Abstract
GRAS genes exist widely and play vital roles in various physiological processes in plants. In this study, to identify Theobroma cacao (T. cacao) GRAS genes involved in environmental stress and phytohormones, we conducted a genome-wide analysis of the GRAS gene family in T. cacao. A total of 46 GRAS genes of T. cacao were identified. Chromosomal distribution analysis showed that all the TcGRAS genes were evenly distributed on ten chromosomes. Phylogenetic relationships revealed that GRAS proteins could be divided into twelve subfamilies (HAM: 6, LISCL: 10, LAS: 1, SCL4/7: 1, SCR: 4, DLT: 1, SCL3: 3, DELLA: 4, SHR: 5, PAT1: 6, UN1: 1, UN2: 4). Of the T. cacao GRAS genes, all contained the GRAS domain or GRAS superfamily domain. Subcellular localization analysis predicted that TcGRAS proteins were located in the nucleus, chloroplast, and endomembrane system. Gene duplication analysis showed that there were two pairs of tandem repeats and six pairs of fragment duplications, which may account for the rapid expansion in T. cacao. In addition, we also predicted the physicochemical properties and cis-acting elements. The analysis of GO annotation predicted that the TcGRAS genes were involved in many biological processes. This study highlights the evolution, diversity, and characterization of the GRAS genes in T. cacao and provides the first comprehensive analysis of this gene family in the cacao genome.
1. Introduction
Abiotic stresses including high temperature, drought, cold, and salt have important effects on plant development and growth. Some transcription factors regulate the transcript levels of their target genes under stress by binding to specific DNA sequences in their target promoters [1,2,3]. Therefore, the regulatory networks of various biological processes can be understood by studying plant transcription factors.
The GRAS transcription factor gene family is named after three originally identified members: gibberellic acid insensitive (GAI), GA1 repressor of GA1 (RGA), and scarecrow (SCR) [4,5,6]. Most GRAS proteins contain only one domain, and the C-terminal region of this domain is highly conserved, which is often referred to as the GRAS domain. It contains five units: SAW, leucine heptapeptide repeat I (LHRI), VHIID, PFYRE, and leucine heptapeptide repeat II (LHRII) [7]. However, some GRAS proteins still contain one domain and another functional domain or two domains [7]. The high variability of the N-terminal region of GRAS proteins determines the regulatory proteins’ specificity [8]. The GRAS transcription factor family has been found in many plants, but the family classification varies slightly in different species [9]. For example, in soybean (Glycine max), 117 GRAS genes are divided into 11 subfamilies: AtSCL4/7, Os19, Os4, HAM, DELLA, DLT, AtPAT1, LISCL, AtSCR, AtSCL3, and AtSHR [10]. In tomato (Solanum lycopersicum), 53 GRAS genes have been identified and classified into 13 subfamilies: HAM, LAS, SCL4/7, SCR, SCL9, SCL28, DELLA, SHR, PAT1, Os4, Os19, GRAS37, and Pt20 [11]. In Brachypodium distachyon (Brachypodium distachyon (Linnaeus) P. Beauvois), 63 GRAS genes are classified into 10 subfamilies: HAM, PAT1, SHR, DELLA, SCL3, SCL4/7, LAS, SCR, DLT, and LISCL [12]. In potato (Solanum tuberosum L.), 52 GRAS genes have been identified and classified into 8 subfamilies: DELLA, LAS, HAM, PATI, SCR, LISCL, SHR, and SCL3 [13]. However, in Populus trichocarpa (P. trichocarpa), 93 GRAS genes are divided into 13 subfamilies: Os19, HAM, Os4, Pt20, DLT, AtSCl3, AtSHR, AtPAT1, AtSCR, AtSCL4/7, AtLAS, DELLA, and LISCL [9]. These studies suggest that members of the GRAS gene family differ in different plants. Their classification reflects their evolutionary history.
The diversity of the GRAS gene family is consistent with its high diversity of functions [14]. To date, functional identification of GRAS transcriptional regulators has occurred mainly in Arabidopsis thaliana and rice. This has shown that they have important roles in the regulation of plant development and growth, including multiple growth regulatory signals and environmental signals such as abiotic/biotic stresses, light, and phytohormones [15]. Sun et al. [16] reviewed the major biological functions of the ten subfamilies of GRAS transcriptional regulators, and the results are reported in Table 1.

Table 1.
Recent advances in biological functions of the GRAS transcription factor gene family.
Although the GRAS gene family has been studied for many years, the mechanisms and evolutionary dynamics of this gene family in woody plants are still not fully understood. Differences in loss and retention of duplicated gene family members between woody and herbaceous species may help in identifying genes with specialized roles in the adaptive evolution of different lineages. The cacao is called “soft gold” because of its high value. The flowers of cacao trees have ornamental value, and cacao is the main ingredient in chocolate and cacao powder [27]. In addition, cacao beans have important uses in the pharmaceutical and cosmetic industries. Cacao is receiving increasing attention for its potential health benefits because it is rich in polyphenols, particularly flavonoids [28]. However, cacao production is hampered for a number of reasons. Therefore, it is of great value to study the cacao tree. The phylogenetic relationship, conserved domain, and collinearity analysis of this family can provide new ideas for further functional analysis. In 2010, Corti et al. [29] carried out the sequencing and assembly study of the T. cacao genome, and since then, researchers have successively identified and analyzed its important gene families, such as the NAC gene family [30], WRKY gene family [31], and the GPX transcription factor family [32]. In 2013, researchers completed an analysis of the metabolome and transcriptome of the cacao tree [33]. Our knowledge about the expansion and diversification of this gene family in plants is presently limited to the herbaceous species Arabidopsis. To date, the GRAS gene family has not been identified and classified in T. cacao.
