes 2017, 8, 230. [Google Scholar] [CrossRef] [PubMed]
- Nurk, S.; Koren, S.; Rhie, A.; Rautiainen, M.; Bzikadze, A.V.; Mikheenko, M.R.; Altemose, N.; Uralsky, L.; Gershamn, A. The complete sequence of a human genome. Science 2022, 376, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Peona, V.; Kutschera, V.E.; Blom, M.P.K.; Irestedt, M.; Suh, A. Satellite DNA evolution in Corvoidea inferred from short and long reads. Mol. Ecol. 2022. [CrossRef] [PubMed]
- Louzada, S.; Lopes, M.; Ferreira, D.; Adega, F.; Escudeiro, A.; Gama-Carvalho, M.; Chaves, R. Decoding the role of satellite DNA in genome architecture and plasticity—An evolutionary and clinical affair. Genes 2020, 11, 72. [Google Scholar] [CrossRef]
- Halbach, R.; Miesen, P.; Joosten, J.; Taşköprü, E.; Rondeel, I.; Pennigns, B.; Vogels, C.; Merkling, S.; Koenraat, C.; Lambrechts, L. An ancient satellite repeat controls gene expression and embryonic development in Aedes aegypti through a highly conserved piRNA. Nature 2020, 580, 274–277. [Google Scholar] [CrossRef]
- Shatskikh, A.S.; Kotov, A.A.; Adashev, V.E.; Bazylev, S.S.; Olenina, L.V. Functional significance of satellite DNAs: Insights from Drosophila. Front. Cell Dev. Biol. 2020, 8, 312. [Google Scholar] [CrossRef]
- Fry, K.; Salser, W. Nucleotide sequences of HS-α satellite DNA from kangaroo rat Dipodomys ordii and characterization of similar sequences in other rodents. Cell 1977, 12, 1069–1084. [Google Scholar] [CrossRef]
- Camacho, J.P.M.; Cabrero, J.; López-León, M.D.; Martín-Pecina, M.; Perfectti, F.; Garrido-Ramos, M.A.; Ruiz-Ruano, F.J. Satellitome comparison of two oedipodine grasshoppers highlights the contingent nature of satellite DNA evolution. BMC Biol. 2022, 20, 36. [Google Scholar]
- de la Herrán, R.; Fontana, F.; Lanfredi, M.; Congiu, L.; Leis, M.; Rossi, R.; Ruiz-Rejón, C.; Ruiz-Rejón, M.; Garrido-Ramos, M.A. Slow rates of evolution and sequence homogenization in an ancient satellite DNA family of sturgeons. Mol. Biol. Evol. 2001, 18, 432–436. [Google Scholar] [CrossRef]
- Mravinac, B.; Plohl, M.; Ugarković, Ð. Sequence of PRAT satellite DNA “frozen” in some coleopteran species. J. Mol. Evol. 2002, 54, 774–783. [Google Scholar] [CrossRef]
- Chaves, R.; Ferreira, D.; Mendes-da-Silva, A.; Meles, S.; Adega, F. FA-SAT is an old satellite DNA frozen in several Bilateria genomes. Genome Biol. Evol. 2017, 9, 3073–3087. [Google Scholar] [CrossRef]
- Escudeiro, A.; Ferreira, D.; Mendes-da-Silva, A.; Heslop-Harrison, J.S.; Adega, F.; Chaves, R. Bovine satellite DNAs–a history of the evolution of complexity and its impact in the Bovidae family. Eur. Zool. J. 2019, 86, 20–37. [Google Scholar] [CrossRef]
