Methodological Changes in the Field of Paleogenetics
Abstract
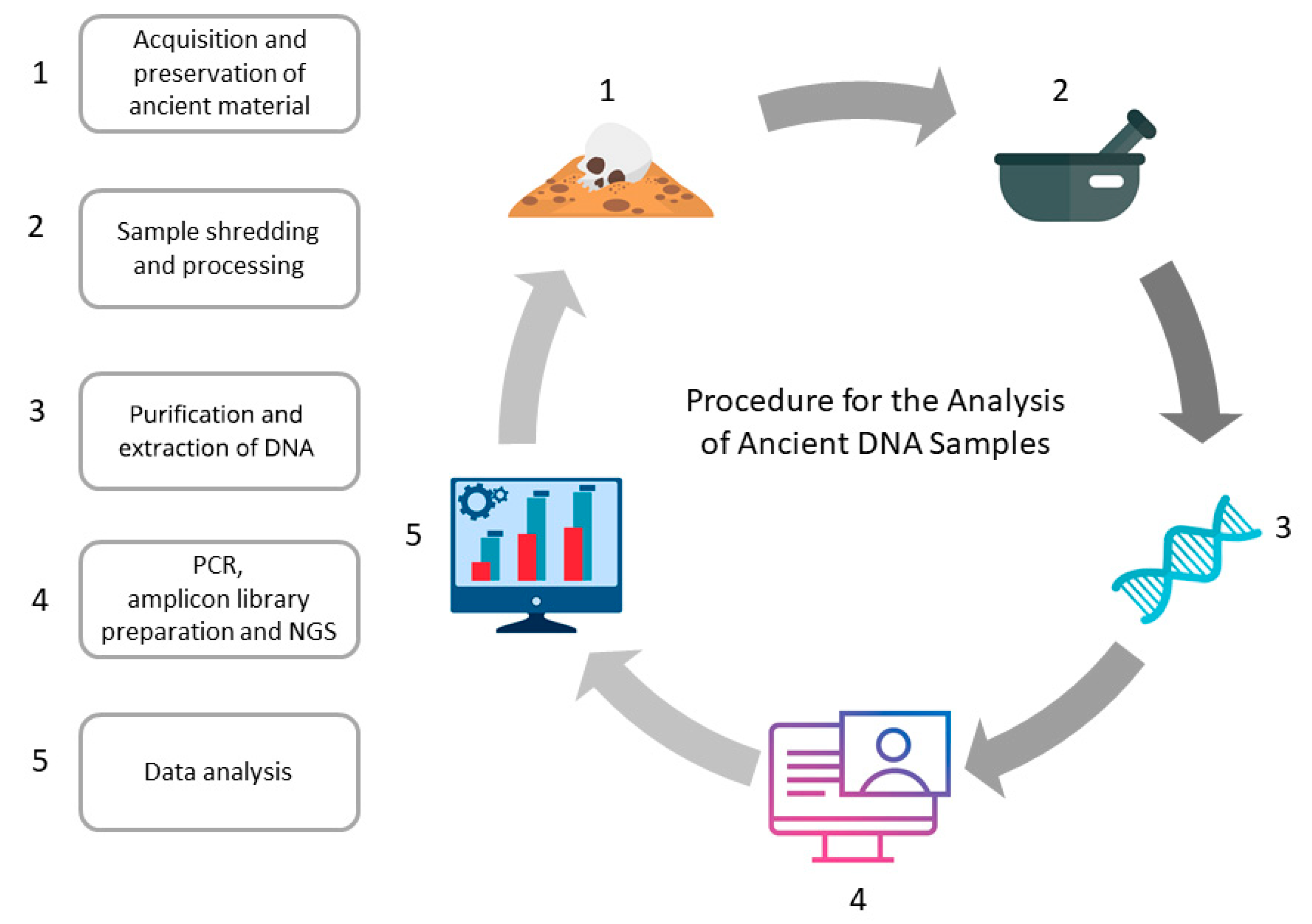
1. Introduction
2. History of aDNA Research
3. Damage of aDNA
4. Materials, Methods and Contamination
5. Extraction of Ancient DNA
6. Amplification
7. Target Enrichment via Hybridization-Based Capture
8. Sequencing
9. Where Paleogenetic and Forensic Sciences Converge
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Paabo, S. Ancient DNA: Extraction, Characterization, Molecular Cloning, and Enzymatic Amplification. Proc. Natl. Acad. Sci. USA 1989, 86, 1939–1943. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, R.; Bowman, B.; Freiberger, M.; Ryder, O.A.; Wilson, A.C. DNA Sequences from the Quagga, an Extinct Member of the Horse Family. Nature 1984, 312, 282–284. [Google Scholar] [CrossRef] [PubMed]
- Ancient DNA|Definition of Ancient DNA by Medical Dictionary. Available online: https://medical-dictionary.thefreedictionary.com/Ancient+DNA (accessed on 6 January 2023).
