Photosynthesis and Salt Exclusion Are Key Physiological Processes Contributing to Salt Tolerance of Canola (Brassica napus L.): Evidence from Physiology and Transcriptome Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Growth Attributes
2.3. Leaf Mineral Nutrients (Na+, K+ mg/g dwt)
2.4. Relative Water Contents (%)
2.5. Hydrogen Peroxide Content (µmol/g f.wt)
2.6. Malondialdehyde (nmol/g f.wt)
2.7. Extraction and Determination of Antioxidant Enzymes
2.7.1. Superoxide Dismutase Activity
2.7.2. Peroxidase Activity
2.7.3. Catalase Activity
2.8. Assessment of Photosystem II (PSII) Structural Stability Using JIP-Test
2.9. Statistical Analysis of Data
2.10. Transcriptome Analysis
2.10.1. Isolation of Total RNA
2.10.2. Next-Generation Sequencing (NGS)
2.10.3. Bioinformatics Analysis
2.11. Validation of NGS Data by qRT-PCR
3. Results
3.1. Growth Attributes
3.2. Relative Water Content
3.3. Ion Accumulation (Shoot K+, Shoot Na+, and Shoot K+/Na+ Ratio)
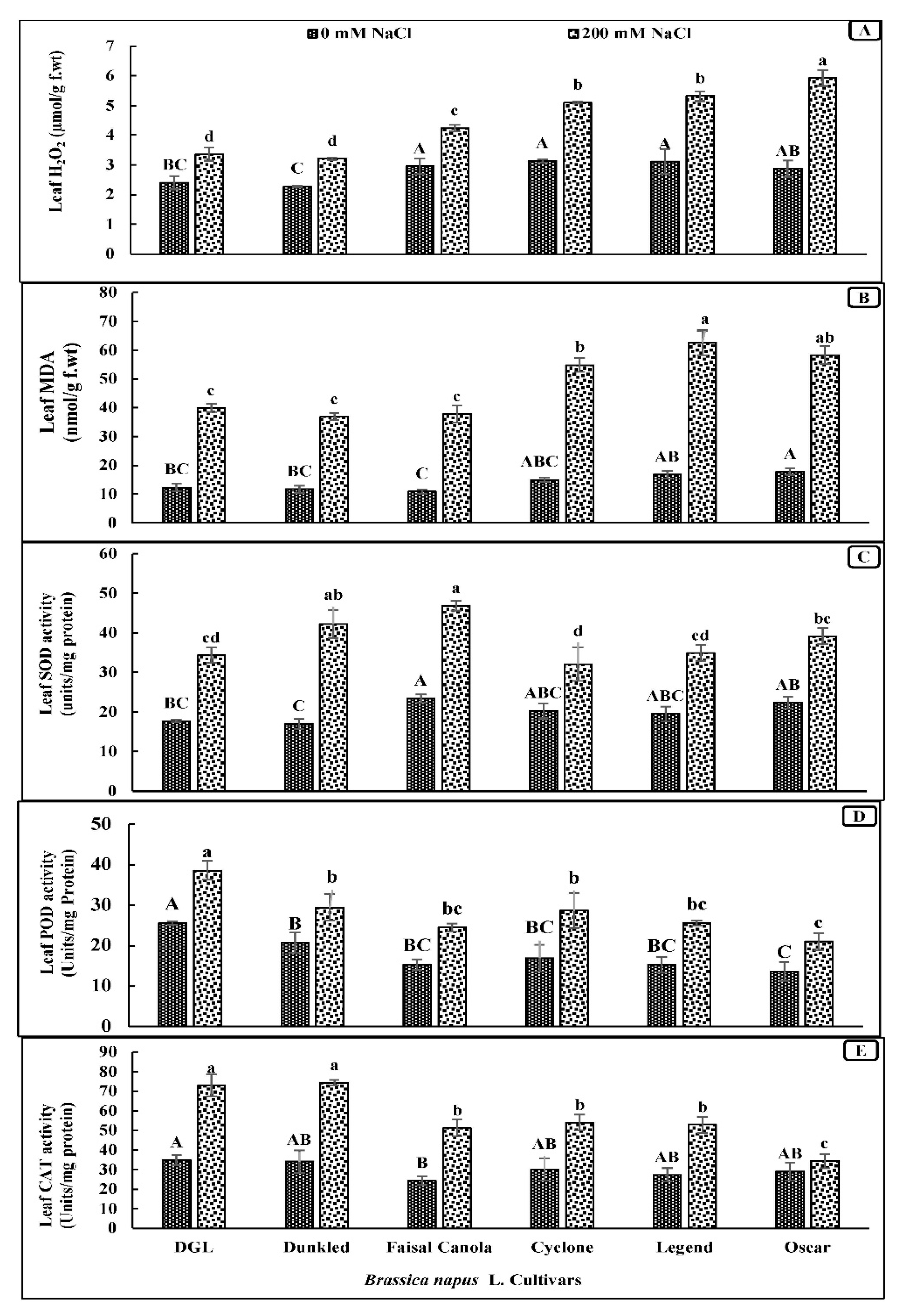
3.4. Leaf Oxidative Stress (H2O2 and MDA)
3.5. Activity of Antioxidants (Leaf SOD, POD, and Catalase)
3.6. Assessment of Photosystem II (PSII) Structural Stability in Canola Using JIP-Test
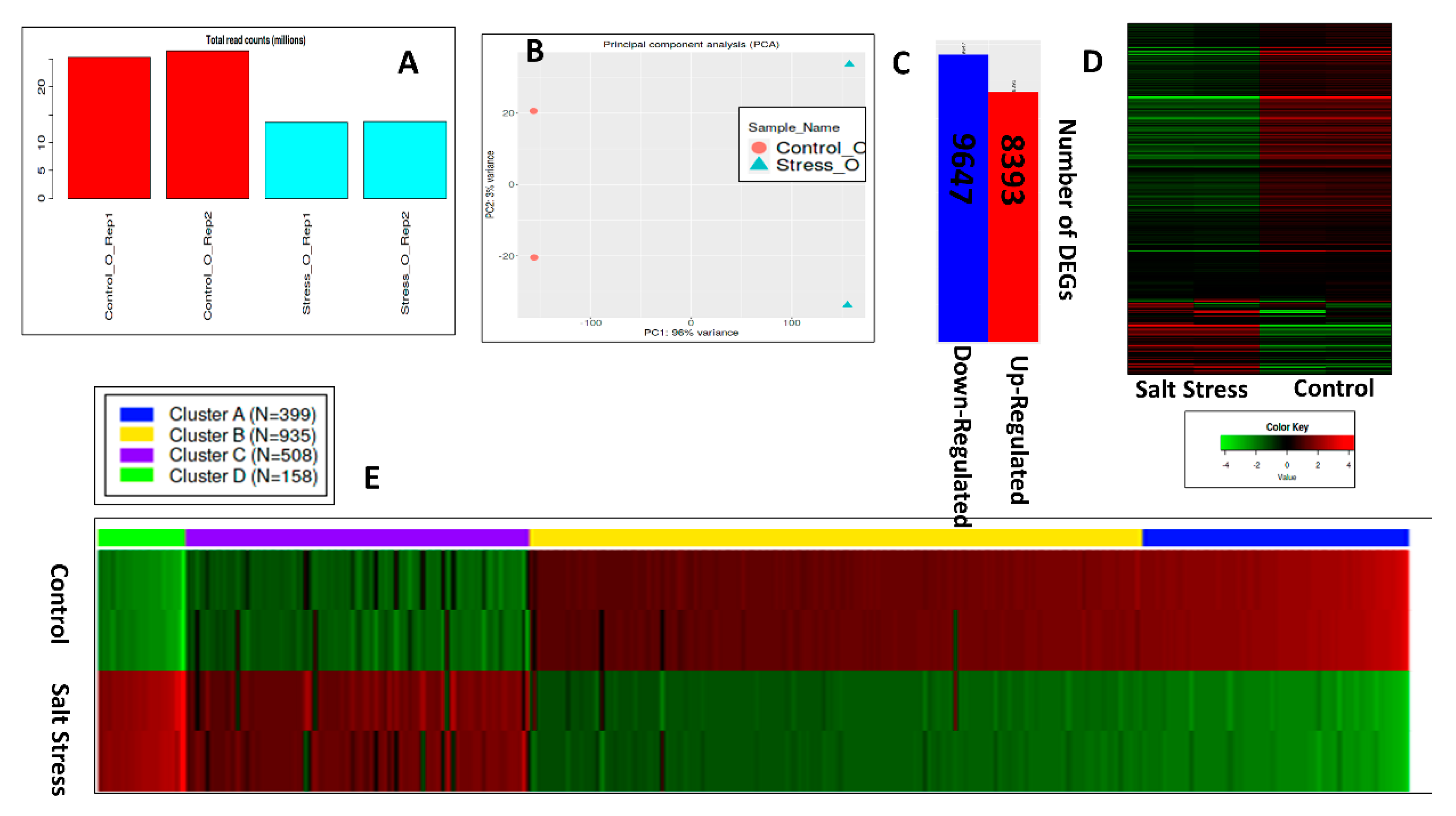
3.7. Transcriptome Analysis of Salt-Sensitive Cultivar Oscar
3.7.1. Raw Data Statistics and Gene Counts
3.7.2. Differential Expression Analysis
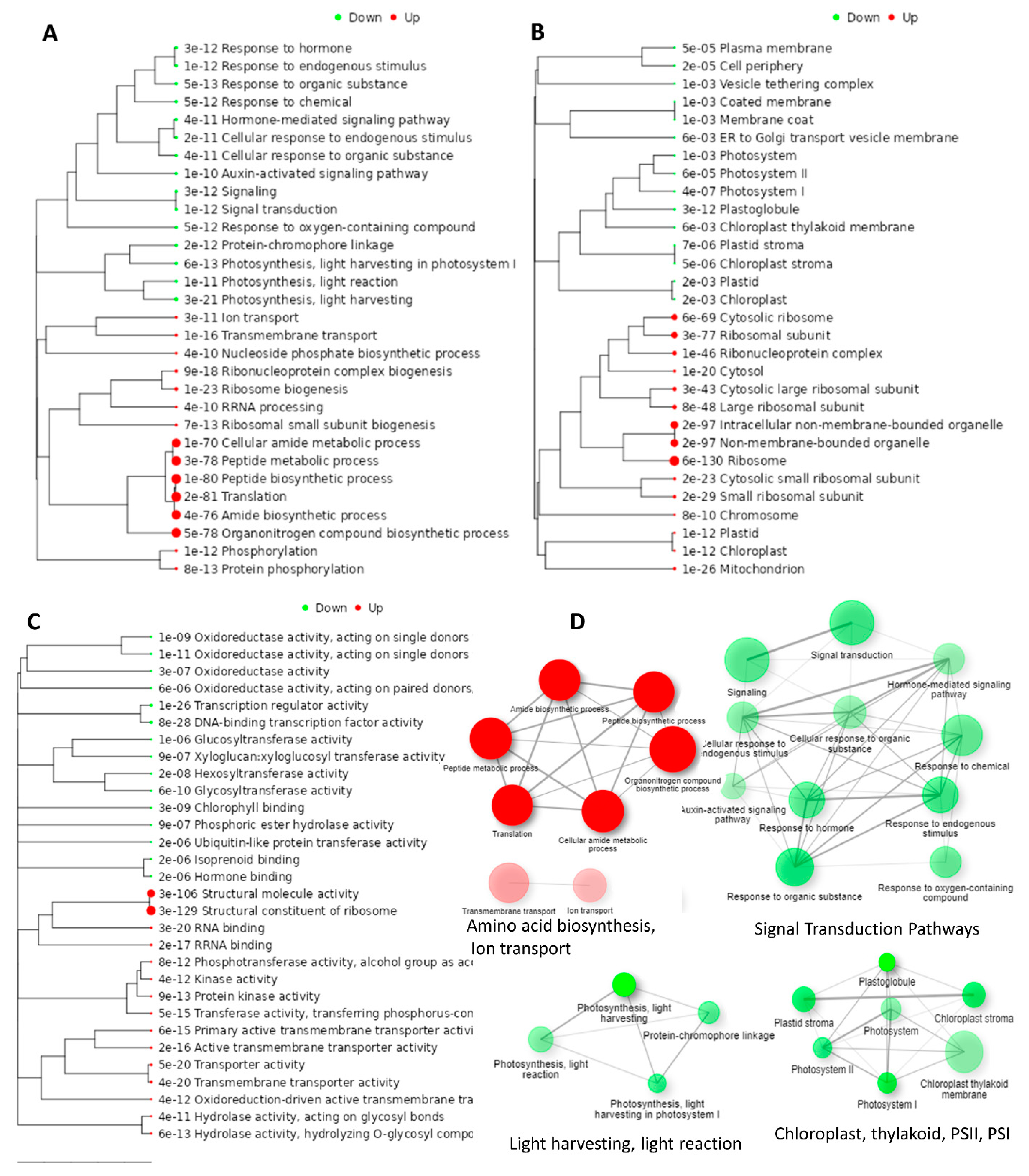
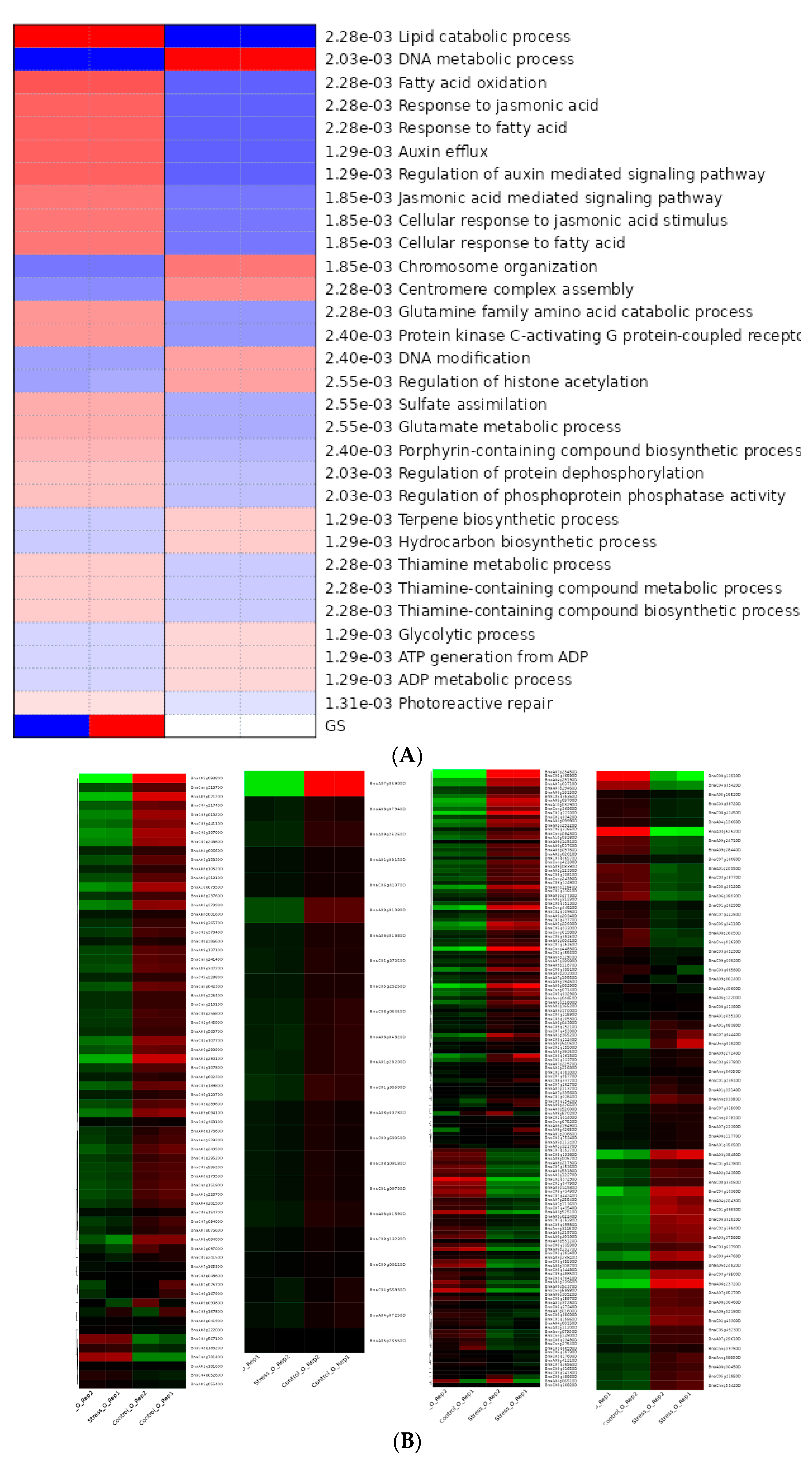
3.7.3. Hierarchical Clustering, GO Terms Analysis of DEGS
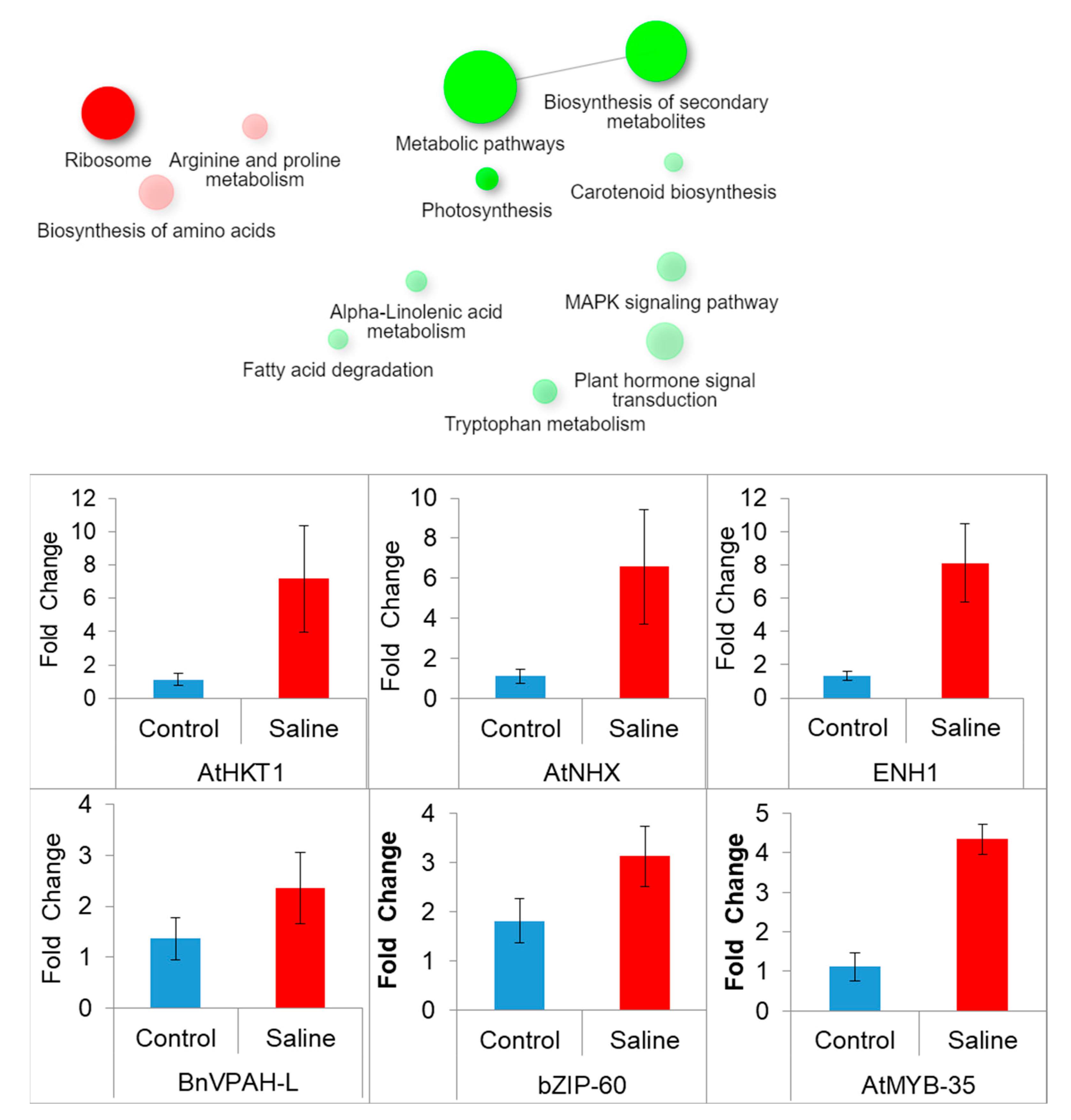
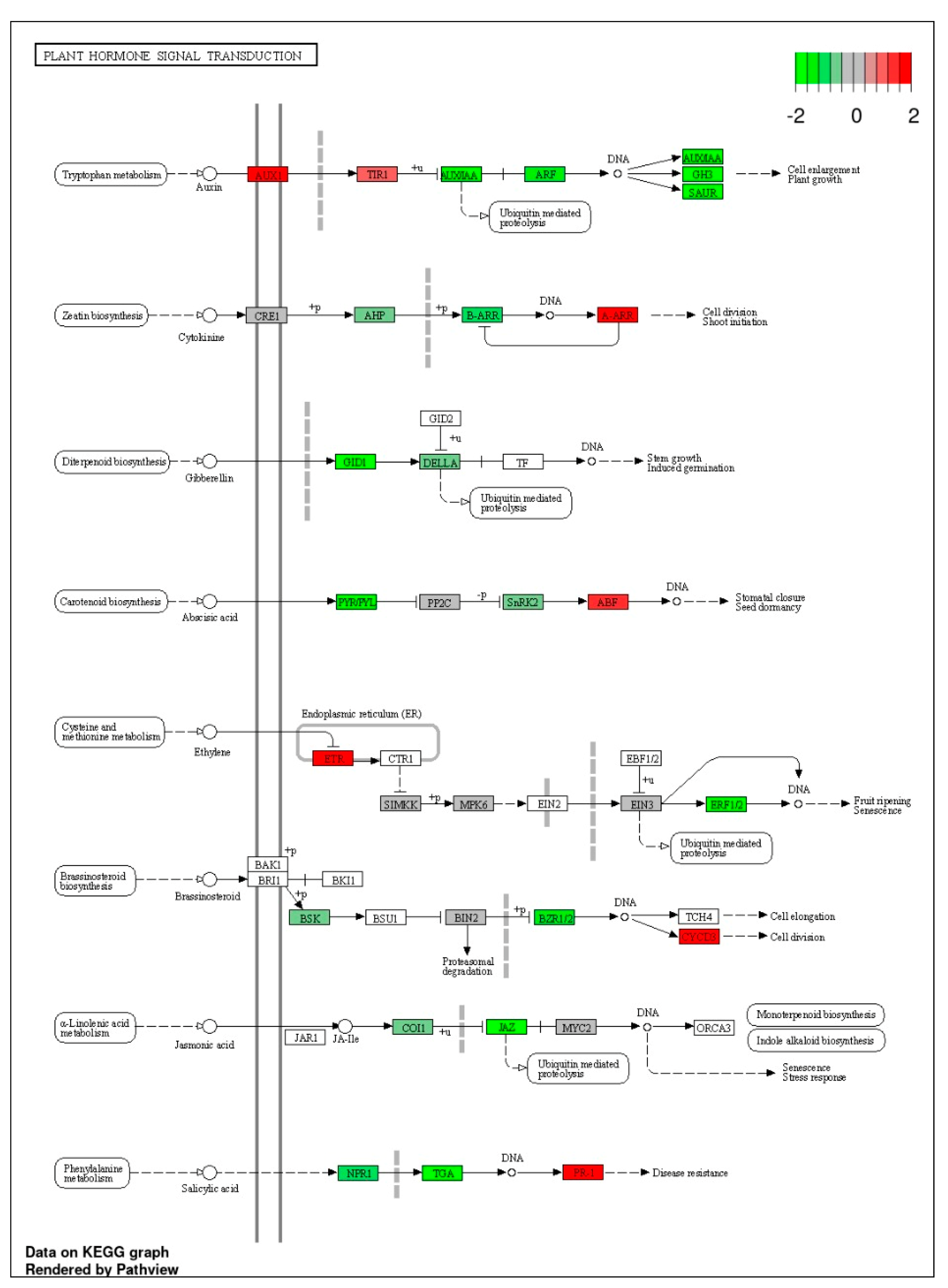
3.7.4. KEGG Pathway Enrichment Analysis
3.8. Validation of Transcriptome Analysis through qRT-PCR
4. Discussion
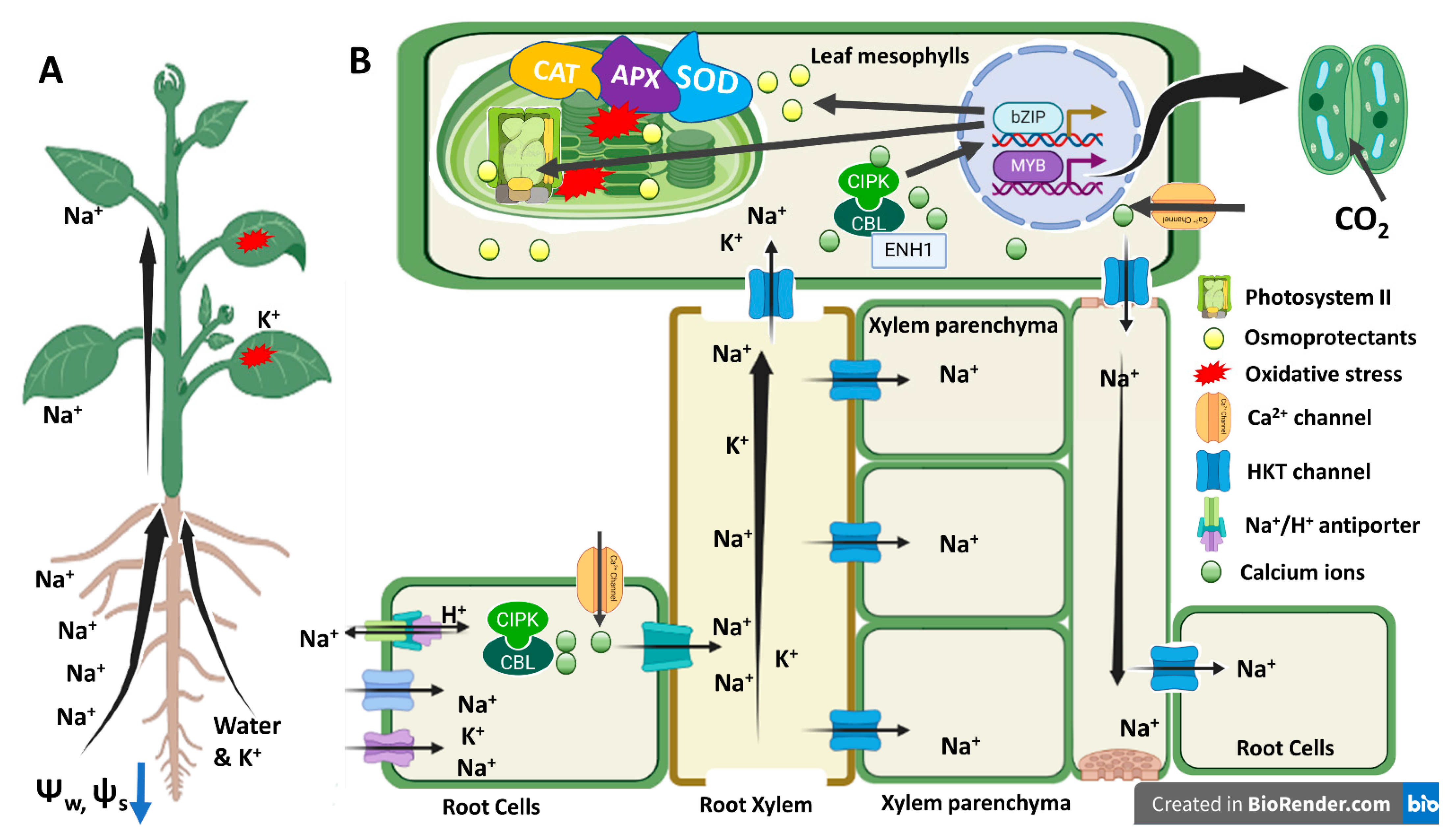
4.1. Physiological Basis of salt Tolerance -Water Status, Ion Exclusion, and Antioxidant Potential
4.2. Activity and Structural Stability of PSII
4.3. Molecular Basis of Salt Tolerance (Transcriptome Analysis)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M.; Athar, H.R.; Harris, P.J.C.; Kwon, T.R. Some prospective strategies for improving crop salt tolerance. Adv. Agron. 2008, 97, 45–110. [Google Scholar] [CrossRef]
- Ashraf, M.; Munns, R. Evolution of Approaches to Increase the Salt Tolerance of Crops. Crit. Rev. Plant Sci. 2022, 41, 1–33. [Google Scholar] [CrossRef]