- Ruiz-Ruano, F.J.; López-León, M.D.; Cabrero, J.; Camacho, J.P.M. High-throughput analysis of the satellitome illuminates satellite DNA evolution. Sci. Rep. 2016, 6, 28333. [Google Scholar] [CrossRef]
- dos Santos, R.Z.; Csalegari, R.M.; Silva, D.M.Z.A.; Ruiz-Ruano, F.J.; Melo, S.; Oliveira, C.; Foresti, F.; Uliano-Silva, M.; Porto-Foresti, F.; Utsunomia, R. A long-term conserved satellite DNA that remains unexpanded in several genomes of Characiformes fish is actively transcribed. Genome Biol. Evol. 2021, 13, evab002. [Google Scholar]
- Ruiz-Ruano, F.J.; Navarro-Domínguez, B.; Camacho, J.P.M.; Garrido-Ramos, M.A. Characterization of the satellitome in lower vascular plants: The case of the endangered fern Vandenboschia speciosa. Ann. Bot. 2019, 123, 587–599. [Google Scholar] [CrossRef]
- Silva, D.M.Z.A.; Utsunomia, R.; Ruiz-Ruano, F.J.; Daniel, S.N.; Porto-Foresti, F.; Hashimoto, D.T.; Oliveira, C.; Camacho, J.P.M.; Foresti, F. High-throughput analysis unveils a highly shared satellite DNA library among three species of fish genus Astyanax. Sci. Rep. 2017, 7, 12726. [Google Scholar] [CrossRef]
- Sader, M.; Vaio, M.; Cauz-Santos, L.A.; Dornelas, M.C.; Vieira, M.L.C.; Melo, N.; Pedrosa-Harand, A. Large vs small genomes in Passiflora: The influence of the mobilome and the satellitome. Planta 2021, 253, 86. [Google Scholar] [CrossRef]
- Lima, L.G.; Ruiz-Ruano, F.J. In-depth Satellitome Analyses of 37 Drosophila species illuminate repetitive DNA evolution in the Drosophila genus. Genome Biol. Evol. 2022, 14, evac064. [Google Scholar] [CrossRef]
- Melo, B.F.; Sidlauskas, B.L.; Near, T.J.; Roxo, F.F.; Ghezelayagh, A.; Ochoa, L.E.; Stiassny, M.L.J.; Arroyaye, J.; Chang, J.; Faircloth, B.C. Accelerated diversification explains the exceptional species richness of tropical characoid fishes. Syst. Biol. 2022, 71, 78–92. [Google Scholar] [CrossRef]
- Utsunomia, R.; Silva, D.M.Z.A.; Ruiz-Ruano, F.J.; Goes, C.A.G.; Melo, S.; Ramos, L.P.; Oliveira, C.; Porto-Foresti, F.; Foresti, F.; Hashimoto, D.T. Satellitome landscape analysis of Megaleporinus macrocephalus (Teleostei, Anostomidae) reveals intense accumulation of satellite sequences on the heteromorphic sex chromosome. Sci. Rep. 2019, 9, 5856. [Google Scholar] [CrossRef] [PubMed]
- Crepaldi, C.; Parise-Maltempi, P.P. Heteromorphic sex chromosomes and their DNA content in fish: An insight through satellite DNA accumulation in Megaleporinus elongatus. Cytogenet. Genome Res. 2020, 160, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Freitas, E.A.; Silva, D.M.Z.A.; Ruiz-Ruano, F.J.; Utsunomia, R.; Araya-Jaime, C.; Oliveira, C.; Camacho, J.P.M.; Foresti, F. Satellite DNA content of B chromosomes in the characid fish Characidium gomesi supports their origin from sex chromosomes. Mol. Genet. Genomics 2020, 295, 195–207. [Google Scholar] [CrossRef]
