The Mitochondrial Genomes of Two Parasitoid Wasps Protapanteles immunis and Parapanteles hyposidrae (Hymenoptera: Braconidae) with Phylogenetic Implications and Novel Gene Rearrangements
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Identification and DNA Extraction
2.2. Next-Generation Sequencing and Assembly
2.3. Mitochondrial Genome Annotation and Analysis
2.4. Phylogenetic Analysis
3. Results
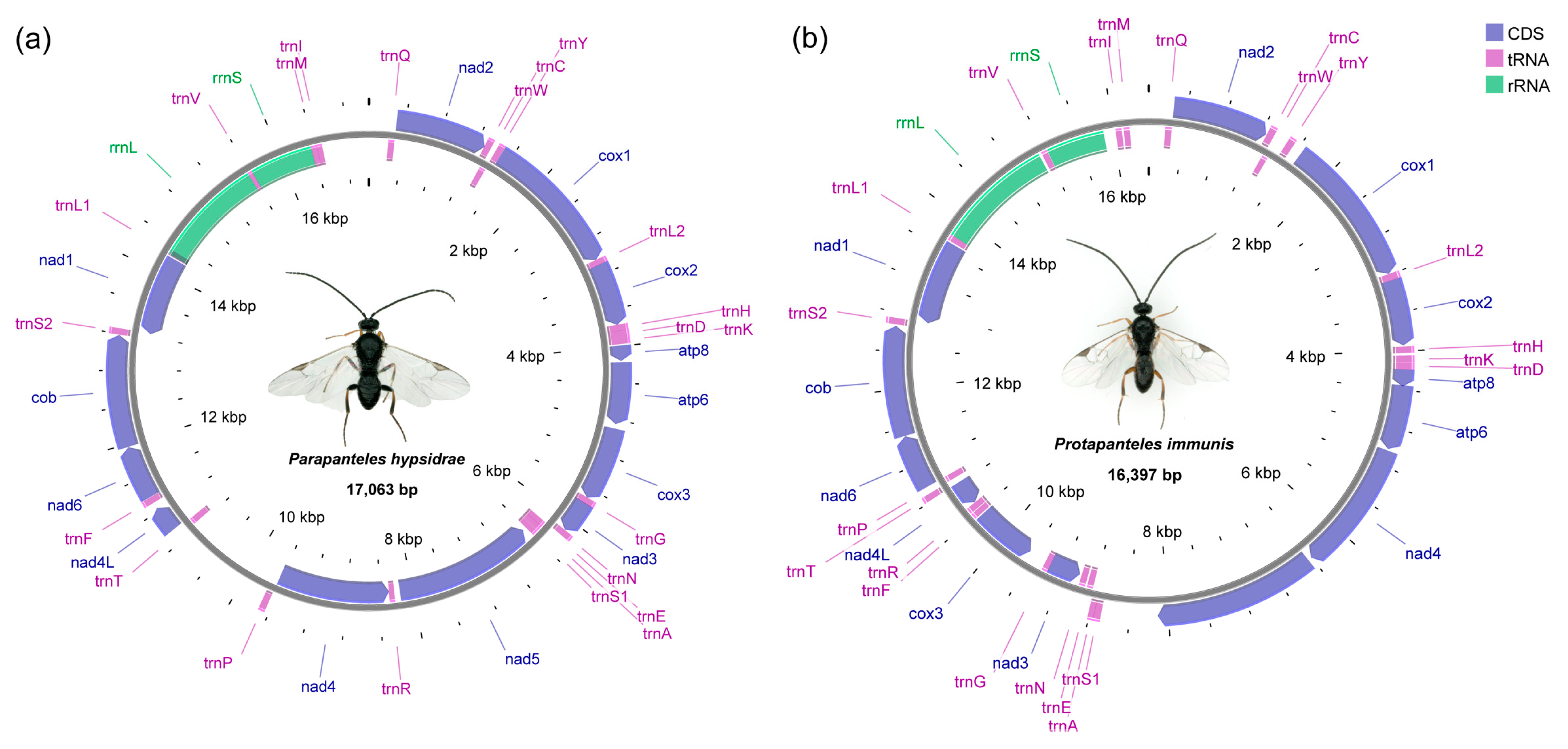
3.1. General Features of Mitochondrial Genomes
3.2. Base Composition, and Codon Usage
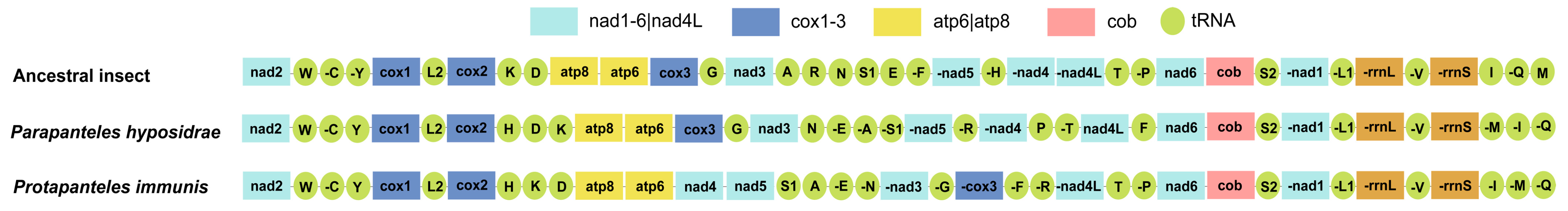
3.3. Gene Rearrangements
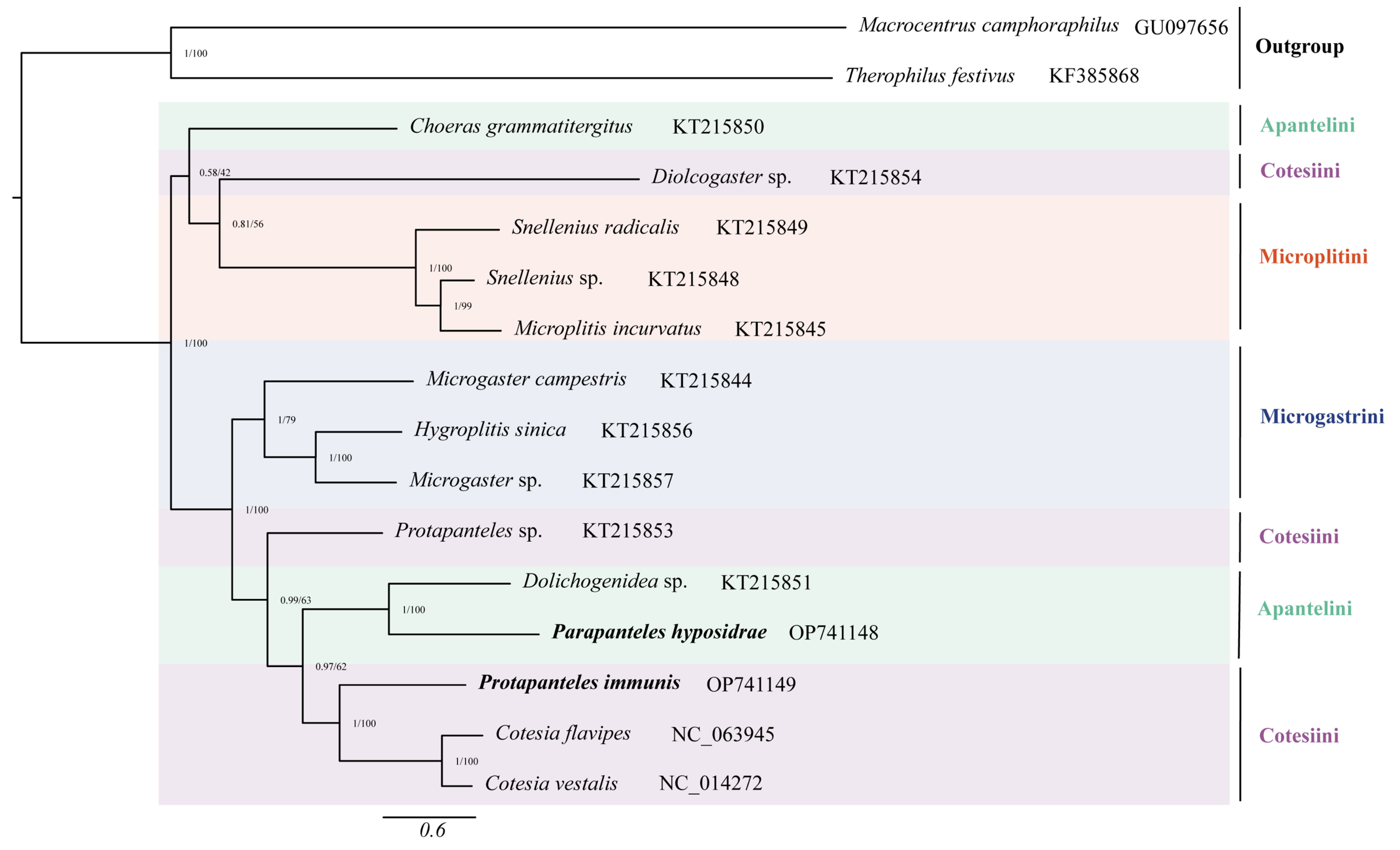
3.4. Phylogenetic Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Whitfield, J.B. Annotated checklist of the Microgastrinae of North America north of Mexico (Hymenoptera: Braconidae). J. Kans. Entomol. Soc. 1995, 68, 245–262. [Google Scholar]
- Banks, J.C.; Whitfield, J.B. Dissecting the ancient rapid radiation of microgastrine wasp genera using additional nuclear genes. Mol. Phylogenet. Evol. 2006, 41, 690–703. [Google Scholar] [CrossRef]
- Fernandez-Triana, J.; Shaw, M.R.; Boudreault, C.; Beaudin, M.; Broad, G.R. Annotated and illustrated world checklist of Microgastrinae parasitoid wasps (Hymenoptera, Braconidae). ZooKeys 2020, 920, 1. [Google Scholar] [CrossRef]
- Whitfield, J.B.; Mardulyn, P.; Austin, A.D.; Dowton, M. Phylogenetic relationships among microgasrinae braconid wasp genera based on data from the 16S, COI and 28S genes and morphology. Syst. Entomol. 2002, 27, 337–359. [Google Scholar] [CrossRef]
- Zhou, X.; Tang, P.; Wu, Q.; Guo, H.; Xiao, Q.; Chen, X.X. Identification of Two Common Larval Parasitic Wasps of Ectropis obliqua and Ectropis grisescens (Lepidoptera: Geometridae). Chin. J. Biol. Control 2022, 1–14. [Google Scholar] [CrossRef]