- Naeem, M.; Iqbal, M.; Shakeel, A.; Ul-Allah, S.; Hussain, M.; Rehman, A.; Zafar, Z.U.; Athar, H.-u.-R.; Ashraf, M. Genetic basis of ion exclusion in salinity stressed wheat: Implications in improving crop yield. Plant Growth Regul. 2020, 92, 479–496. [Google Scholar] [CrossRef]
- Athar, H.-u.-R.; Zafar, Z.U.; Ashraf, M. Glycinebetaine Improved Photosynthesis in Canola under Salt Stress: Evaluation of Chlorophyll Fluorescence Parameters as Potential Indicators. J. Agron. Crop Sci. 2015, 201, 428–442. [Google Scholar] [CrossRef]
- Khalid, A.; Athar, H.-u.-R.; Zafar, Z.U.; Akram, A.; Hussain, K.; Manzoor, H.; Al-Qurainy, F.; Ashraf, M. Photosynthetic capacity of canola (Brassica napus L.) plants as affected by glycinebetaine under salt stress. J. Appl. Bot. Food Qual. 2015, 88, 78–86. [Google Scholar] [CrossRef]
- Ogbaga, C.C.; Stepien, P.; Athar, H.-U.-R.; Ashraf, M. Engineering Rubisco activase from thermophilic cyanobacteria into high-temperature sensitive plants. Crit. Rev. Biotechnol. 2018, 38, 559–572. [Google Scholar] [CrossRef]
- Simkin, A.J.; López-Calcagno, P.E.; Raines, C.A. Feeding the world: Improving photosynthetic efficiency for sustainable crop production. J. Exp. Bot. 2019, 70, 1119–1140. [Google Scholar] [CrossRef]
- Lei, Y.; Xu, Y.; Hettenhausen, C.; Lu, C.; Shen, G.; Zhang, C.; Li, J.; Song, J.; Lin, H.; Wu, J. Comparative analysis of alfalfa (Medicago sativa L.) leaf transcriptomes reveals genotype-specific salt tolerance mechanisms. BMC Plant Biol. 2018, 18, 35. [Google Scholar] [CrossRef]
- Arfan, M.; Athar, H.R.; Ashraf, M. Does exogenous application of salicylic acid through the rooting medium modulate growth and photosynthetic capacity in two differently adapted spring wheat cultivars under salt stress? J. Plant Physiol. 2007, 164, 685–694. [Google Scholar] [CrossRef]
- Shahzadi, A.K.; Bano, H.; Ogbaga, C.C.; Ayyaz, A.; Parveen, R.; Zafar, Z.U.; Athar, H.-U.-R.; Ashraf, M. Coordinated impact of ion exclusion, antioxidants and photosynthetic potential on salt tolerance of ridge gourd [Luffa acutangula (L.) Roxb.]. Plant Physiol. Biochem. 2021, 167, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Smillie, R.M.; Nott, R. Salt Tolerance in Crop Plants Monitored by Chlorophyll Fluorescence In Vivo. Plant Physiol. 1982, 70, 1049–1054. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.J.; Huang, W.; Wang, X.J.; Evrard, A.; SchmÖCkel, S.M.; Zafar, Z.U.; Tester, M. A novel protein kinase involved in Na+ exclusion revealed from positional cloning. Plant Cell Environ. 2013, 36, 553–568. [Google Scholar] [CrossRef] [PubMed]