In this study, we identified 46 GRAS gene family members and conducted a comprehensive genome-wide analysis of the GRAS gene family of the cacao tree, including gene structure, domain analysis, intron/exon, chromosome location, subcellular localization, and cis-acting elements of GRAS genes. In addition, we analyzed the phylogenetic relationship of GRAS proteins between T. cacao and A. thaliana. Furthermore, we performed the gene duplication pattern of T. cacao GRAS proteins, and we analyzed a syntenic analysis of GRAS proteins among T. cacao, A. thaliana, P. trichocarpa, and Sesamum indicum (S. indicum). The results of this study lay the foundation for further studies of the biological function of genes in T. cacao and provide a reference for subsequent molecular mechanisms.
2. Materials and Methods
2.1. Identification of GRAS Gene Family in T. cacao
The genome file, protein file, coding sequences (CDS), and annotation files of T. cacao were obtained from Ensembl Plants (http://plants.ensembl.org/index.html, accessed on 1 June 2022). The Hidden Markov Model (HMM) profile of the GRAS protein domain was downloaded from the Pfam protein family database (release 35.0; http://pfam.xfam.org/, accessed on 1 June 2022) under the accession number ‘PF03514’ [34,35].
The HMM model of HMMER (version 3.1b2) was used to screen GRAS protein candidate members of T. cacao twice to determine the final target members. Firstly, the downloaded HMM profile was employed using the HMMER v3.3.2 program to search for proteins containing target GRAS domains as the initial filtering results, and ClustalW (version 2.1) was used to perform multiple sequence alignment for the initial target proteins [36]. Secondly, to expand the filtering scope, we constructed a new HMM model with e-value < 1 × 10−20. The new model was used to filter second target proteins using HMMER (version 3.3.2), with e-value < 0.05. The two results were combined and used as the final candidate proteins. Finally, the NCBI Conserved Domain Search (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi, accessed on 6 June 2022), Pfam Batch Sequence Search (http://pfam.xfam.org/search#tabview=tab1, accessed on 6 June 2022), and the SMART program (http://smart.embl.de/smart/batch.pl, accessed on 6 June 2022) [37] were used to verify the existence of the GRAS domain in each candidate protein sequence. After combining all results, 46 GRAS genes were obtained from the T. cacao genome.
2.2. Physicochemical Properties and Subcellular Localization Analyses of TcGRAS Genes
The online software ProtParam (https://web.expasy.org/protparam, accessed on 7 June 2022) and Compute pI/Mw (https://web.expasy.org/compute_pi, accessed on 7 June 2022) in the Expasy web server were used to analyze the physicochemical properties of the 46 GRAS proteins identified, including the theoretical isoelectric point (pI), molecular weight (MW), instability index, and aliphatic index [38]. Amino acid (aa) numbers and open reading frame (ORF) lengths were obtained on the ORFfinder website. (https://www.ncbi.nlm.nih.gov/orffinder, accessed on 7 June 2022). The subcellular localization (SL) of TcGRAS proteins was predicted by the BUSCA online program (https://busca.biocomp.unibo.it, accessed on 7 June 2022).
2.3. Phylogenetic Analysis and Classification of TcGRAS Genes
To provide family classification of GRAS genes and understand their phylogenetic relationships, a rooted neighbor-joining (NJ) phylogenetic tree between T. cacao (TcGRAS) and Arabidopsis GRAS proteins was built using the MEGA 11 software (version 11.0.11) [39,40]. The TcGRAS genes were classified according to their phylogenetic relationship with A.thaliana GRAS members. We obtained Arabidopsis GRAS protein sequences from TAIR (https://www.Arabidopsis.org, accessed on 10 June 2022) [9,41]. Both families of protein sequences were aligned by Muscle [42] in MEGA 11 software (version 11.0.11) under the default parameters. The maximum likelihood (ML) method was used with the following parameters: 1000 iterations for the bootstrap method, the Poisson model, and use all sites. In addition, an individual phylogenetic tree of TcGRAS genes was constructed in the same way and visualized using online software iTOL (http://itol.embl.de/, accessed on 10 June 2022) [43].
2.4. Gene Structure and Conserved Motif Analyses of TcGRAS Genes
The conserved motifs of the TcGRAS proteins were predicted by using the online program MEME (https://meme-suite.org/meme/tools/meme, accessed on 22 June 2022) with the following settings: maximum number of motifs 15, minimum motif width 6, maximum motif width 50, and any number of repetitions [44]. The domain analyses of TcGRAS proteins were performed under the Gene Structure Display Server 2.0 program. The gene structure view function of TBtools (version 0.665) was used to obtain conserved motifs and gene structures.
2.5. Chromosomal Mapping and Cis-Acting Regulatory Analyses of TcGRAS Genes
The online program MG2C (http://mg2c.iask.in/mg2c_v2.1, accessed on 15 June 2022) was used to predict the chromosomal position of TcGRAS genes. All the identified genes were mapped to 10 chromosomes according to the location information of the chromosome by TBtools. The upstream 2000 bp sequences of TcGRAS genes’ CDS were extracted by TBtools software (version 1.098696), and then submitted to the online software PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html, accessed on 15 June 2022) [45] to predict cis-acting elements, including light-responsive elements, abscisic acid-responsive elements, MeJA-responsive elements, low-temperature-responsive elements, defense- and stress-responsive elements, gibberellin-responsive elements, and auxin-responsive elements, after filtering and screening [45]. The Simple BioSequence Gene Viewer function of TBtools software (version 1.098696) was used to visualize the cis-acting elements.