- Wang, Z.Q.; Zhou, X.G.; Xiao, Q.; Tang, P.; Chen, X.X. The Potential of Parapanteles hyposidrae and Protapanteles immunis (Hymenoptera: Braconidae) as Biocontrol Agents for the Tea Grey Geometrid Ectropis grisescens (Lepidoptera). Insects 2022, 13, 937. [Google Scholar] [CrossRef]
- Cameron, S.L.; Johnson, K.P.; Whiting, M.F. The mitochondrial genome of the screamer louse Bothriometopus (Phthiraptera: Ischnocera): Effects of extensive gene rearrangements on the evolution of the genome. J. Mol. Evol. 2007, 65, 589–604. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect Mitochondrial Genomics: Implications for Evolution and Phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef] [PubMed]
- Ballard, J.W.O. Comparative genomics of mitochondrial DNA in members of the Drosophila melanogaster subgroup. J. Mol. Evol. 2000, 51, 48–63. [Google Scholar] [CrossRef]
- Dowton, M.; Austin, A.D. Evolutionary dynamics of a mitochondrial rearrangement “hot spot” in the hymenoptera. Mol. Biol. Evol. 1999, 16, 298–309. [Google Scholar] [CrossRef]
- Dowton, M.; Castro, L.R.; Campbell, S.L.; Bargon, S.D.; Austin, A.D. Frequent mitochondrial gene rearrangements at the hymenopteran nad3-nad5 junction. J. Mol. Evol. 2003, 56, 517–526. [Google Scholar] [CrossRef]
- Shao, R.; Campbell, N.J.H.; Barker, S.C. Numerous gene rearrangements in the mitochondrial genome of the wallaby louse, Heterodoxus macropus (Phthiraptera). Mol. Biol. Evol. 2001, 18, 858–865. [Google Scholar] [CrossRef]
- Shao, R.F.; Campbell, N.J.H.; Schmidt, E.R.; Barker, S.C. Increased rate of gene rearrangement in the mitochondrial genomes of three orders of hemipteroid insects. Mol. Biol. Evol. 2001, 18, 1828–1832. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Zhang, L.F.; Lu, T.; Ma, J.N.; Zeng, C.J.; Yue, B.S.; Zhang, X.Y. Mitochondrial genomes of blister beetles (Coleoptera, Meloidae) and two large intergenic spacers in Hycleus genera. BMC Genom. 2017, 18, 15. [Google Scholar] [CrossRef]