- Ulfat, M.; Athar, H.-u.-R.; Khan, Z.-D.; Kalaji, H.M. RNAseq Analysis Reveals Altered Expression of Key Ion Transporters Causing Differential Uptake of Selective Ions in Canola (Brassica napus L.) Grown under NaCl Stress. Plants 2020, 9, 891. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Pang, J.; Zhang, F.; Sun, L.; Yang, L.; Fu, T.; Guo, L.; Siddique, K.H.M. Salt-responsive transcriptome analysis of canola roots reveals candidate genes involved in the key metabolic pathway in response to salt stress. Sci. Rep. 2022, 12, 1666. [Google Scholar] [CrossRef]
- Long, W.; Zou, X.; Zhang, X. Transcriptome Analysis of Canola (Brassica napus) under Salt Stress at the Germination Stage. PLoS ONE 2015, 10, e0116217. [Google Scholar] [CrossRef]
- Mohamed, I.A.A.; Shalby, N.; El-Badri, A.M.; Batool, M.; Wang, C.; Wang, Z.; Salah, A.; Rady, M.M.; Jie, K.; Wang, B.; et al. RNA-seq analysis revealed key genes associated with salt tolerance in rapeseed germination through carbohydrate metabolism, hormone, and MAPK signaling pathways. Ind. Crops Prod. 2022, 176, 114262. [Google Scholar] [CrossRef]
- Zhao, S.; Fung-Leung, W.-P.; Bittner, A.; Ngo, K.; Liu, X. Comparison of RNA-Seq and Microarray in Transcriptome Profiling of Activated T Cells. PLoS ONE 2014, 9, e78644. [Google Scholar] [CrossRef]
- Athar, H.-u.-R.; Ashraf, M.; Wahid, A.; Jamil, A. Inducing salt tolerance in canola (Brassica napus L.) by exogenous application of glycinebetaine and proline: Response at the initial growth stages. Pak. J. Bot. 2009, 41, 1311–1319. [Google Scholar]
- Allen, S.E.; Grimshaw, H.M.; Rowland, A.P. Chemical analysis. In Methods in Plant Ecology, 2nd ed.; Moore, P.D., Chapman, S.B., Eds.; Blackwell Scientific Publication: Oxford, UK, 1986; pp. 258–344. [Google Scholar]
- Velikova, V.; Yordanov, I.; Edreva, A. Oxidative stress and some antioxidant systems in acid rain-treated bean plants: Protective role of exogenous polyamines. Plant Sci. 2000, 151, 59–66. [Google Scholar] [CrossRef]