2.6. Gene Duplication and Synteny Analyses of TcGRAS Genes
The ‘MCScanX’ function of the TBtools software with default parameters was used to predict gene duplications of TcGRAS genes. MCScanX Diamond output was used to calculate the replication events of the T. cacao genome. The Duplicate_gene_classifier program in MCScanX (https://github.com/wyp1125/MCScanX, accessed on 22 June 2022) was used to analyze the duplication type of each TcGRAS gene. KaKs_calculator software (version 2.0) [46] was used to calculate the Ka/Ks ratio of tandem repeat gene pairs in the TcGRAS gene, with the following parameters: method of calculation: YN, and genetic code Table 1 (Standard code). The Advanced Circos function of TBtools software (version 1.098696) was used to visualize WGD or segment duplications. The synteny of TcGRAS genes with the GRAS genes of A. thaliana, P. trichocarpa, and S. indicum was visualized by the One-Step MCScanX function of TBtools software. The Dual Systeny Plot for the MCScanX function of TBtools software (version 1.098696) was used to visualize the synteny.
2.7. GO Annotation Analyses of T. cacao TcGRAS Genes
The DAVID online program was used to annotate TcGRAS genes. The official gene sample lists of TcGRAS genes were uploaded to the program. The analysis included three parts: molecular function, cell components, and biological processes. The R programming language (version 4.1.3) was used to visualize the GO annotation analysis [47].
3. Results
3.1. Identification of GRAS Members in T. cacao
A total of 70 GRAS protein candidates were obtained from the initial filtering. After the second filtering, 53 candidate proteins were obtained. Finally, 46 GRAS genes were identified by redefining conserved domains and deleting repeats (Supplementary File S1). The identified genes were named from TcGRAS1 to TcGRAS46 according to their chromosomal position. The number of amino acids (aa), average molecular weight (MW), theoretical pI, instability index, and aliphatic index of identified TcGRAS genes were statistically analyzed (Table 2). The number of amino acids of the TcGRAS genes ranged from 347 (TcGRAS21) to 1659 (TcGRAS22) aa, and the molecular weight ranged from 39,709.90 to 191,183.51 Da. The results showed that 44 GRAS proteins were acidic with pI values less than 6.5. Two (TcGRAS2 and TcGRAS19) were neutral, with pI between 6.5 and 7.5. The results of the instability index analysis showed that most TcGRAS proteins were unstable, except for TcGRAS8, TcGRAS12, TcGRAS24, and TcGRAS33. Prediction of the subcellular localization of TcGRAS proteins by the online software BUSCA tool revealed that 41 TcGRAS proteins were mainly located in the nucleus, 4 in the chloroplasts, and only 1 in the endomembrane system.

Table 2.
Physicochemical properties and subcellular localization analyses of the GRAS gene family in T. cacao.
3.2. Phylogenetic Analysis of TcGRAS and AtGRAS
To explore the evolutionary relationship of GRAS proteins between T. cacao and A. thaliana, we performed a multiple sequence alignment of 46 TcGRAS proteins and 34 AtGRAS proteins, and then constructed an unrooted phylogenetic tree using the MEGA 11 software (Figure 1). According to the homology of GRAS proteins in A. thaliana, 46 TcGRAS proteins were divided into 10 clades, which were HAM, LISCL, LAS, SCL4/7, SCR, DLT, SCL3, DELLA, SHR, and PAT1. It is notable that 5 of the 46 TcGRAS proteins were not classified as any of these subfamilies; therefore, we grouped TcGRAS22 as UN1 and TcGRAS11, TcGRAS13, TcGRAS17, and TcGRAS33 as UN2. The largest clade was subgroup LISCL, which contained ten TcGRAS members (TcGRAS8, TcGRAS24, TcGRAS25, TcGRAS27, TcGRAS39, TcGRAS40, TcGRAS41, TcGRAS42, TcGRAS43, and TcGRAS46), whereas subgroups UN1, DLT, SCL4/7, and LAS only had one member. Subgroups UN1 and UN2 only contained T. cacao members, meaning that these genes may have been specialized during the evolutionary process.
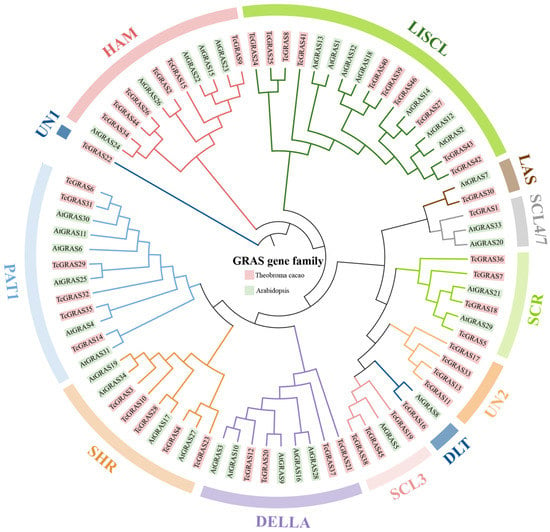
Figure 1.
Phylogenetic tree of GRAS genes in T. cacao and A. thaliana. The GRAS genes in T. cacao and A. thaliana are shown in red and green, respectively. The tree branched the GRAS proteins into different subgroups, illustrated by different colored clusters within the clade. TcGRAS are divided into twelve subgroups according to the subgrouping of A. thaliana. The phylogenetic tree was constructed using the maximum likelihood (ML) method with 1000 bootstrap replications of the MEGA 11 software.
3.3. Gene Structure, Conserved Motifs, and Domain Analyses of TcGRAS Genes
To understand the structural diversity and similarity of GRAS gene family members in the cacao tree, we used the Gene Structure View function of the TBtools software to construct a triad map of the evolutionary tree, gene structure, and motif of GRAS gene family members, as shown in Figure 2. We first performed an individual phylogenetic tree using an NJ method consistent with the phylogenetic analysis between TcGRAS and AtGRAS (Figure 2A).