- Barlow, A.; Cahill, J.A.; Hartmann, S.; Theunert, C.; Xenikoudakis, G.; Fortes, G.G.; Paijmans, J.L.A.; Rabeder, G.; Frischauf, C.; Grandal-d’Anglade, A.; et al. Partial Genomic Survival of Cave Bears in Living Brown Bears. Nat. Ecol. Evol. 2018, 2, 1563–1570. [Google Scholar] [CrossRef] [PubMed]
- Park, S.D.E.; Magee, D.A.; McGettigan, P.A.; Teasdale, M.D.; Edwards, C.J.; Lohan, A.J.; Murphy, A.; Braud, M.; Donoghue, M.T.; Liu, Y.; et al. Genome Sequencing of the Extinct Eurasian Wild Aurochs, Bos Primigenius, Illuminates the Phylogeography and Evolution of Cattle. Genome Biol. 2015, 16, 234. [Google Scholar] [CrossRef] [PubMed]
- Palkopoulou, E.; Mallick, S.; Skoglund, P.; Enk, J.; Rohland, N.; Li, H.; Omrak, A.; Vartanyan, S.; Poinar, H.; Götherström, A.; et al. Complete Genomes Reveal Signatures of Demographic and Genetic Declines in the Woolly Mammoth. Curr. Biol. 2015, 25, 1395–1400. [Google Scholar] [CrossRef]
- Wielgus, K.; Danielewski, M.; Walkowiak, J. Svante Pääbo, Reader of the Neanderthal Genome. Acta Physiol. 2022, 237, e13902. [Google Scholar] [CrossRef]
- Mathieson, I.; Lazaridis, I.; Rohland, N.; Mallick, S.; Patterson, N.; Roodenberg, S.A.; Harney, E.; Stewardson, K.; Fernandes, D.; Novak, M.; et al. Genome-Wide Patterns of Selection in 230 Ancient Eurasians. Nature 2015, 528, 499–503. [Google Scholar] [CrossRef]
- Klunk, J.; Vilgalys, T.P.; Demeure, C.E.; Cheng, X.; Shiratori, M.; Madej, J.; Beau, R.; Elli, D.; Patino, M.I.; Redfern, R.; et al. Evolution of Immune Genes Is Associated with the Black Death. Nature 2022, 611, 312–319. [Google Scholar] [CrossRef]
- Benton, M.L.; Abraham, A.; LaBella, A.L.; Abbot, P.; Rokas, A.; Capra, J.A. The Influence of Evolutionary History on Human Health and Disease. Nat. Rev. Genet. 2021, 22, 269–283. [Google Scholar] [CrossRef]
- Quintana-Murci, L. Understanding Rare and Common Diseases in the Context of Human Evolution. Genome Biol. 2016, 17, 225. [Google Scholar] [CrossRef]
- Harney, É.; May, H.; Shalem, D.; Rohland, N.; Mallick, S.; Lazaridis, I.; Sarig, R.; Stewardson, K.; Nordenfelt, S.; Patterson, N.; et al. Ancient DNA from Chalcolithic Israel Reveals the Role of Population Mixture in Cultural Transformation. Nat. Commun. 2018, 9, 3336. [Google Scholar] [CrossRef] [PubMed]
- Brandt, G.; Haak, W.; Adler, C.J.; Roth, C.; Szécsényi-Nagy, A.; Karimnia, S.; Möller-Rieker, S.; Meller, H.; Ganslmeier, R.; Friederich, S.; et al. Ancient DNA Reveals Key Stages in the Formation of Central European Mitochondrial Genetic Diversity. Science 2013, 342, 257–261. [Google Scholar] [CrossRef] [PubMed]
- Murchie, T.J.; Monteath, A.J.; Mahony, M.E.; Long, G.S.; Cocker, S.; Sadoway, T.; Karpinski, E.; Zazula, G.; MacPhee, R.D.E.; Froese, D.; et al. Collapse of the Mammoth-Steppe in Central Yukon as Revealed by Ancient Environmental DNA. Nat. Commun. 2021, 12, 7120. [Google Scholar] [CrossRef] [PubMed]
- Ottoni, C.; Bekaert, B.; Decorte, R. DNA Degradation: Current Knowledge and Progress in DNA Analysis. In Taphonomy of Human Remains: Forensic Analysis of the Dead and the Depositional Environment; John Wiley & Sons, Ltd.: New York, NY, USA, 2017; pp. 65–80. [Google Scholar]
- Pääbo, S.; Wilson, A.C. Polymerase Chain Reaction Reveals Cloning Artefacts. Nature 1988, 334, 387–388. [Google Scholar] [CrossRef]
- Gitschier, J. Imagine: An Interview with Svante Pääbo. PLoS Genet. 2008, 4, e1000035. [Google Scholar] [CrossRef]
- Willerslev, E.; Cooper, A. Ancient DNA. Proc. R. Soc. B Biol. Sci. 2005, 272, 3–16. [Google Scholar] [CrossRef]
- Jones, E.D.; Bösl, E. Ancient Human DNA: A History of Hype (Then and Now). J. Soc. Archaeol. 2021, 21, 236–255. [Google Scholar] [CrossRef]
- Poinar, H.N.; Schwarz, C.; Qi, J.; Shapiro, B.; MacPhee, R.D.E.; Buigues, B.; Tikhonov, A.; Huson, D.M.; Tomsho, L.P.; Auch, A.; et al. Metagenomics to Paleogenomics: Large-Scale Sequencing of Mammoth DNA. Science 2006, 311, 392–394. [Google Scholar] [CrossRef]
- Rasmussen, M.; Li, Y.; Lindgreen, S.; Pedersen, J.S.; Albrechtsen, A.; Moltke, I.; Metspalu, M.; Metspalu, E.; Kivisild, T.; Gupta, R.; et al. Ancient Human Genome Sequence of an Extinct Palaeo-Eskimo. Nature 2010, 463, 757–762. [Google Scholar] [CrossRef]
- Green, R.E.; Krause, J.; Briggs, A.W.; Maricic, T.; Stenzel, U.; Kircher, M.; Patterson, N.; Li, H.; Zhai, W.; Fritz, M.H.Y.; et al. A Draft Sequence of the Neandertal Genome. Science 2010, 328, 710–722. [Google Scholar] [CrossRef]