- Heath, R.L.; Packer, L. Photoperoxidation in isolated chloroplasts: I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 1968, 125, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Stewart, R.R.C.; Bewley, J.D. Lipid Peroxidation Associated with Accelerated Aging of Soybean Axes. Plant Physiol. 1980, 65, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Castillo, F.J.; Penel, C.; Greppin, H. Peroxidase Release Induced by Ozone in Sedum album Leaves: Involvement of Ca. Plant Physiol. 1984, 74, 846–851. [Google Scholar] [CrossRef]
- Aebi, H. [13] Catalase in vitro. In Methods in Enzymology; Packer, L., Ed.; Elsevier: Amsterdam, The Netherlands, 1984; Volume 105, pp. 121–126. [Google Scholar]
- Çiçek, N.; Oukarroum, A.; Strasser, R.J.; Schansker, G. Salt stress effects on the photosynthetic electron transport chain in two chickpea lines differing in their salt stress tolerance. Photosynth. Res. 2017, 136, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Snedecor, G.W.; Cochran, W.G. Statistical Methods; Iowa State University Press: Ames, IA, USA, 1980. [Google Scholar]
- Wan, C.Y.; Wilkins, T.A. A Modified Hot Borate Method Significantly Enhances the Yield of High-Quality RNA from Cotton (Gossypium hirsutum L.). Anal. Biochem. 1994, 223, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2013, 30, 923–930. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Ashraf, M.; McNeilly, T. Salinity tolerance in Brassica oilseeds. Crit. Rev. Plant Sci. 2004, 23, 157–174. [Google Scholar] [CrossRef]
- Athar, H.R.; Ashraf, M. Strategies for Crop Improvement Against Salinity and Drought Stress: An Overview. In Salinity and Water Stress: Improving Crop Efficiency; Ashraf, M., Ozturk, M., Athar, H.R., Lieth, H., Kratochwil, A., Eds.; Tasks for Vegetation Science 34; Springer: Dordrecht, The Netherlands, 2009; Volume 44, pp. 1–16. [Google Scholar]
- Ulfat, M.; Athar, H.U.R.; Ashraf, M.; Akram, N.A.; Jamil, A. Appraisal of physiological and biochemical selection criteria for evaluation of salt tolerance in canola (Brassica napus L.). Pak. J. Bot. 2007, 39, 1593–1608. [Google Scholar]
- Jones, M.; Osmond, C.; Turner, N. Accumulation of solutes in leaves of sorghum and sunflower in response to water deficits. Funct. Plant Biol. 1980, 7, 193–205. [Google Scholar] [CrossRef]
- Nandwal, A.S.; Hooda, A.; Datta, D. Effect of Substrate Moisture and Potassium on Water Relations and C, N and K Distribution in Vigna radiata. Biol. Plant. 1998, 41, 149–153. [Google Scholar] [CrossRef]