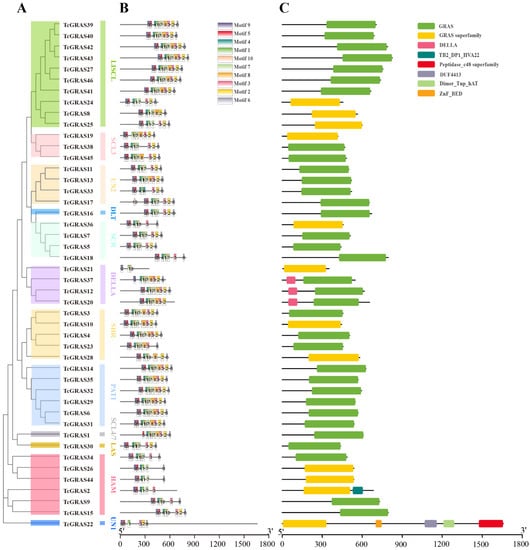
Figure 2.
Phylogenetic relationship, conserved motifs, and gene structure of TcGRAS genes. (A) An unrooted phylogenetic tree of TcGRAS proteins was constructed using MEGA 11 software under the ML method with 1000 bootstrap replicates. (B) Conserved TcGRAS proteins’ motifs performed by MEME. The colored boxes refer to motifs. The black lines refer to non-conserved sequences. The scale bar is 300 amino acids. (C) The domain analyses of TcGRAS proteins were performed under the Gene Structure Display Server 2.0 program. The domains are displayed in different colored boxes.
To further understand the characteristics of the GRAS gene families in T. cacao and the conserved motifs shared among different subfamilies, we used the Multiple Expectation Maximization for Motif Elicitation program to find the conserved motifs. A total of 10 conserved motifs were predicted and named Motif 1–10 (Figure 2B and Supplementary File S2). Early sequence analysis indicated that the GRAS proteins typically share a variable N terminus and a highly conserved C terminus. In this study, we found that the C-terminal regions contained a highly conserved domain (Motif 6). Three proteins did not contain this conserved motif, including TcGRAS2, TcGRAS20, and TcGRAS21, and we hypothesized that the C-terminal region of these GRAS proteins was truncated, lacking part of the GRAS domain.
We used the Gene Structure Display Server 2.0 program to construct a domain analysis of TcGRAS proteins (Figure 2C). We found a total of eight types of conserved domains. All the GRAS genes contain the GRAS domain or GRAS superfamily domain. In addition, the domains of GRAS members in DELLA also have the DELLA superfamily. The TcGRAS2 gene has a GRAS superfamily and TB2_DP1_HVA22 superfamily domain. The TcGRAS22 gene has a GRAS superfamily, ZnF_BED, DUF4413, Dimer_Tnp_hAT, and Peptidase_c48 superfamily domain.
3.4. Chromosomal Mapping and Cis-Acting Regulatory Analyses of TcGRAS Genes
The location of TcGRAS genes was obtained from genome annotation files. A total of 46 TcGRAS genes were randomly distributed on 10 chromosomes and were named from TcGRAS1 to TcGRAS46 according to their positions on the chromosomes (Figure 3A). Chr01 and Chr04 had the largest number (nine, 19.57%) of TcGRAS genes, followed by Chr09 with eight members (17.39%). In contrast, Chr05, Chr06, and Chr10 contained only two TcGRAS genes each (4.35%). Chr04 contained seven subgroups of TcGRAS genes, as shown in Figure 3B, while Chr05, Chr06, and Chr10 contained only two subgroups each. Subgroup DLT was only observed on Chr03, subgroup SCL4/7 was only observed on Chr01, and subgroup LAS was only observed on Chr07.
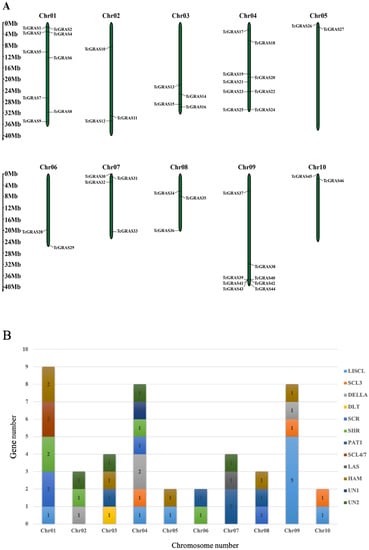
Figure 3.
(A) Physical distribution of TcGRAS genes among 10 chromosomes. (B) Number of TcGRAS subfamilies on each chromosome.
Cis-acting elements regulate transcription initiation and transcription activity by binding to transcription factors. To explore the promoter function of TcGRAS genes, we extracted 2000 bp sequences upstream of the transcription start site. Then, we submitted these to the online program PlantCARE. Seven types of important cis-acting elements were obtained after sorting and screening, including light-responsive element, abscisic acid-responsive element, MeJA-responsive element, defense and stress-responsive element, low-temperature-responsive element, auxin-responsive element and gibberellin-responsive element. The light-responsive element was found in all promoter regions of TcGRAS genes. In addition, more than half of the 46 GRAS genes had the abscisic acid-responsive element, MeJA-responsive element, and gibberellin-responsive element. Compared with the MADS-Box transcription factor family in cacao tree, the GRAS gene family contains significantly more light-responsive elements, abscisic acid-responsive elements, and MeJA-responsive elements. The distribution of these cis-acting elements is shown in Supplementary File S3.