- Reich, D.; Green, R.E.; Kircher, M.; Krause, J.; Patterson, N.; Durand, E.Y.; Viola, B.; Briggs, A.W.; Stenzel, U.; Johnson, P.L.F.; et al. Genetic History of an Archaic Hominin Group from Denisova Cave in Siberia. Nature 2010, 468, 1053–1060. [Google Scholar] [CrossRef] [PubMed]
- Kjær, K.H.; Pedersen, M.W.; de Sanctis, B.; de Cahsan, B.; Korneliussen, T.S.; Michelsen, C.S.; Sand, K.K.; Jelavić, S.; Ruter, A.H.; Schmidt, A.M.A.; et al. A 2-Million-Year-Old Ecosystem in Greenland Uncovered by Environmental DNA. Nature 2022, 612, 283–291. [Google Scholar] [CrossRef]
- van der Valk, T.; Pečnerová, P.; Díez-del-Molino, D.; Bergström, A.; Oppenheimer, J.; Hartmann, S.; Xenikoudakis, G.; Thomas, J.A.; Dehasque, M.; Sağlıcan, E.; et al. Million-Year-Old DNA Sheds Light on the Genomic History of Mammoths. Nature 2021, 591, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Lawlor, D.A.; Dickel, C.D.; Hauswirth, W.W.; Parham, P. Ancient HLA Genes from 7500-Year-Old Archaeological Remains. Nature 1991, 349, 785–788. [Google Scholar] [CrossRef]
- Poinar, H.N.; Hofreiter, M.; Spaulding, W.G.; Martin, P.S.; Stankiewicz, B.A.; Bland, H.; Evershed, R.P.; Possnert, G.; Pääbo, S. Molecular Coproscopy: Dung and Diet of the Extinct Ground Sloth Nothrotheriops Shastensis. Science 1998, 281, 402–406. [Google Scholar] [CrossRef]
- Willerslev, E.; Hansen, A.J.; Binladen, J.; Brand, T.B.; Gilbert, M.T.P.; Shapiro, B.; Bunce, M.; Wiuf, C.; Gilichinsky, D.A.; Cooper, A. Diverse Plant and Animal Genetic Records from Holocene and Pleistocene Sediments. Science 2003, 300, 791–795. [Google Scholar] [CrossRef] [PubMed]
- Haak, W.; Forster, P.; Bramanti, B.; Matsumura, S.; Brandt, G.; Tänzer, M.; Villems, R.; Renfrew, C.; Gronenborn, D.; Alt, K.W.; et al. Ancient DNA from the First European Farmers in 7500-Year-Old Neolithic Sites. Science 2005, 310, 1016–1018. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.; Drautz, D.I.; Ratan, A.; Pusey, B.; Qi, J.; Lesk, A.M.; Tomsho, L.P.; Packard, M.D.; Zhao, F.; Sher, A.; et al. Sequencing the Nuclear Genome of the Extinct Woolly Mammoth. Nature 2008, 456, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Green, R.E.; Malaspinas, A.S.; Krause, J.; Briggs, A.W.; Johnson, P.L.F.; Uhler, C.; Meyer, M.; Good, J.M.; Maricic, T.; Stenzel, U.; et al. A Complete Neandertal Mitochondrial Genome Sequence Determined by High-Throughput Sequencing. Cell 2008, 134, 416–426. [Google Scholar] [CrossRef]
- Bos, K.I.; Schuenemann, V.J.; Golding, G.B.; Burbano, H.A.; Waglechner, N.; Coombes, B.K.; McPhee, J.B.; Dewitte, S.N.; Meyer, M.; Schmedes, S.; et al. A Draft Genome of Yersinia Pestis from Victims of the Black Death. Nature 2011, 478, 506–510. [Google Scholar] [CrossRef]
- Meyer, M.; Kircher, M.; Gansauge, M.T.; Li, H.; Racimo, F.; Mallick, S.; Schraiber, J.G.; Jay, F.; Prüfer, K.; de Filippo, C.; et al. A High-Coverage Genome Sequence from an Archaic Denisovan Individual. Science 2012, 338, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.; Graefen, A.; Ball, M.; Matzas, M.; Boisguerin, V.; Maixner, F.; Leidinger, P.; Backes, C.; Khairat, R.; Forster, M.; et al. New Insights into the Tyrolean Iceman’s Origin and Phenotype as Inferred by Whole-Genome Sequencing. Nat. Commun. 2012, 3, 698. [Google Scholar] [CrossRef] [PubMed]
- Warinner, C.; Rodrigues, J.F.M.; Vyas, R.; Trachsel, C.; Shved, N.; Grossmann, J.; Radini, A.; Hancock, Y.; Tito, R.Y.; Fiddyment, S.; et al. Pathogens and Host Immunity in the Ancient Human Oral Cavity. Nat. Genet. 2014, 46, 336–344. [Google Scholar] [CrossRef] [PubMed]
- King, T.E.; Fortes, G.G.; Balaresque, P.; Thomas, M.G.; Balding, D.; Delser, P.M.; Neumann, R.; Parson, W.; Knapp, M.; Walsh, S.; et al. Identification of the Remains of King Richard III. Nat. Commun. 2014, 5, 5631. [Google Scholar] [CrossRef]
- Prüfer, K.; Racimo, F.; Patterson, N.; Jay, F.; Sankararaman, S.; Sawyer, S.; Heinze, A.; Renaud, G.; Sudmant, P.H.; de Filippo, C.; et al. The Complete Genome Sequence of a Neanderthal from the Altai Mountains. Nature 2014, 505, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Prüfer, K.; de Filippo, C.; Grote, S.; Mafessoni, F.; Korlević, P.; Hajdinjak, M.; Vernot, B.; Skov, L.; Hsieh, P.; Peyrégne, S.; et al. A High-Coverage Neandertal Genome from Vindija Cave in Croatia. Science 2017, 358, 655–658. [Google Scholar] [CrossRef]
- Margaryan, A.; Lawson, D.J.; Sikora, M.; Racimo, F.; Rasmussen, S.; Moltke, I.; Cassidy, L.M.; Jørsboe, E.; Ingason, A.; Pedersen, M.W.; et al. Population Genomics of the Viking World. Nature 2020, 585, 390–396. [Google Scholar] [CrossRef]