- Stornioli, J.H.F.; Goes, C.A.G.; Calegari, R.M.; dos Santos, R.Z.; Giglio, L.M.; Foresti, F.; Oliveira, C.; Penitente, M.; Porto-Foresti, F.; Utsunomia, R. The B Chromosomes of Prochilodus lineatus (Teleostei, Characiformes) Are Highly Enriched in Satellite DNAs. Cells 2021, 10, 1527. [Google Scholar] [CrossRef] [PubMed]
- Goes, C.A.G.; dos Santos, R.Z.; Aguiar, W.R.C.; Alves, D.C.V.; Silva, D.M.Z.A.; Foresti, F.; Oliveira, C.; Utsunomia, R.; Porto-Foresti, F. Revealing the satellite DNA history in Psalidodon and Astyanax characid fish by comparative satellitomics. Front. Genet. 2022, 13, 1567. [Google Scholar] [CrossRef] [PubMed]
- Kretschmer, R.; Goes, C.A.G.; Bertollo, L.A.C.; Ezaz, T.; Porot-Foresti, F.; Toma, G.A.; Utsunomia, R.; Cioffi, M.B. Satellitome analysis illuminates the evolution of ZW sex chromosomes of Triportheidae fishes (Teleostei: Characiformes). Chromosoma 2022, 131, 29–45. [Google Scholar] [CrossRef]
- Eschmeyer, W.; Fong, R. Eschmeyer’s Catalog of Fishes: Genera, Species, References; California Academy of Sciences: San Francisco, CA, USA, 2022. [Google Scholar]
- Ortí, G.; Sivasundar, A.; Dietz, K.; Jégu, M. Phylogeny of the Serrasalmidae (Characiformes) based on mitochondrial DNA sequences. Genet. Mol. Biol. 2008, 31, 343–351. [Google Scholar] [CrossRef]
- Nico, L.; Jegu, M.; Andrade, M.C. Serrasalmidae—Piranhas and Pacus. 2017. [Google Scholar]
- Kolmann, M.A.; Hughes, L.C.; Hernandez, L.P.; Arcila, D.; Betancur, R.R.; Sabaj, M.H.; López-Fernandez, H.; Ortí, G. Phylogenomics of piranhas and pacus (Serrasalmidae) uncovers how dietary convergence and parallelism obfuscate traditional morphological taxonomy. Syst. Biol. 2021, 70, 576–592. [Google Scholar] [CrossRef]
- Adriano, M.P.W.; Bertollo, L.A.C.; Moreira-Filho, O. Karyotypic characterization of Myleus micans (Lütken, 1875)(Pisces, Characidae, Serrasalminae). Caryologia 2006, 59, 125–130. [Google Scholar] [CrossRef]
- Cestari, M.M.; Galetti, P.M., Jr. Chromosome studies of Serrasalmus spilopleura (Characidae, Serrasalminae) from the Parana-Paraguay rivers: Evolutionary and cytotaxonomic considerations. Copeia 1992, 1992, 108–112. [Google Scholar] [CrossRef]
- Favarato, R.M.; Ribeiro, L.B.; Campos, A.; Porto, J.I.R.; Nakayama, C.M.; Ota, R.P.; Feldberg, E. Comparative cytogenetics of Serrasalmidae (Teleostei: Characiformes): The relationship between chromosomal evolution and molecular phylogenies. PLoS ONE 2021, 16, e0258003. [Google Scholar] [CrossRef]
- Nakayama, C.M.; Rebelo Porto, J.I.; Feldberg, E. A comparative cytogenetic study of five piranha species (Serrasalmus, Serrasalminae) from the Amazon basin. Genetica 2002, 114, 231–236. [Google Scholar] [CrossRef]