3.5. Gene Duplication and Syntenic Analysis of TcGRAS Genes
Genome-wide analysis of cacao tree gene replication by MCScanX software revealed 2148 tandem duplicated genes in the cacao tree genome, while only 2 pairs of tandem duplicated genes were present among 46 TcGRAS genes (Figure 4). The analysis showed that one pair of tandem duplication genes (TcGRAS24 and TcGRAS25) was located on Chr04, and another pair (TcGRAS42 and TcGRAS43) on Chr09. In addition, the substitution ratio of non-synonymous (Ka) to synonymous (Ks) mutations (Ka/Ks) of two pairs were calculated (Table 3). The Ka/Ks values of both pairs were more than 1, indicating that these genes were positively selected over the course of evolution, and the novel protein functions may be beneficial to the survival and reproduction of T. cacao.
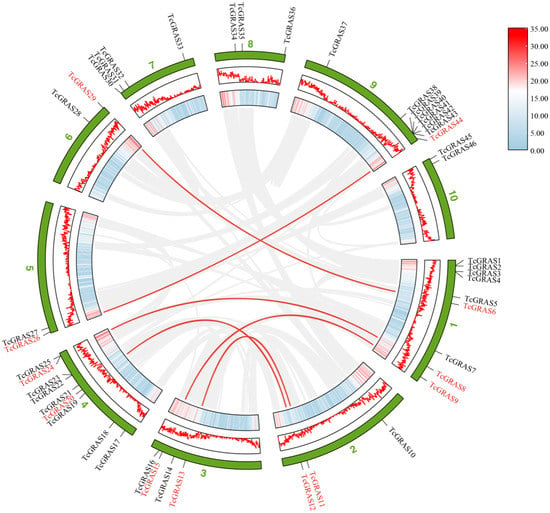
Figure 4.
Schematic diagram of the duplication patterns of the TcGRAS genes. The red lines show segmental duplications of TcGRAS gene pairs. The gray lines show segmental duplications of all gene pairs in the T. cacao genome. The first ring from outside indicates the chromosomal localization of 46 putative GRAS genes in T. cacao. The second and third rings from outside represent the density of genes on the chromosomes. The blue-to-red scale bar on the right indicates the number of SNPs within 1 Mb window size.

Table 3.
Tandem duplication in TcGRAS genes and corresponding Ka, Ks, and Ka/Ks values.
The MCScanX showed that there were 2767 segmental duplications in the genome of T. cacao, and only 6 pairs of fragmental duplicated genes were predicted out of 46 identified TcGRAS genes. The Advanced Circos function of the TBtools software was used to visualize the segmental duplication of GRAS genes on 10 chromosomes, as shown in Figure 4. Chr01 contained three duplicated genes, and Chr02, Chr03, and Chr04 contained two duplicated genes, while Chr05, Chr06, and Chr09 each contained only one duplicated gene. However, Chr07, Chr08, and Chr10 did not contain any segmental duplicated genes.
The syntenic analyses of TcGRAS genes with the GRAS genes of A. thaliana, P. trichocarpa, and S. indicum were separately analyzed to find homologous gene pairs (Figure 5). A total of 36 GRAS genes of T. cacao had a syntenic relationship with the GRAS genes of A. thaliana (16), P. trichocarpa (32), and S. indicum (30). Some genes had multiple syntenic relationships with other closely related species. Therefore, a total number of 25 (Supplementary File S4), 77 (Supplementary File S5), and 48 (Supplementary File S6) GRAS genes of A. thaliana, P. trichocarpa, and S. indicum, respectively, had synteny with 36 GRAS genes. Furthermore, it was found that 13 GRAS genes existed in these 4 plants at the same time (Figure 6). Two homologous GRAS genes existed in T. cacao and S. indicum rather than in P. trichocarpa and A. thaliana. Similarly, T. cacao, P. trichocarpa, and S. indicum had 13 homologous TcGRAS genes that did not exist in A. thaliana, T. cacao, P. trichocarpa, and A. thaliana had 1 homologous TcGRAS gene that did not exist in S. indicum, and T. cacao, S. indicum, and A. thaliana had 2 homologous GRAS genes that did not exist in P. trichocarpa. Five homologous GRAS genes existed in T. cacao and P. trichocarpa but did not exist in A. thaliana and S. indicum.
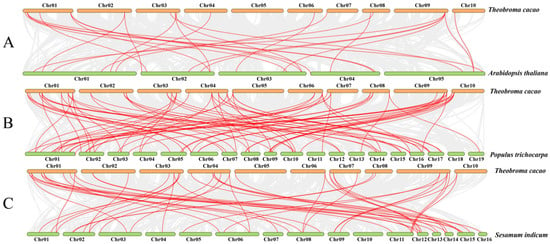
Figure 5.
Visualization of the syntenic analysis. Synteny of the GRAS genes in T. cacao with the GRAS genes of A. thaliana (A), T. cacao (B), and S. indicum (C) was visualized by MCScanX analysis of TBtools software. Gray lines between the genomes show all synteny blocks, and red lines between the genomes indicate the synteny between the genes.
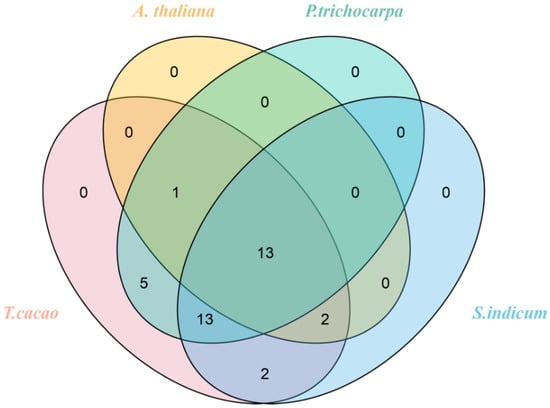
Figure 6.
Venn diagram of the identical and different GRAS genes among T. cacao, A. thaliana, P. trichocarpa, and S. indicum.