- Mafessoni, F.; Grote, S.; de Filippo, C.; Slon, V.; Kolobova, K.A.; Viola, B.; Markin, S.V.; Chintalapati, M.; Peyrégne, S.; Skov, L.; et al. A High-Coverage Neandertal Genome from Chagyrskaya Cave. Proc. Natl. Acad. Sci. USA 2020, 117, 15132–15136. [Google Scholar] [CrossRef]
- Bailly, V.; Verly, W.G. Possible Roles of F-Elimination and 6-Elimination Reactions in the Repair of DNA Containing AP (Apurinic/Apyrimidinic) Sites in Mammalian Cells. Biochem. J. 1988, 253, 553–559. [Google Scholar] [CrossRef]
- Overballe-Petersen, S.; Orlando, L.; Willerslev, E. Next-Generation Sequencing Offers New Insights into DNA Degradation. Trends Biotechnol. 2012, 30, 364–368. [Google Scholar] [CrossRef]
- Lindahl, T.; Karlström, O. Heat-Induced Depyrimidination of Deoxyribonucleic Acid in Neutral Solution. Biochemistry 1973, 12, 5151–5154. [Google Scholar] [CrossRef]
- Lindahl, T.; Nyberg, B. Rate of Depurination of Native Deoxyribonucleic Acid. Biochemistry 1972, 11, 3610–3618. [Google Scholar] [CrossRef]
- Shapiro, R. Damage to DNA Caused by Hydrolysis. In Chromosome Damage and Repair; Springer: New York, NY, USA, 1981; pp. 3–18. [Google Scholar]
- Poulakakis, N.; Tselikas, A.; Bitsakis, I.; Mylonas, M.; Lymberakis, P. Ancient DNA and the Genetic Signature of Ancient Greek Manuscripts. J. Archaeol. Sci. 2007, 34, 675–680. [Google Scholar] [CrossRef]
- Briggs, A.W.; Stenzel, U.; Johnson, P.L.F.; Green, R.E.; Kelso, J.; Prüfer, K.; Meyer, M.; Krause, J.; Ronan, M.T.; Lachmann, M.; et al. Patterns of Damage in Genomic DNA Sequences from a Neandertal. Proc. Natl. Acad. Sci. USA 2007, 104, 14616–14621. [Google Scholar] [CrossRef] [PubMed]
- Dabney, J.; Meyer, M.; Pääbo, S. Ancient DNA Damage. Cold Spring Harb. Perspect. Biol. 2013, 5, a012567. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, T. Instability and Decay of the Primary Structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef] [PubMed]
- Pääbo, S.; Gifford, J.A.; Wilson, A.C. Mitochondrial DNA Sequences from a 7000-Year Old Brain. Nucleic Acids Res. 1988, 16, 9775–9787. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, M.T.P.; Menez, L.; Janaway, R.C.; Tobin, D.J.; Cooper, A.; Wilson, A.S. Resistance of Degraded Hair Shafts to Contaminant DNA. Forensic Sci. Int. 2006, 156, 208–212. [Google Scholar] [CrossRef]
- Gilbert, M.T.P.; Wilson, A.S.; Bunce, M.; Hansen, A.J.; Willerslev, E.; Shapiro, B.; Higham, T.F.G.; Richards, M.P.; O’Connell, T.C.; Tobin, D.J.; et al. Ancient Mitochondrial DNA from Hair. Curr. Biol. 2004, 14, R463–R464. [Google Scholar] [CrossRef]
- Wolinsky, H. History in a Single Hair. EMBO Rep. 2010, 11, 427–430. [Google Scholar] [CrossRef]
- Bonnichsen, R.; Hodges, L.; Ream, W.; Field, K.G.; Kirner, D.L.; Selsor, K.; Taylor, R.E. Methods for the Study of Ancient Hair: Radiocarbon Dates and Gene Sequences from Individual Hairs. J. Archaeol. Sci. 2001, 28, 775–785. [Google Scholar] [CrossRef]
- Wadsworth, C.; Procopio, N.; Anderung, C.; Carretero, J.M.; Iriarte, E.; Valdiosera, C.; Elburg, R.; Penkman, K.; Buckley, M. Comparing Ancient DNA Survival and Proteome Content in 69 Archaeological Cattle Tooth and Bone Samples from Multiple European Sites. J. Proteom. 2017, 158, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Latham, K.E.; Miller, J.J. DNA Recovery and Analysis from Skeletal Material in Modern Forensic Contexts. Forensic Sci. Res. 2019, 4, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Gamba, C.; Jones, E.R.; Teasdale, M.D.; McLaughlin, R.L.; Gonzalez-Fortes, G.; Mattiangeli, V.; Domboróczki, L.; Kovári, I.; Pap, I.; Anders, A.; et al. Genome Flux and Stasis in a Five Millennium Transect of European Prehistory. Nat. Commun. 2014, 5, 5257. [Google Scholar] [CrossRef] [PubMed]
- Mann, A.E.; Sabin, S.; Ziesemer, K.; Vågene, Å.J.; Schroeder, H.; Ozga, A.T.; Sankaranarayanan, K.; Hofman, C.A.; Fellows Yates, J.A.; Salazar-García, D.C.; et al. Differential Preservation of Endogenous Human and Microbial DNA in Dental Calculus and Dentin. Sci. Rep. 2018, 8, 9822. [Google Scholar] [CrossRef]
- Zhang, D.; Xia, H.; Chen, F.; Li, B.; Slon, V.; Cheng, T.; Yang, R.; Jacobs, Z.; Dai, Q.; Massilani, D.; et al. Denisovan DNA in Late Pleistocene Sediments from Baishiya Karst Cave on the Tibetan Plateau. Science 2020, 370, 584–587. [Google Scholar] [CrossRef]
- Llamas, B.; Valverde, G.; Fehren-Schmitz, L.; Weyrich, L.S.; Cooper, A.; Haak, W. From the Field to the Laboratory: Controlling DNA Contamination in Human Ancient DNA Research in the High-Throughput Sequencing Era. Sci. Technol. Archaeol. Res. 2017, 3, 1–14. [Google Scholar] [CrossRef]