- Nirchio, M.; Fenocchio, A.S.; Swarca, A.C.; Pérez, J.E.; Grandado, A.; Estrada, A.; Ron, E. Cytogenetic characterization of hybrids offspring between Colossoma macropomum (Cuvier, 1818) and Piaractus brachypomus (Cuvier, 1817) from Caicara del Orinoco, Venezuela. Caryologia 2003, 56, 405–411. [Google Scholar] [CrossRef][Green Version]
- Ribeiro, L.B.; Matoso, D.A.; Feldberg, E. Chromosome mapping of repetitive sequences in four Serrasalmidae species (Characiformes). Genet. Mol. Biol. 2014, 37, 46–53. [Google Scholar] [CrossRef]
- Valladão, G.M.R.; Gallani, S.U.; Pilarski, F. South American fish for continental aquaculture. Rev. Aquac. 2018, 10, 351–369. [Google Scholar] [CrossRef]
- Fernandes, E.M.; Almeida, L.C.F.; Hashimoto, D.T.; Lattanzi, G.R.; Gervaz, W.R.; Leonardo, A.F.; Neto, R.V.R. Survival of purebred and hybrid Serrasalmidae under low water temperature conditions. Aquaculture 2018, 497, 97–102. [Google Scholar] [CrossRef]
- Foresti, F.; Almeida-Toledo, L.F.; Toledo-Filho, S.A. Polymorphic nature of nucleolus organizer regions in fishes. Cytogenet. Genome Res. 1981, 31, 137–144. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Novák, P.; Ávila Robledillo, L.; Koblížková, A.; Vrbová, I.; Neumann, P.; Macas, J. TAREAN: A computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucleic Acids Res. 2017, 45, e111. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Smit, A.F.A.; Hubley, R.; Green, P. 1996–2010. RepeatMasker Open-3.0. 2017. [Google Scholar]
- Pinkel, D.; Straume, T.; Gray, J.W. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc. Natl. Acad. Sci. USA 1986, 83, 2934–2938. [Google Scholar] [CrossRef] [PubMed]
- Sumner, A.T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 1972, 75, 304–306. [Google Scholar] [CrossRef] [PubMed]
- Meyne, J.; Baker, R.J.; Hobart, H.H.; Hsu, T.C.; Ryder, O.A.; Ward, O.G.; Wiley, J.E.; Wurster-Hill, D.H.; Yates, T.L.; Moyzes, R.K. Distribution of non-telomeric sites of the (TTAGGG) n telomeric sequence in vertebrate chromosomes. Chromosoma 1990, 99, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Almeida-Toledo, L.F.; Viegas-Péquignot, E.; Coutinho-Barbosa, A.C.; Foresti, F.; Niveleau, A.; Toledo-Filho, S.A. Localization of 5-methylcytosine in metaphase chromosomes of diploid and triploid pacu fish, Piaractus mesopotamicus (Pisces, Characiformes). Cytogenet. Genome Res. 1998, 83, 21–24. [Google Scholar] [CrossRef]
- Yunis, J.J.; Yasmineh, W.G. Heterochromatin, Satellite DNA, and Cell Function: Structural DNA of eucaryotes may support and protect genes and aid in speciation. Science 1971, 174, 1200–1209. [Google Scholar] [CrossRef]