- Premachandra, G.S.; Hahn, D.T.; Rhodes, D.; Joly, R.J. Leaf water relations and solute accumulation in two grain sorghum lines exhibiting contrasting drought tolerance. J. Exp. Bot. 1995, 46, 1833–1841. [Google Scholar] [CrossRef]
- Suriya-arunroj, D.; Supapoj, N.; Toojinda, T.; Vanavichit, A. Relative leaf water content as an efficient method for evaluating rice cultivars for tolerance to salt stress. Science Asia 2004, 30, 411–415. [Google Scholar] [CrossRef]
- Jha, D.; Shirley, N.; Tester, M.; Roy, S.J. Variation in salinity tolerance and shoot sodium accumulation in Arabidopsis ecotypes linked to differences in the natural expression levels of transporters involved in sodium transport. Plant Cell Environ. 2010, 33, 793–804. [Google Scholar] [CrossRef]
- Munns, R.; James, R.A. Screening methods for salinity tolerance: A case study with tetraploid wheat. Plant Soil 2003, 253, 201–218. [Google Scholar] [CrossRef]
- Ali, Q.; Athar, H.-U.-R.; Ashraf, M. Ion transport in four canola cultivars as influenced by salt stress. Pak. J. Bot. 2006, 38, 1703–1708. [Google Scholar]
- Ashraf, M. Biotechnological approach of improving plant salt tolerance using antioxidants as markers. Biotechnol. Adv. 2009, 27, 84–93. [Google Scholar] [CrossRef]
- Sreenivasulu, N.; Grimm, B.; Wobus, U.; Weschke, W. Differential response of antioxidant compounds to salinity stress in salt-tolerant and salt-sensitive seedlings of foxtail millet (Setaria italica). Physiol. Plant. 2000, 109, 435–442. [Google Scholar] [CrossRef]
- Kalaji, H.M.; Govindjee; Bosa, K.; Kościelniak, J.; Żuk-Gołaszewska, K. Effects of salt stress on photosystem II efficiency and CO2 assimilation of two Syrian barley landraces. Environ. Exp. Bot. 2011, 73, 64–72. [Google Scholar] [CrossRef]
- Dąbrowski, P.; Baczewska, A.H.; Pawluśkiewicz, B.; Paunov, M.; Alexantrov, V.; Goltsev, V.; Kalaji, M.H. Prompt chlorophyll a fluorescence as a rapid tool for diagnostic changes in PSII structure inhibited by salt stress in Perennial ryegrass. J. Photochem. Photobiol. B Biol. 2016, 157, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Qiu, N.; Lu, Q.; Wang, B.; Kuang, T. Does salt stress lead to increased susceptibility of photosystem II to photoinhibition and changes in photosynthetic pigment composition in halophyte Suaeda salsa grown outdoors? Plant Sci. 2002, 163, 1063–1068. [Google Scholar] [CrossRef]