- Cooper, A.; Poinar, H.N. Ancient DNA: Do It Right or Not at All. Science 2000, 289, 1139. [Google Scholar] [CrossRef]
- Orlando, L.; Allaby, R.; Skoglund, P.; der Sarkissian, C.; Stockhammer, P.W.; Ávila-Arcos, M.C.; Fu, Q.; Krause, J.; Willerslev, E.; Stone, A.C.; et al. Ancient DNA Analysis. Nat. Rev. Methods Prim. 2021, 1, 14. [Google Scholar] [CrossRef]
- Poinar, H.N. The Top 10 List: Criteria of Authenticity for DNA from Ancient and Forensic Samples. Int. Congr. Ser. 2003, 1239, 575–579. [Google Scholar] [CrossRef]
- Marchi, N.; Winkelbach, L.; Schulz, I.; Brami, M.; Hofmanová, Z.; Blöcher, J.; Reyna-Blanco, C.S.; Diekmann, Y.; Thiéry, A.; Kapopoulou, A.; et al. The Genomic Origins of the World’s First Farmers. Cell 2022, 185, 1842−1859.e18. [Google Scholar] [CrossRef] [PubMed]
- Rohland, N.; Hofreiter, M. Ancient DNA Extraction from Bones and Teeth. Nat. Protoc. 2007, 2, 1756–1762. [Google Scholar] [CrossRef] [PubMed]
- Xavier, C.; Eduardoff, M.; Bertoglio, B.; Amory, C.; Berger, C.; Casas-Vargas, A.; Pallua, J.; Parson, W. Evaluation of DNA Extraction Methods Developed for Forensic and Ancient DNA Applications Using Bone Samples of Different Age. Genes 2021, 12, 146. [Google Scholar] [CrossRef] [PubMed]
- Nieves-Colón, M.A.; Ozga, A.T.; Pestle, W.J.; Cucina, A.; Tiesler, V.; Stanton, T.W.; Stone, A.C. Comparison of Two Ancient DNA Extraction Protocols for Skeletal Remains from Tropical Environments. Am. J. Phys. Anthropol. 2018, 166, 824–836. [Google Scholar] [CrossRef] [PubMed]
- Höss, M.; Pääbo, S. DNA Extraction from Pleistocene Bones by a Silica-Based Purification Method. Nucleic Acids Res. 1993, 21, 3913–3914. [Google Scholar] [CrossRef]
- Zeyland, J.; Wolko; Bocianowski, J.; Szalata, M.; Słomski, R.; Dzieduszycki, A.M.; Ryba, M.; Przystałowska, H.; Lipiński, D. Complete Mitochondrial Genome of Wild Aurochs (Bos Primigenius) Reconstructed from Ancient DNA. Pol. J. Vet. Sci. 2013, 16, 265–273. [Google Scholar] [CrossRef]
- Juras, A. 2012. Etnogeneza Słowian w Świetle Badań Kopalnego DNA. Ph.D. Thesis, Adam Mickiewicz University, Poznan, Poland.
- Loreille, O.M.; Diegoli, T.M.; Irwin, J.A.; Coble, M.D.; Parsons, T.J. High Efficiency DNA Extraction from Bone by Total Demineralization. Forensic Sci. Int. Genet. 2007, 1, 191–195. [Google Scholar] [CrossRef]
- Dabney, J.; Meyer, M. Extraction of Highly Degraded DNA from Ancient Bones and Teeth. In Methods in Molecular Biology; Humana Press Inc.: New York, NY, USA, 2019; Volume 1963, pp. 25–29. [Google Scholar]
- Gamba, C.; Hanghøj, K.; Gaunitz, C.; Alfarhan, A.H.; Alquraishi, S.A.; Al-Rasheid, K.A.S.; Bradley, D.G.; Orlando, L. Comparing the Performance of Three Ancient DNA Extraction Methods for High-Throughput Sequencing. Mol. Ecol. Resour. 2016, 16, 459–469. [Google Scholar] [CrossRef]
- Boessenkool, S.; Hanghøj, K.; Nistelberger, H.M.; der Sarkissian, C.; Gondek, A.T.; Orlando, L.; Barrett, J.H.; Star, B. Combining Bleach and Mild Predigestion Improves Ancient DNA Recovery from Bones. Mol. Ecol. Resour. 2017, 17, 742–751. [Google Scholar] [CrossRef]
- Yang, D.Y.; Eng, B.; Waye, J.S.; Dudar, J.C.; Saunders, S.R. Improved DNA Extraction from Ancient Bones Using Silica-Based Spin Columns. Am. J. Phys. Anthropol. 1998, 105, 539–543. [Google Scholar] [CrossRef]
- Wales, N.; Kistler, L. Extraction of Ancient DNA from Plant Remains. In Methods in Molecular Biology; Humana Press Inc.: New York, NY, USA, 2019; Volume 1963, pp. 45–55. [Google Scholar]
- Tuross, N. The Biochemistry of Ancient DNA in Bone. Experientia 1994, 50, 530–535. [Google Scholar] [CrossRef] [PubMed]
- Brzobohatá, K.; Drozdová, E.; Smutný, J.; Zeman, T.; Beňuš, R. Comparison of Suitability of the Most Common Ancient DNA Quantification Methods. Genet. Test. Mol. Biomark. 2017, 21, 265–271. [Google Scholar] [CrossRef]
- Veeramah, K.R.; Hammer, M.F. The Impact of Whole-Genome Sequencing on the Reconstruction of Human Population History. Nat. Rev. Genet. 2014, 15, 149–162. [Google Scholar] [CrossRef] [PubMed]
- Lipiński, D.; Przystałowska, H.; Szalata, M.; Zeyland, J.; Wielgus, K.; Frąckowiak, H.; Dzieduszycki, A.M.; Ryba, M.S.; Słomski, R. Biotechnology in the Restoration of Extinct Animal Species. An Analysis of Genomic and Mitochondrial DNA of Aurochs, BioTechnologia 2011, 92, 13–21. [Google Scholar]