- Cohen, S.; Agmon, N.; Sobol, O.; Segal, D. Extrachromosomal circles of satellite repeats and 5S ribosomal DNA in human cells. Mob. DNA 2010, 1, 11. [Google Scholar] [CrossRef]
- Costa, M.S.; Silva, H.C.M.; Soares, S.C.; Favarato, R.M.; Feldberg, E.; Gomes, A.L.S.; Artoni, R.F.; Matoso, D.A. A Perspective of Molecular Cytogenomics, Toxicology, and Epigenetics for the Increase of Heterochromatic Regions and Retrotransposable Elements in Tambaqui (Colossoma macropomum) Exposed to the Parasiticide Trichlorfon. Animals 2022, 12, 1945. [Google Scholar] [CrossRef]
- Cioffi, M.C.; Martins, C.; Bertollo, L.A.C. Chromosome spreading of associated transposable elements and ribosomal DNA in the fish Erythrinus erythrinus. Implications for genome change and karyoevolution in fish. BMC Evol. Biol. 2010, 10, 271. [Google Scholar] [CrossRef]
- Henikoff, S.; Ahmad, S.; Malik, H.S. The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 2001, 293, 1098–1102. [Google Scholar] [CrossRef]
- Gao, D.; Gill, N.; Kim, H.; Walling, J.G.; Zhang, W.; Fan, C.; Yu, Y.; Ma, J.; SanMiguel, P.; Jiang, N. A lineage-specific centromere retrotransposon in Oryza brachyantha. Plant J. 2009, 60, 820–831. [Google Scholar] [CrossRef]
- Zhang, H.; Koblízková, A.; Kai, W.; Zhiyun, G.; Oliveira, L.; Torres, G.A.; Wu, Y.; Zhang, W.; Novák, P.; Buell, C.R. Boom-bust turnovers of megabase-sized centromeric DNA in Solanum species: Rapid evolution of DNA sequences associated with centromeres. Plant Cell 2014, 26, 1436–1447. [Google Scholar] [CrossRef]
- Gong, Z.; Yufen, W.; Koblizkova, A.; Torres, G.A.; Kai, W.; Iowene, M.; Neumann, P.; Zhang, W.; Novák, P.; Buell, C.R. Repeatless and repeat-based centromeres in potato: Implications for centromere evolution. Plant Cell 2012, 24, 3559–3574. [Google Scholar] [CrossRef]
- Melters, D.P.; Bradnam, K.R.; Telis, N.; May, M.R.; Graham, R.J.; Robert, S.; Paul, P.; John, E. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 2013, 14, R10. [Google Scholar] [CrossRef]
- Elder, J.F., Jr.; Turner, B.J. Concerted evolution of repetitive DNA sequences in eukaryotes. Q. Rev. Biol. 1995, 70, 297–320. [Google Scholar] [CrossRef]
- Ohta, T.; Dover, G.A. The cohesive population genetics of molecular drive. Genetics 1984, 108, 501–521. [Google Scholar] [CrossRef]
- Dover, G.A. DNA turnover and the molecular clock. J. Mol. Evol. 1987, 26, 47–58. [Google Scholar] [CrossRef]
- Meštrović, N.; Castagnone-Sereno, P.; Plohl, M. Interplay of selective pressure and stochastic events directs evolution of the MEL172 satellite DNA library in root-knot nematodes. Mol. Biol. Evol. 2006, 23, 2316–2325. [Google Scholar] [CrossRef]