3.6. GO Annotation of T.cacao TcGRAS Proteins
To understand TcGRAS protein function in different biological processes, we performed a GO annotation analysis of the TcGRAS genes (Figure 7), and the GO numbers are shown in Supplementary File S7. The analysis of the cellular composition showed that most of the TcGRAS proteins were mainly concentrated in the nucleus. The analysis of biological processes showed that the TcGRAS genes were involved in many biological processes. The large portions of GRAS proteins were involved in transcriptional regulation. Otherwise, some TcGRAS proteins were involved in the negative regulation of biological processes, for example, negative regulation of seed germination and the gibberellic acid-mediated signaling pathway. In addition, some TcGRAS genes also respond to abiotic stresses and regulate plant organ development. The analysis of the molecular functions of TcGRAS genes revealed that they had functions in transcription factor activity and sequence-specific DNA binding.
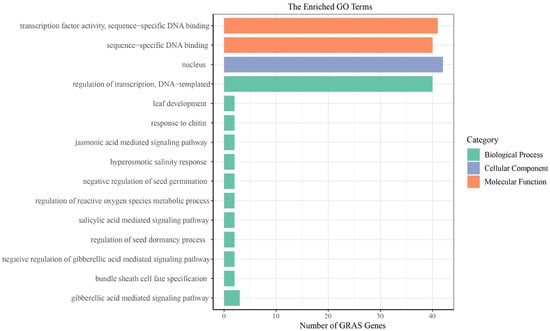
Figure 7.
GO annotation of TcGRAS proteins. Green represents biological processes, purple represents cellular components, and orange represents molecular functions.
4. Discussion
In this study, we provided the first comprehensive analysis of the GRAS gene family in T. cacao. Based on the latest genome sequences and annotation files, we identified 46 GRAS genes distributed across 10 chromosomes in the T. cacao genome. These 46 TcGRAS genes were classified into 12 subgroups (HAM, LISCL, LAS, SCL4/7, SCR, DLT, SCL3, DELLA, SHR, PAT1, UN1, and UN2) according to their phylogenetic relationship with A. thaliana. We found that the GRAS gene family members were unevenly distributed among subgroups; for instance, the subgroups of UN1 and UN2 only contained T. cacao members, and the member number of subgroups of SCR and SCL3 in T. cacao was more than that in A. thaliana. During the evolution of gene families, the gene structure changes in response to environmental changes to acquire new functions. The structural analysis of TcGRAS genes according to phylogenetic relationships showed that different subgroups had different gene structures and conserved motifs, while the same subgroup had similar motifs and gene structures, which meant that members of the same subgroup had similar functions. Since T. cacao and A. thaliana were exposed to different environments during their evolutionary processes, the number of GRAS genes in their subgroups became different as GRAS genes differentiated.
By analyzing the intron/exon structure of the TcGRAS genes, we found that majority of these genes were free of introns, which was similar to the observed lack of introns in Arabidopsis and rice GRAS genes [8]. A previous study showed that ancestors of each eukaryote had intron-rich genes and that extensive loss and insertion of introns from most genes may have occurred due to selective pressure, with gene duplication accelerating this process [48,49]. Nevertheless, some GRAS genes have evolved different intron/exon structures, indicating that they likely evolved new specialized functions to adapt to their environment.
Tandem and segmental duplications are thought to be the main mechanisms contributing to the expansions of gene families in plants [50]. Both tandemly and segmentally duplicated genes that have been retained in plant genomes play important roles in adaptive responses to environmental stimuli [51,52]. The collinearity analysis in our study showed that there were two pairs of tandem duplication and six pairs of segmental duplication events in the T. cacao GRAS gene family, and this might play an important role in the GRAS family expansion in T. cacao.
The cis-acting elements play a vital role in regulating gene expression during plant growth and development [53]. The promoter analysis showed that the light-responsive element was found in all promoter regions of TcGRAS genes. In addition, more than half of the 46 GRAS genes had the abscisic acid-responsive element, MeJA-responsive element, and gibberellin-responsive element, which made it possible to study the function of these genes in the future.
To further analyze the function of GRAS transcription factors in T. cacao, we studied the end of the genotype affected by functional diversity after GO enrichment analysis, and the results showed that the majority of cacao GRAS proteins play an important role in many different biological processes, including abiotic stresses and plant organ development.
5. Conclusions
In this study, we identified and systematically analyzed the GRAS gene family in T. cacao. Based on the genomic data of the cacao tree, we finally identified 46 GRAS genes using double HMM profiles. These 46 GRAS genes were distributed on 10 chromosomes and phylogenetically divided into 12 subfamilies, with highly similar gene structures and conserved motifs within the same subfamily. Cis-acting element analysis indicated that GRAS genes may be involved in various abiotic stress responses. In addition, we found that tandem and segmental duplications contribute to the expansions of the GRAS gene family. A further syntenic analysis showed that the functions of TcGRAS genes might be speculated from the function of GRAS genes in other plants. Through GO analysis, we found that most of the TcGRAS genes were involved in transcriptional regulation. In summary, the results provide information for further research of the TcGRAS genes’ function and lay the foundation for further investigation.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes14010057/s1, Supplementary File S1: GRAS gene sequences identified in T. cacao in this study; Supplementary File S2: The ten conserved motifs in T. cacao identified in this study; Supplementary File S3: The predicted cis-regulatory elements in promoters of TcGRAS genes; Supplementary File S4: One-to-one orthologous relationship of GRAS genes of T. cacao and A. thaliana; Supplementary File S5: One-to-one orthologous relationship of GRAS genes of T. cacao and P. trichocarpa; Supplementary File S6: One-to-one orthologous relationship of GRAS genes of T. cacao and S. indicum; Supplementary File S7: The GO annotation of T. cacao TcGRAS proteins.