- Lee, P.L.M.; Prys-Jones, R.P. Extracting DNA from Museum Bird Eggs, and Whole Genome Amplification of Archive DNA. Mol. Ecol. Resour. 2008, 8, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Delamoye, M.; Duverneuil, C.; Riva, K.; Leterreux, M.; Taieb, S.; de Mazancourt, P. False Homozygosities at Various Loci Revealed by Discrepancies between Commercial Kits: Implications for Genetic Databases. Forensic Sci. Int. 2004, 143, 47–52. [Google Scholar] [CrossRef]
- Harder, M.; Renneberg, R.; Meyer, P.; Krause-Kyora, B.; von Wurmb-Schwark, N. STR-Typing of Ancient Skeletal Remains: Which Multiplex-PCR Kit Is the Best. Croat. Med. J. 2012, 53, 416–422. [Google Scholar] [CrossRef]
- Tucker, V.C.; Kirkham, A.J.; Hopwood, A.J. Forensic Validation of the PowerPlex® ESI 16 STR Multiplex and Comparison of Performance with AmpFlSTR® SGM Plus. Int. J. Leg. Med. 2012, 126, 345–356. [Google Scholar] [CrossRef]
- Bini, C.; Cilli, E.; Sarno, S.; Traversari, M.; Fontani, F.; Boattini, A.; Pelotti, S.; Luiselli, D. Twenty-Seven Y-Chromosome Short Tandem Repeats Analysis of Italian Mummies of the 16th and 18th Centuries: An Interdisciplinary Research. Front. Genet. 2021, 12, 1629. [Google Scholar] [CrossRef]
- Gaudin, M.; Desnues, C. Hybrid Capture-Based Next Generation Sequencing and Its Application to Human Infectious Diseases. Front. Microbiol. 2018, 9, 2924. [Google Scholar] [CrossRef]
- Soares, A.E.R. Hybridization Capture of Ancient DNA Using RNA Baits. Methods Mol. Biol. 2019, 1963, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.; Scholl, U.I.; Ji, W.; Liu, T.; Tikhonova, I.R.; Zumbo, P.; Nayir, A.; Bakkaloǧlu, A.; Özen, S.; Sanjad, S.; et al. Genetic Diagnosis by Whole Exome Capture and Massively Parallel DNA Sequencing. Proc. Natl. Acad. Sci. USA 2009, 106, 19096–19101. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, M.L.; Buenrostro, J.D.; Valdiosera, C.; Schroeder, H.; Allentoft, M.E.; Sikora, M.; Rasmussen, M.; Gravel, S.; Guillén, S.; Nekhrizov, G.; et al. Pulling out the 1%: Whole-Genome Capture for the Targeted Enrichment of Ancient DNA Sequencing Libraries. Am. J. Hum. Genet. 2013, 93, 852–864. [Google Scholar] [CrossRef] [PubMed]
- Enk, J.M.; Devault, A.M.; Kuch, M.; Murgha, Y.E.; Rouillard, J.M.; Poinar, H.N. Ancient Whole Genome Enrichment Using Baits Built from Modern DNA. Mol. Biol. Evol. 2014, 31, 1292–1294. [Google Scholar] [CrossRef]
- Eduardoff, M.; Xavier, C.; Strobl, C.; Casas-Vargas, A.; Parson, W. Optimized MtDNA Control Region Primer Extension Capture Analysis for Forensically Relevant Samples and Highly Compromised MtDNA of Different Age and Origin. Genes 2017, 8, 237. [Google Scholar] [CrossRef]
- Loreille, O.; Ratnayake, S.; Bazinet, A.L.; Stockwell, T.B.; Sommer, D.D.; Rohland, N.; Mallick, S.; Johnson, P.L.F.; Skoglund, P.; Onorato, A.J.; et al. Biological Sexing of a 4000-Year-Old Egyptian Mummy Head to Assess the Potential of Nuclear DNA Recovery from the Most Damaged and Limited Forensic Specimens. Genes 2018, 9, 135. [Google Scholar] [CrossRef]
- Armbrecht, L.; Hallegraeff, G.; Bolch, C.J.S.; Woodward, C.; Cooper, A. Hybridisation Capture Allows DNA Damage Analysis of Ancient Marine Eukaryotes. Sci. Rep. 2021, 11, 3220. [Google Scholar] [CrossRef]
- Mohandesan, E.; Speller, C.F.; Peters, J.; Uerpmann, H.P.; Uerpmann, M.; de Cupere, B.; Hofreiter, M.; Burger, P.A. Combined Hybridization Capture and Shotgun Sequencing for Ancient DNA Analysis of Extinct Wild and Domestic Dromedary Camel. Mol. Ecol. Resour. 2017, 17, 300–313. [Google Scholar] [CrossRef]
- Schuenemann, V.J.; Bos, K.; DeWitte, S.; Schmedes, S.; Jamieson, J.; Mittnik, A.; Forrest, S.; Coombes, B.K.; Wood, J.W.; Earn, D.J.D.; et al. Targeted Enrichment of Ancient Pathogens Yielding the PPCP1 Plasmid of Yersinia Pestis from Victims of the Black Death. Proc. Natl. Acad. Sci. USA 2011, 108, E746–E752. [Google Scholar] [CrossRef]
- Feldman, M.; Harbeck, M.; Keller, M.; Spyrou, M.A.; Rott, A.; Trautmann, B.; Scholz, H.C.; Päffgen, B.; Peters, J.; McCormick, M.; et al. A High-Coverage Yersinia Pestis Genome from a Sixth-Century Justinianic Plague Victim. Mol. Biol. Evol. 2016, 33, 2911–2923. [Google Scholar] [CrossRef]
- Spyrou, M.A.; Tukhbatova, R.I.; Wang, C.C.; Valtueña, A.A.; Lankapalli, A.K.; Kondrashin, V.V.; Tsybin, V.A.; Khokhlov, A.; Kühnert, D.; Herbig, A.; et al. Analysis of 3800-Year-Old Yersinia Pestis Genomes Suggests Bronze Age Origin for Bubonic Plague. Nat. Commun. 2018, 9, 2234. [Google Scholar] [CrossRef] [PubMed]