- Mravinac, B.; Plohl, M.; Ugarković, Ð. Preservation and high sequence conservation of satellite DNAs suggest functional constraints. J. Mol. Evol. 2005, 61, 542–550. [Google Scholar] [CrossRef]
- Plohl, M.; Luchetti, A.; Meštrović, N.; Mantovani, B. Satellite DNAs between selfishness and functionality: Structure, genomics and evolution of tandem repeats in centromeric (hetero) chromatin. Gene 2008, 409, 72–82. [Google Scholar] [CrossRef]

| satDNA | RUL | A + T (%) | Abundance | Divergence (%) |
|---|---|---|---|---|
| PmeSat01-508 | 508 | 58.6 | 0.00938 | 8.10 |
| PmeSat02-143 | 143 | 55.9 | 0.00837 | 10.74 |
| PmeSat03-1068 | 1068 | 58.8 | 0.00752 | 11.85 |
| PmeSat04-118 | 118 | 62.7 | 0.00322 | 10.75 |
| PmeSat05-247 | 247 | 61.9 | 0.00192 | 18.62 |
| PmeSat06-1853 | 1853 | 56.1 | 0.00173 | 8.38 |
| PmeSat07-42 | 42 | 52.3 | 0.00112 | 15.06 |
| PmeSat08-177 | 177 | 65.5 | 0.00092 | 14.96 |
| PmeSat09-696 | 696 | 66.6 | 0.00076 | 7.88 |
| PmeSat10-21 | 21 | 76.1 | 0.00069 | 16.97 |
| PmeSat11-1242 | 1242 | 58.5 | 0.00064 | 1.82 |
| PmeSat12-72 | 72 | 68.0 | 0.00053 | 6.72 |
| PmeSat13-92 | 92 | 73.9 | 0.00051 | 4.75 |
| PmeSat14-956 | 956 | 63.2 | 0.00051 | 1.48 |
| PmeSat15-157 | 157 | 50.3 | 0.00051 | 5.05 |
| PmeSat16-42 | 42 | 45.2 | 0.00044 | 12.12 |
| PmeSat17-65 | 65 | 63.0 | 0.00038 | 6.26 |
| PmeSat18-67 | 67 | 65.6 | 0.00036 | 9.11 |
| PmeSat19-30 | 30 | 56.6 | 0.00035 | 10.15 |
| PmeSat20-142 | 142 | 57.0 | 0.00033 | 13.53 |
| PmeSat21-54 | 54 | 51.8 | 0.00031 | 10.04 |
| PmeSat22-28 | 28 | 50.0 | 0.00030 | 10.87 |
| PmeSat23-38 | 38 | 63.1 | 0.00026 | 8.25 |
| PmeSat24-83 | 83 | 54.2 | 0.00023 | 9.93 |
| PmeSat25-6 | 6 | 50.0 | 0.00021 | 23.98 |
| PmeSat26-81 | 81 | 64.1 | 0.00021 | 7.68 |
| PmeSat27-102 | 102 | 59.8 | 0.00019 | 11.35 |
| PmeSat28-398 | 398 | 57.2 | 0.00017 | 6.54 |
| PmeSat29-30 | 30 | 60.0 | 0.00016 | 13.14 |
| PmeSat30-33 | 33 | 63.6 | 0.00006 | 5.67 |
| satDNA | RUL | A + T (%) | Abundance | Divergence (%) |
|---|---|---|---|---|
| CmaSat01-144 | 144 | 56.2 | 0.01389 | 6.62 |
| CmaSat02-543 | 543 | 58.0 | 0.00534 | 13.07 |
| CmaSat03-177 | 177 | 64.9 | 0.00515 | 4.27 |
| CmaSat04-141 | 141 | 57.4 | 0.00464 | 5.22 |
| CmaSat05-247 | 247 | 63.1 | 0.00447 | 26.04 |
| CmaSat06-3168 | 3168 | 57.0 | 0.00399 | 14.69 |
| CmaSat07-525 | 525 | 57.1 | 0.00318 | 1.99 |
| CmaSat08-285 | 177 | 58.5 | 0.00280 | 8.90 |
| CmaSat09-42 | 42 | 54.7 | 0.00166 | 12.92 |
| CmaSat10-43 | 43 | 48.8 | 0.00158 | 19.89 |
| CmaSat11-234 | 1242 | 57.2 | 0.00130 | 5.46 |
| CmaSat12-1846 | 1846 | 66.3 | 0.00103 | 5.91 |
| CmaSat13-2220 | 2220 | 60.0 | 0.00100 | 12.00 |
| CmaSat14-170 | 170 | 56.4 | 0.00095 | 0.03 |
| CmaSat15-21 | 21 | 76.1 | 0.00089 | 17.19 |