- Kalaji, H.M.; Račková, L.; Paganová, V.; Swoczyna, T.; Rusinowski, S.; Sitko, K. Can chlorophyll-a fluorescence parameters be used as bio-indicators to distinguish between drought and salinity stress in Tilia cordata Mill? Environ. Exp. Bot. 2018, 152, 149–157. [Google Scholar] [CrossRef]
- Ceusters, N.; Valcke, R.; Frans, M.; Claes, J.E.; Van den Ende, W.; Ceusters, J. Performance Index and PSII Connectivity Under Drought and Contrasting Light Regimes in the CAM Orchid Phalaenopsis. Front. Plant Sci. 2019, 10, 1012. [Google Scholar] [CrossRef]
- Ali, Q.; Athar, H.-u.-R.; Ashraf, M. Modulation of growth, photosynthetic capacity and water relations in salt stressed wheat plants by exogenously applied 24-epibrassinolide. Plant Growth Regul. 2008, 56, 107–116. [Google Scholar] [CrossRef]
- Xiong, X.; Wei, Y.-Q.; Chen, J.-H.; Liu, N.; Zhang, Y.-J. Transcriptome analysis of genes and pathways associated with salt tolerance in alfalfa under non-uniform salt stress. Plant Physiol. Biochem. 2020, 151, 323–333. [Google Scholar] [CrossRef]
- Wang, W.; Pang, J.; Zhang, F.; Sun, L.; Yang, L.; Zhao, Y.; Yang, Y.; Wang, Y.; Siddique, K.H.M. Integrated transcriptomics and metabolomics analysis to characterize alkali stress responses in canola (Brassica napus L.). Plant Physiol. Biochem. 2021, 166, 605–620. [Google Scholar] [CrossRef]
- Barkla, B.J.; Vera-Estrella, R.; Pantoja, O. Progress and challenges for abiotic stress proteomics of crop plants. Proteomics 2013, 13, 1801–1815. [Google Scholar] [CrossRef]
- Sandhu, D.; Cornacchione, M.V.; Ferreira, J.F.S.; Suarez, D.L. Variable salinity responses of 12 alfalfa genotypes and comparative expression analyses of salt-response genes. Sci. Rep. 2017, 7, 42958. [Google Scholar] [CrossRef]
- Li, M.; Zhang, K.; Long, R.; Sun, Y.; Kang, J.; Zhang, T.; Cao, S. iTRAQ-based comparative proteomic analysis reveals tissue-specific and novel early-stage molecular mechanisms of salt stress response in Carex rigescens. Environ. Exp. Bot. 2017, 143, 99–114. [Google Scholar] [CrossRef]