Author Contributions
Conceptualization, S.H.; methodology, S.H.; software, S.H. and Q.Z.; formal analysis, S.H.; investigation, S.H.; resources, S.H. and J.C.; writing—original draft preparation, S.H.; writing—review and editing, Y.G.; visualization, S.H. and J.M.; supervision, S.H., C.W. and J.D.; project administration, Y.G.; funding acquisition, Y.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (31370669).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article or Supplementary Files.
Acknowledgments
We thank Qianqian Zhang (Chinese Institute for Brain Research, College of Biological Sciences and Technology, Beijing Forestry University) and Ang Dong (Center for Computational Biology, College of Biological Sciences and Technology, Beijing Forestry University) for the technical support. This research was supported by the National Natural Science Foundation of China (31370669).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Riano-Pachon, D.M.; Ruzicic, S.; Dreyer, I.; Mueller-Roeber, B. PlnTFDB: An integrative plant transcription factor database. BMC Bioinform. 2007, 8, 42. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Sun, Y.; Xue, J.; Jia, X.; Li, R. Genome-wide characterization and expression analysis of GRAS gene family in pepper (Capsicum annuum L.). PeerJ 2018, 6, e4796. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Xuan, A.; Bu, C.; Ci, D.; Tian, M.; Zhang, D. Osmotic stress-responsive promoter upstream transcripts (PROMPTs) act as carriers of MYB transcription factors to induce the expression of target genes in Populus simonii. Plant Biotechnol. J. 2019, 17, 164–177. [Google Scholar] [CrossRef] [PubMed]
- Laurenzio, L.D.; Wysocka-Diller, J.; Malamy, J.E.; Pysh, L.; Helariutta, Y.; Freshour, K.; Hahn, M.G.; Feldmann, K.A.; Benfey, P.N. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 1996, 86, 423–433. [Google Scholar] [CrossRef]
- Peng, J.; Carol, P.; Richards, D.E.; King, K.E.; Cowling, R.J.; Murphy, G.P.; Harberd, P.H. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 1997, 11, 3194–3205. [Google Scholar] [CrossRef]
- Silverstone, A.L.; Ciampaglio, C.N.; Sun, T. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 1998, 10, 155–169. [Google Scholar] [CrossRef]
- Bolle, C. The role of GRAS proteins in plant signal transduction and development. Planta 2004, 218, 683–692. [Google Scholar] [CrossRef]
- Tian, C.; Wan, P.; Sun, S.; Li, J.; Chen, M. Genome-wide analysis of the GRAS gene family in Rice and Arabidopsis. Plant Mol. Biol. 2004, 54, 519–532. [Google Scholar] [CrossRef]
- Liu, X.; Widmer, A. Genome-wide comparative analysis of the GRAS gene family in Populus, Arabidopsis and Rice. Plant Mol. Biol. Rep. 2014, 32, 1129–1145. [Google Scholar] [CrossRef]
- Wang, T.; Yu, T.; Fu, J.; Su, H.; Chen, J.; Zhou, Y.; Chen, M.; Guo, J.; Ma, Y.; Wei, W.; et al. Genome-wide analysis of the GRAS gene family and functional identification of GmGRAS37 in drought and salt tolerance. Front. Plant Sci. 2020, 11, 604690. [Google Scholar] [CrossRef]
- Huang, W.; Xian, Z.; Kang, X.; Tang, N.; Li, Z. Genome-wide identification, phylogeny and expression analysis of GRAS gene family in tomato. BMC Plant Biol. 2015, 15, 209. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.J.; Song, N.; Peng, W.Y.; Yang, Y.; Qiu, T.; Huang, C.T.; Dai, L.Y.; Wang, B. Genome identification and expression analysis of GRAS family related to development, hormone and pathogen stress in Brachypodium distachyon. Front. Sustain. Food Syst. 2021, 5, 675177. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, N.; Zhu, X.; Yang, J.; Li, S.; Che, Y.; Liu, W.; Si, H. Identification and expression analysis of StGRAS gene family in potato (Solanum tuberosum L.). Comput. Biol. Chem. 2019, 80, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Duan, X.; Jiang, H.; Sun, Y.; Tang, Y.; Yuan, Z.; Guo, J.; Liang, W.; Chen, L.; Yin, J.; et al. Genome- wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006, 141, 1167–1184. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, S.; Oldroyd, G.E. GRAS-domain transcription factors that regulate plant development. Plant Signal. Behav. 2009, 4, 698–700. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Jones, W.T.; Rikkerink, E.H. GRAS proteins: The versatile roles of intrinsically disordered proteins in plant signalling. Biochem. J. 2012, 442, 1–12. [Google Scholar] [CrossRef]
- Stuurman, J.; Jäggi, F.; Kuhlemeier, C. Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells. Genes Dev. 2002, 16, 2213–2218. [Google Scholar] [CrossRef]
- Morohashi, K.; Minami, M.; Takase, H.; Hotta, Y.; Hiratsuka, K. Isolation and characterization of a novel GRAS gene that regulates meiosis-associated gene expression. J. Biol. Chem. 2003, 278, 20865–20873. [Google Scholar] [CrossRef]
- Schumacher, K.; Schmitt, T.; Rossberg, M.; Schmitz, G.; Theres, K. The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc. Natl. Acad. Sci. USA 1999, 96, 290–295. [Google Scholar] [CrossRef]
- Ma, H.; Liang, D.; Shuai, P.; Xia, X.; Yin, W. The salt- and drought-inducible poplar GRAS protein SCL7 confers salt and drought tolerance in Arabidopsis thaliana. J. Exp. Bot. 2010, 61, 4011–4019. [Google Scholar] [CrossRef] [PubMed]