| CmaSat16-955 | 955 | 63.2 | 0.00076 | 0.73 |
| CmaSat17-1946 | 1946 | 53.5 | 0.00060 | 7.25 |
| CmaSat18-30 | 30 | 56.6 | 0.00058 | 9.28 |
| CmaSat19-38 | 38 | 65.7 | 0.00055 | 6.71 |
| CmaSat20-72 | 72 | 65.2 | 0.00054 | 3.10 |
| CmaSat21-28 | 28 | 53.5 | 0.00051 | 10.39 |
| CmaSat22-68 | 68 | 63.2 | 0.00049 | 5.11 |
| CmaSat23-898 | 898 | 59.9 | 0.00046 | 12.74 |
| CmaSat24-327 | 327 | 52.5 | 0.00046 | 3.34 |
| CmaSat25-6 | 6 | 50.0 | 0.00046 | 20.53 |
| CmaSat26-50 | 50 | 64.0 | 0.00043 | 4.40 |
| CmaSat27-394 | 394 | 55.5 | 0.00041 | 4.28 |
| CmaSat28-183 | 183 | 53.5 | 0.00035 | 4.23 |
| CmaSat29-34 | 34 | 70.5 | 0.00034 | 17.93 |
| CmaSat30-919 | 919 | 67.2 | 0.00034 | 14.39 |
| CmaSat31-54 | 54 | 53.7 | 0.00034 | 8.78 |
| CmaSat32-237 | 237 | 59.4 | 0.00032 | 16.73 |
| CmaSat33-66 | 66 | 65.1 | 0.00032 | 6.47 |
| CmaSat34-101 | 101 | 60.3 | 0.00031 | 6.93 |
| CmaSat35-30 | 30 | 53.3 | 0.00029 | 11.94 |
| CmaSat36-1250 | 1250 | 59.9 | 0.00024 | 2.49 |
| CmaSat37-51 | 51 | 62.7 | 0.00021 | 2.66 |
| CmaSat38-30 | 30 | 63.3 | 0.00019 | 11.07 |
| CmaSat39-192 | 192 | 61.9 | 0.00015 | 7.47 |
| CmaSat40-932 | 932 | 51.0 | 0.00014 | 2.00 |
| CmaSat41-263 | 263 | 57.7 | 0.00014 | 9.78 |
| CmaSat42-83 | 83 | 60.2 | 0.00013 | 10.92 |
| CmaSat43-62 | 62 | 61.2 | 0.00012 | 7.20 |
| CmaSat44-324 | 324 | 68.2 | 0.00011 | 9.01 |
| CmaSat45-289 | 289 | 62.2 | 0.00010 | 4.82 |
| CmaSat46-30 | 30 | 43.3 | 0.00009 | 6.48 |
| P. mesopotamicus | C. macropomum | Similarity (%) | Classification |
|---|---|---|---|
| PmeSat02-143 | CmaSat04-141 | 78.3 | SF |
| PmeSat02-143 | CmaSat01-144 | 67.5 | SF |
| PmeSat07-42 | CmaSat09-42 | 97.6 | SV |
| PmeSat05-247 | CmaSat05-247 | 72.5 | SF |
| PmeSat05-247 | CmaSat32-237 | 59.7 | SF |
| PmeSat08-177 | CmaSat03-177 | 87 | V |
| PmeSat10-21 | CmaSat15-21 | 100 | SV |
| PmeSat11-1242 | CmaSat36-1250 | 87 | V |
| PmeSat12-72 | CmaSat20-72 | 97.2 | SV |
| PmeSat14-956 | CmaSat16-955 | 91.4 | V |
| PmeSat15-170 | CmaSat14-157 | 73.5 | SF |
| PmeSat17-65 | CmaSat22-68 | 95.5 | SV |
| PmeSat18-67 | CmaSat33-66 | 98.5 | SV |
| PmeSat19-30 | CmaSat18-30 | 100 | SV |
| PmeSat21-54 | CmaSat31-54 | 98.1 | SV |
| PmeSat22-28 | CmaSat21-28 | 96.4 | SV |
| PmeSat23-38 | CmaSat19-38 | 81.5 | V |
| PmeSat25-6 | CmaSat25-6 | 100 | SV |
| PmeSat27-102 | CmaSat34-101 | 97 | SV |
| PmeSat28-398 | CmaSat27-398 | 91.2 | V |
| PmeSat29-30 | CmaSat38-30 | 90 | V |
| Cma | Pme | Mma | Mel | Cgo | Pli | Tau |
|---|---|---|---|---|---|---|
| CmaSat09-42 | PmeSat07-42 | MmaSat07-42 | MelSat08-42 | CgoSat10-42 | PliSat12-42 | TauSat06-42 |
| CmaSat15-21 | PmeSat10-21 | - | - | - | PliSat17-21 | - |
| CmaSat18-30 | PmeSat19-30 | - | MelSat25-30 | - | PliSat19-30 | - |