- Kaundal, R.; Duhan, N.; Acharya, B.R.; Pudussery, M.V.; Ferreira, J.F.S.; Suarez, D.L.; Sandhu, D. Transcriptional profiling of two contrasting genotypes uncovers molecular mechanisms underlying salt tolerance in alfalfa. Sci. Rep. 2021, 11, 5210. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhu, J.; Zhang, Y.; Fan, F.; Li, W.; Wang, F.; Zhong, W.; Wang, C.; Yang, J. Comparative transcriptome analysis reveals molecular response to salinity stress of salt-tolerant and sensitive genotypes of indica rice at seedling stage. Sci. Rep. 2018, 8, 2085. [Google Scholar] [CrossRef] [PubMed]
- Quan, W.; Liu, X.; Wang, H.; Chan, Z. Physiological and transcriptional responses of contrasting alfalfa (Medicago sativa L.) varieties to salt stress. Plant Cell Tissue Organ Cult. (PCTOC) 2016, 126, 105–115. [Google Scholar] [CrossRef]
- Ismail, A.M.; Horie, T. Genomics, Physiology, and Molecular Breeding Approaches for Improving Salt Tolerance. Annu. Rev. Plant Biol. 2017, 68, 405–434. [Google Scholar] [CrossRef]
- Kobayashi, N.I.; Yamaji, N.; Yamamoto, H.; Okubo, K.; Ueno, H.; Costa, A.; Tanoi, K.; Matsumura, H.; Fujii-Kashino, M.; Horiuchi, T.; et al. OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. Plant J. 2017, 91, 657–670. [Google Scholar] [CrossRef]
- El Mahi, H.; Pérez-Hormaeche, J.; De Luca, A.; Villalta, I.; Espartero, J.; Gámez-Arjona, F.; Fernández, J.L.; Bundó, M.; Mendoza, I.; Mieulet, D.; et al. A Critical Role of Sodium Flux via the Plasma Membrane Na+/H+ Exchanger SOS1 in the Salt Tolerance of Rice. Plant Physiol. 2019, 180, 1046–1065. [Google Scholar] [CrossRef]
- Zhu, J.; Fan, Y.; Li, C.; Shabala, S.; Zhao, C.; Hong, Y.; Lv, C.; Guo, B.; Xu, R.; Zhou, M. Candidate genes for salinity tolerance in barley revealed by RNA-seq analysis of near-isogenic lines. Plant Growth Regul. 2020, 92, 571–582. [Google Scholar] [CrossRef]
- Shen, Q.; Fu, L.; Su, T.; Ye, L.; Huang, L.; Kuang, L.; Wu, L.; Wu, D.; Chen, Z.-H.; Zhang, G. Calmodulin HvCaM1 Negatively Regulates Salt Tolerance via Modulation of HvHKT1s and HvCAMTA4. Plant Physiol. 2020, 183, 1650–1662. [Google Scholar] [CrossRef]
- Wang, J.; Jiao, J.; Zhou, M.; Jin, Z.; Yu, Y.; Liang, M. Physiological and Transcriptional Responses of Industrial Rapeseed (Brassica napus) Seedlings to Drought and Salinity Stress. Int. J. Mol. Sci. 2019, 20, 5604. [Google Scholar] [CrossRef]


| Source of Variation | Mean Squares | |||
|---|---|---|---|---|
| Cultivar | Salinity | Cultivar × Salinity | Error | |
| Degree of Freedom (df) | 5 | 1 | 5 | 36 |
| Attributes | ||||
| Shoot Fresh Weight | 240.71 *** | 11,249.359 *** | 61.29 *** | 6.643 |