- Mamanova, L.; Coffey, A.J.; Scott, C.E.; Kozarewa, I.; Turner, E.H.; Kumar, A.; Howard, E.; Shendure, J.; Turner, D.J. Target-Enrichment Strategies for next-Generation Sequencing. Nat. Methods 2010, 7, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Chitnis, N.; Monos, D.; Dinh, A. Next-Generation Sequencing Technologies: An Overview. Hum. Immunol. 2021, 82, 801–811. [Google Scholar] [CrossRef]
- Mardis, E.R. Next-Generation Sequencing Platforms. Annu. Rev. Anal. Chem. 2013, 6, 287–303. [Google Scholar] [CrossRef]
- Carøe, C.; Gopalakrishnan, S.; Vinner, L.; Mak, S.S.T.; Sinding, M.H.S.; Samaniego, J.A.; Wales, N.; Sicheritz-Pontén, T.; Gilbert, M.T.P. Single-Tube Library Preparation for Degraded DNA. Methods Ecol. Evol. 2018, 9, 410–419. [Google Scholar] [CrossRef]
- Gansauge, M.T.; Meyer, M. Single-Stranded DNA Library Preparation for the Sequencing of Ancient or Damaged DNA. Nat. Protoc. 2013, 8, 737–748. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, X.; Tang, P.S.; O’Leary, G.M.; Zhang, M. Targeted Sequencing of Both DNA Strands Barcoded and Captured Individually by RNA Probes to Identify Genome-Wide Ultra-Rare Mutations. Sci. Rep. 2017, 7, 3356. [Google Scholar] [CrossRef] [PubMed]
- Bentley, D.R.; Balasubramanian, S.; Swerdlow, H.P.; Smith, G.P.; Milton, J.; Brown, C.G.; Hall, K.P.; Evers, D.J.; Barnes, C.L.; Bignell, H.R.; et al. Accurate Whole Human Genome Sequencing Using Reversible Terminator Chemistry. Nature 2008, 456, 53–59. [Google Scholar] [CrossRef]
- Gansauge, M.T.; Gerber, T.; Glocke, I.; Korlević, P.; Lippik, L.; Nagel, S.; Riehl, L.M.; Schmidt, A.; Meyer, M. Single-Stranded DNA Library Preparation from Highly Degraded DNA Using T4 DNA Ligase. Nucleic Acids Res. 2017, 45, e79. [Google Scholar] [CrossRef]
- Barlow, A.; Fortes, G.M.G.; Dalen, L.; Pinhasi, R.; Gasparyan, B.; Rabeder, G.; Frischchauf, C.; Paijmans, J.L.A.; Hofreiter, M. Massive Influence of DNA Isolation and Library Preparation Approaches on Palaeogenomic Sequencing Data. BioRxiv 2016, 075911. [Google Scholar] [CrossRef]
- Sandoval-Velasco, M.; Lundstrøm, I.K.C.; Wales, N.; Ávila-Arcos, M.C.; Schroeder, H.; Gilbert, M.T.P. Relative Performance of Two DNA Extraction and Library Preparation Methods on Archaeological Human Teeth Samples. Sci. Technol. Archaeol. Res. 2017, 3, 80–88. [Google Scholar] [CrossRef]
- Zavala, E.I.; Thomas, J.T.; Sturk-Andreaggi, K.; Daniels-Higginbotham, J.; Meyers, K.K.; Barrit-Ross, S.; Aximu-Petri, A.; Richter, J.; Nickel, B.; Berg, G.E.; et al. Ancient DNA Methods Improve Forensic DNA Profiling of Korean War and World War II Unknowns. Genes 2022, 13, 129. [Google Scholar] [CrossRef] [PubMed]
- Pluta, D.; Lebioda, A.; Jonkisz, A.; Dobosz, T. First Successful DNA Isolation and Profiling from Bone Using an Approach That Is Non-Destructive toward Bone Surface. Arch. Med. Sądowej i Kryminol. 2016, 66, 65–70. [Google Scholar] [CrossRef]
| Year of Discovery | Methods | Sample Material | Result | References |
|---|---|---|---|---|
| 1984 | Molecular cloning, Sanger’s sequencing | Dried muscle tissue, quagga specimen | Two sequenced mitochondrial DNA fragments (117 and 112 bp). First recovered aDNA. | [2] |
| 1988 | PCR | Dried muscle, quagga specimen | Detected cloning artefacts previously unnoticed in [2] with PCR. | [16] |
| 1988 | Molecular cloning, PCR | Numerous different ancient samples | Comparing the usefulness of molecular cloning and PCR in aDNA research. | [1] |
| 1991 | PCR | Human brain tissue, 6990–8130 years old | Sequenced fragments of 6 nuclear genes. | [26] |
| 1998 | PCR | Coprolite | Amplification of DNA from ancient feces. Analysis of the diet of the specimen and identification of species of the specimen. | [27] |
| 2003 | PCR | Sediment | First analysis of environmental aDNA | [28] |
| 2005 | PCR | Bones, teeth | Intact stretches of mitochondrial DNA from 24 Neolithic skeletons. | [29] |
| 2006 | NGS | Woolly mammoth’s mandible | 28 million bp sequenced, 13 million bp were endogenous. First use of NGS in paleogenetics. Analyses of the metagenomic nature of ancient remains. | [20] |
| 2008 | NGS | Woolly mammoth’s hair | 4.17 billion bp sequenced, 3.3 billion of which were endogenous | [30] |
| 2008 | NGS | Neanderthal bone | Fully sequenced Neanderthal mitochondrial genome | [31] |
| 2010 | NGS | 21 Neanderthal bones, 3 selected for further analysis | First sequenced Neanderthal genome (1.2× coverage), evidence for Neanderthals interbreeding with anatomically modern humans | [22] |
| 2010 | NGS | Finger bone | Discovery of Denisovans and sequenced Denisovan genome | [23] |