| CmaSat20-72 | PmeSat12-72 | - | - | - | PliSat15-75 | TauSat19-76 |
| CmaSat21-28 | PmeSat22-28 | MmaSat41-29 | MelSat48-29 | CgoSat26-29 | - | TauSat16-29 |
| CmaSat22-68 | PmeSat17-65 | MmaSat84-65 | MelSat39-65 | - | PliSat36-68 | TauSat12-66 |
| CmaSat31-54 | PmeSat21-54 | MmaSat38-54 | - | - | - | - |
| CmaSat33-66 | PmeSat18-67 | MmaSat27-67 | MelSat12-67 | - | PliSat16-67 | - |
| CmaSat35-30 | - | MmaSat57-30 | MelSat58-31 | - | PliSat30-31 | - |
| CmaSat38-30 | PmeSat29-30 | - | MelSat54-30 | - | PliSat32-30 | - |
| SatDNA | C. macropomum | P. mesopotamicus | Position |
|---|---|---|---|
| PmeSat02-143 | C | C | c/t |
| PmeSat05-247 | C | C | c |
| PmeSat08-177 | C | C | t |
| PmeSat11-1242 | C | C | t |
| PmeSat12-72 | C | C | t |
| PmeSat14-956 | C | C | c |
| PmeSat15-170 | C | C | c |
| PmeSat17-65 | M | M | - |
| PmeSat18-67 | NC | NC | nc |
| PmeSat21-54 | C | C | t |
| PmeSat27-102 | NC | NC | nc |
| PmeSat28-398 | NC | NC | nc |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goes, C.A.G.; dos Santos, N.; Rodrigues, P.H.d.M.; Stornioli, J.H.F.; Silva, A.B.d.; dos Santos, R.Z.; Vidal, J.A.D.; Silva, D.M.Z.d.A.; Artoni, R.F.; Foresti, F.; et al. The Satellite DNA Catalogues of Two Serrasalmidae (Teleostei, Characiformes): Conservation of General satDNA Features over 30 Million Years. Genes 2023, 14, 91. https://doi.org/10.3390/genes14010091
Goes CAG, dos Santos N, Rodrigues PHdM, Stornioli JHF, Silva ABd, dos Santos RZ, Vidal JAD, Silva DMZdA, Artoni RF, Foresti F, et al. The Satellite DNA Catalogues of Two Serrasalmidae (Teleostei, Characiformes): Conservation of General satDNA Features over 30 Million Years. Genes. 2023; 14(1):91. https://doi.org/10.3390/genes14010091
Chicago/Turabian StyleGoes, Caio Augusto Gomes, Natalia dos Santos, Pedro Henrique de Mira Rodrigues, José Henrique Forte Stornioli, Amanda Bueno da Silva, Rodrigo Zeni dos Santos, Jhon Alex Dziechciarz Vidal, Duílio Mazzoni Zerbinato de Andrade Silva, Roberto Ferreira Artoni, Fausto Foresti, and et al. 2023. "The Satellite DNA Catalogues of Two Serrasalmidae (Teleostei, Characiformes): Conservation of General satDNA Features over 30 Million Years" Genes 14, no. 1: 91. https://doi.org/10.3390/genes14010091
APA StyleGoes, C. A. G., dos Santos, N., Rodrigues, P. H. d. M., Stornioli, J. H. F., Silva, A. B. d., dos Santos, R. Z., Vidal, J. A. D., Silva, D. M. Z. d. A., Artoni, R. F., Foresti, F., Hashimoto, D. T., Porto-Foresti, F., & Utsunomia, R. (2023). The Satellite DNA Catalogues of Two Serrasalmidae (Teleostei, Characiformes): Conservation of General satDNA Features over 30 Million Years. Genes, 14(1), 91. https://doi.org/10.3390/genes14010091