| Root Fresh Weight | 2.105 ** | 292.152 *** | 2.604 *** | 0.485 |
| Shoot Dry Weight | 2.256 *** | 83.136 *** | 1.234 *** | 0.15 |
| Root Dry Weight | 0.510 *** | 10.303 *** | 0.063 *** | 0.009 |
| Relative Water Content | 36.555 * | 2585.238 *** | 64.675 ** | 14.069 |
| Shoot Potassium Content | 29.10 ** | 2324.88 *** | 21.815 * | 6.573 |
| Shoot Sodium Content | 59.68 *** | 7272.54 *** | 60.669 *** | 4.764 |
| Na+/K+ Ratio | 3.228 ns | 1390.26 *** | 2.453 ns | 1.965 |
| Leaf Hydrogen Peroxide | 4.04 *** | 36.02 *** | 1.39 *** | 0.13 |
| Leaf Malondialdehyde | 403.63 *** | 14,145.74 *** | 154.22 *** | 13.58 |
| Leaf Superoxide Dismutase | 98.11 *** | 3989.43 *** | 52.02 ** | 13.92 |
| Leaf Peroxidase | 215.92 *** | 1218.06 *** | 8.45 ns | 17.35 |
| Leaf Catalase | 655.72 *** | 8545.44 *** | 310.73 *** | 52.81 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gul, H.S.; Ulfat, M.; Zafar, Z.U.; Haider, W.; Ali, Z.; Manzoor, H.; Afzal, S.; Ashraf, M.; Athar, H.-u.-R. Photosynthesis and Salt Exclusion Are Key Physiological Processes Contributing to Salt Tolerance of Canola (Brassica napus L.): Evidence from Physiology and Transcriptome Analysis. Genes 2023, 14, 3. https://doi.org/10.3390/genes14010003
Gul HS, Ulfat M, Zafar ZU, Haider W, Ali Z, Manzoor H, Afzal S, Ashraf M, Athar H-u-R. Photosynthesis and Salt Exclusion Are Key Physiological Processes Contributing to Salt Tolerance of Canola (Brassica napus L.): Evidence from Physiology and Transcriptome Analysis. Genes. 2023; 14(1):3. https://doi.org/10.3390/genes14010003
Chicago/Turabian StyleGul, Hafiza Saima, Mobina Ulfat, Zafar Ullah Zafar, Waseem Haider, Zain Ali, Hamid Manzoor, Shehrooz Afzal, Muhammad Ashraf, and Habib-ur-Rehman Athar. 2023. "Photosynthesis and Salt Exclusion Are Key Physiological Processes Contributing to Salt Tolerance of Canola (Brassica napus L.): Evidence from Physiology and Transcriptome Analysis" Genes 14, no. 1: 3. https://doi.org/10.3390/genes14010003
APA StyleGul, H. S., Ulfat, M., Zafar, Z. U., Haider, W., Ali, Z., Manzoor, H., Afzal, S., Ashraf, M., & Athar, H.-u.-R. (2023). Photosynthesis and Salt Exclusion Are Key Physiological Processes Contributing to Salt Tolerance of Canola (Brassica napus L.): Evidence from Physiology and Transcriptome Analysis. Genes, 14(1), 3. https://doi.org/10.3390/genes14010003