| 2010 | NGS | Hair | First sequenced ancient human genome (Paleo-Inuit) | [21] |
| 2011 | NGS | Teeth, bones | First fully sequenced genome of ancient bacterial pathogen | [32] |
| 2012 | NGS | Finger bone | First high coverage (30×) of Denisovan genome, use of single-stranded library preparation. | [33] |
| 2012 | NGS | Bone from the mummy of Tyrolean Iceman | Genome of Tyrolean Iceman fully sequenced, analysis of phenotype and metagenome | [34] |
| 2014 | NGS | Ancient calcified dental plaque | First high-resolution taxonomic and proteomic analysis of ancient oral microbiome from calcified dental plaque | [35] |
| 2014 | NGS | Bones | Identification of English king Richard III | [36] |
| 2014 | NGS | Toe phalanx | High-quality sequence of Neanderthal woman genome (coverage ~50×) | [37] |
| 2015 | NGS | - | Analysis of 230 ancient Eurasian genomes to determine genome-wide patterns of selection | [8] |
| 2015 | NGS | Molar tooth, soft tissue | Complete high-quality two woolly mammoth genomes, analysis of demographic history | [6] |
| 2015 | NGS | Auroch bone | 6750-year-old auroch genome, analysis of domestication process and its impact on the genome | [5] |
| 2017 | NGS | Bone | High-coverage genome (30×) of Neanderthal from Vindija Cave, analysis of gene flow between Neanderthals, Denisovans and anatomically modern humans | [38] |
| 2020 | NGS | - | Sequencing of 442 genomes from archaeological sites across Europe and Greenland to understand the expansion of the Scandinavian population during the Viking Age | [39] |
| 2020 | NGS | Finger bone | High coverage (27×) sequencing of a Neanderthal from Chagyrskaya Cave. Detection of selection patterns in Neanderthal lineage | [40] |
| 2021 | NGS | Loessal permafrost silts | Analysis of ancient sedimentary DNA from a period of 30,000 years from the central Yukon in Canada. | [14] |
| 2021 | NGS | Mammoth molars | Previous record for the oldest sequenced genome (older than 1 million years). | [25] |
| 2022 | NGS | Sediment | Current record holder for the oldest sequenced DNA | [24] |
| Criterion | Explanation |
|---|---|
| Physically isolated work area | All work on low-copy number DNA should be carried out in an isolated laboratory where no other genetic research is performed. |
| PCR control amplifications | Test laboratory environment for contamination. |
| Test the molecular behavior | Check the PCR products for unusual results. aDNA is heavily fragmented, so longer fragments should be increasingly rarer. |
| Quantitation | Check the number of starting templates. If below 1000, sporadic contamination cannot be ruled out. |
| Reproducibility | Results from the same sample material should be repeatable. |
| Clone | After sequencing, the PCR product should be cloned and sequenced in multiple copies to determine the ratio of exogenous sequences and sequencing errors resulting from aDNA damage. |
| Independent replication | The results should be reproduced in another independent laboratory. |
| Biochemical preservation | Survival of other ancient biomolecules makes the survival of aDNA more believable. |
| Associated remains | If target DNA sequences also survive in associated faunal material, it may be used as supporting evidence. |
| Phylogenetic sense | Reproducible sequences should be placed in a phylogenetic tree with other known haplotypes. |
| Damage patterns | The DNA sequences should show specific damage patterns: a high degree of fragmentation and a high concentration of substitutions on the ends of the fragments (C>T on 5′ and G>A on 3′). |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Danielewski, M.; Żuraszek, J.; Zielińska, A.; Herzig, K.-H.; Słomski, R.; Walkowiak, J.; Wielgus, K. Methodological Changes in the Field of Paleogenetics. Genes 2023, 14, 234. https://doi.org/10.3390/genes14010234
Danielewski M, Żuraszek J, Zielińska A, Herzig K-H, Słomski R, Walkowiak J, Wielgus K. Methodological Changes in the Field of Paleogenetics. Genes. 2023; 14(1):234. https://doi.org/10.3390/genes14010234
Chicago/Turabian StyleDanielewski, Mikołaj, Joanna Żuraszek, Aleksandra Zielińska, Karl-Heinz Herzig, Ryszard Słomski, Jarosław Walkowiak, and Karolina Wielgus. 2023. "Methodological Changes in the Field of Paleogenetics" Genes 14, no. 1: 234. https://doi.org/10.3390/genes14010234
APA StyleDanielewski, M., Żuraszek, J., Zielińska, A., Herzig, K.-H., Słomski, R., Walkowiak, J., & Wielgus, K. (2023). Methodological Changes in the Field of Paleogenetics. Genes, 14(1), 234. https://doi.org/10.3390/genes14010234