- Helariutta, Y.; Fukaki, H.; Wysocka-Diller, J.; Nakajima, K.; Jung, J.; Sena, G.; Hauser, M.T.; Benfey, P.N. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 2000, 101, 555–567. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Andersson-Gunneras, S.; Gaboreanu, I.; Hertzberg, M.; Tucker, M.R.; Zheng, B.; Lesniewska, J.; Mellerowicz, E.J.; Laux, T.; Sandberg, G.; et al. Reduced expression of the SHORT-ROOT gene increases the rates of growth and development in hybrid poplar and Arabidopsis. PLoS ONE 2011, 6, e28878. [Google Scholar] [CrossRef] [PubMed]
- Tong, H.; Jin, Y.; Liu, W.; Li, F.; Fang, J.; Yin, Y.; Qian, Q.; Zhu, L.; Chu, C. DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 2009, 58, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.O.; Chang, K.S.; Kim, I.A.; Lee, M.H.; Lee, S.A.; Song, S.K.; Lee, M.M.; Lim, J. Funneling of gibberellin signaling by the GRAS transcription regulator SCARECROW-LIKE 3 in the Arabidopsis root. Proc. Natl. Acad. Sci. USA 2011, 108, 2166–2171. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Lee, L.; Xia, K.; Yan, Y.; Yu, H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 2010, 19, 884–894. [Google Scholar]
- Bolle, C.; Koncz, C.; Chua, N.H. PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev. 2000, 14, 1269–1278. [Google Scholar] [CrossRef]
- Mustiga, G.M.; Gezan, S.A.; Phillips-Mora, W.; Arciniegas-Leal, A.; Mata-Quirós, A.; Motamayor, J.C. Phenotypic description of Theobroma cacao L. for yield and vigor traits from 34 hybrid families in Costa Rica based on the genetic basis of the parental population. Front. Plant Sci. 2018, 9, 808. [Google Scholar] [CrossRef]
- Corti, R.; Flammer, A.J.; Hollenberg, N.K.; Lüscher, T.F. Cocoa and cardiovascular health. Circulation 2009, 119, 1433–1441. [Google Scholar] [CrossRef]
- Argout, X.; Salse, J.; Aury, J.M.; Guiltinan, M.J.; Droc, G.; Gouzy, J.; Allegre, M.; Chaparro, C.; Legavre, T.; Maximova, S.N. The genome of Theobroma cacao. Nat. Genet. 2011, 3, 101–108. [Google Scholar] [CrossRef]
- Shen, S.; Zhang, Q.; Shi, Y.; Sun, Z.; Zhang, Q.; Hou, S.; Wu, R.; Jiang, L.; Zhao, X.; Guo, Y. Genome-wide analysis of the NAC Domain transcription factor gene family in Theobroma cacao. Genes 2019, 11, 35. [Google Scholar] [CrossRef]
- Silva Monteiro de Almeida, D.; Oliveira Jordão do Amaral, D.; Del-Bem, L.E.; Bronze Dos Santos, E.; Santana Silva, R.J.; Peres Gramacho, K.; Vincentz, M.; Micheli, F. Genome-wide identification and characterization of cacao WRKY transcription factors and analysis of their expression in response to witches’ broom disease. PLoS ONE 2017, 12, e0187346. [Google Scholar] [CrossRef] [PubMed]
- Martins Alves, A.M.; Pereira Menezes Reis, S.; Peres Gramacho, K.; Micheli, F. The glutathione peroxidase family of Theobroma cacao: Involvement in the oxidative stress during witches’ broom disease. Int. J. Biol. Macromol. 2020, 164, 3698–3708. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wu, B.; Yan, L.; Qin, X.; Lai, J. Metabolome and transcriptome profiling of Theobroma cacao provides insights into the molecular basis of pod color variation. J. Plant Res. 2021, 134, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Wang, T.; Xu, Z.; Sun, L.; Zhang, Q. Genome-wide analysis of the GRAS gene family in Prunus mume. Mol. Gen. Genomics 2015, 290, 303–317. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Letunic, I.; Copley, R.R.; Schmidt, S.; Ciccarelli, F.D.; Doerks, T.; Schultz, J.; Ponting, C.P.; Bork, P. SMART 4.0: Towards genomic data integration. Nucleic Acids Res. 2004, 32, D142–D144. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef]
- Pysh, L.D.; Wysocka-Diller, J.W.; Camilleri, C.; Bouchez, D.; Benfey, P.N. The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 1999, 18, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Letunic, B. Interactive Tree of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.Y.; Li, W.F.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.P.; Zhang, Y.B.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A Toolkit Incorporating Gamma-Series Methods and Sliding Window Strategies. Genom. Proteom. Bioinf. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Yu, G. Using ggtree to visualize data on tree-like structures. Curr. Protoc. Bioinform. 2020, 69, e96. [Google Scholar] [CrossRef]
- Roy, S.W.; Penny, D. Patterns of intron loss and gain in plants: Intron loss-dominated evolution and genome-wide comparison of O. sativa and A. thaliana. Mol. Biol. Evol. 2007, 24, 171–181. [Google Scholar] [CrossRef]
- Rogozin, I.B.; Carmel, L.; Csuros, M.; Koonin, E.V. Origin and evolution of spliceosomal introns. Biol. Direct. 2012, 7, 11. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Hanada, K.; Zou, C.; Lehti-Shiu, M.D.; Shinozaki, K.; Shiu, S.H. Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 2008, 148, 993–1003. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.Y.; Ma, Z.; Ramachandran, S. Evolutionary history and stress regulation of the lectin superfamily in higher plants. BMC Evol. Biol. 2010, 10, 79. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.L.; Geisler, M. Genome-wide computational identification of biologically significant cis-regulatory elements and associated transcription factors from rice. Plants 2019, 8, 441. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).