- Li, Q.; Wei, S.J.; Tang, P.; Wu, Q.; Shi, M.; Sharkey, M.J.; Chen, X.X. Multiple Lines of Evidence from Mitochondrial Genomes Resolve Phylogenetic Relationships of Parasitic Wasps in Braconidae. Genome Biol. Evol. 2016, 8, 2651–2662. [Google Scholar] [CrossRef]
- Wei, S.J.; Shi, M.; Sharkey, M.J.; van Achterberg, C.; Chen, X.X. Comparative mitogenomics of Braconidae (Insecta: Hymenoptera) and the phylogenetic utility of mitochondrial genomes with special reference to Holometabolous insects. BMC Genom. 2010, 11, 16. [Google Scholar] [CrossRef]
- Zheng, X.Y.; Cao, L.J.; Chen, P.Y.; Chen, X.X.; van Achterberg, K.; Hoffmann, A.A.; Liu, J.X.; Wei, S.J. Comparative mitogenomics and phylogenetics of the stinging wasps (Hymenoptera: Aculeata). Mol. Phylogenet. Evol. 2021, 159, 9. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S.; Fast, Q.C. Available online: https://qubeshub.org/resources/fastqc (accessed on 30 December 2021).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Crampton-Platt, A.; Timmermans, M.; Gimmel, M.L.; Kutty, S.N.; Cockerill, T.D.; Khen, C.V.; Vogler, A.P. Soup to Tree: The Phylogeny of Beetles Inferred by Mitochondrial Metagenomics of a Bornean Rainforest Sample. Mol. Biol. Evol. 2015, 32, 2302–2316. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Peng, Y.; Leung, H.C.M.; Yiu, S.M.; Chin, F.Y.L. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Juhling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Putz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11 Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Calcott, B.; Ho, S.Y.W.; Guindon, S. PartitionFinder: Combined Selection of Partitioning Schemes and Substitution Models for Phylogenetic Analyses. Mol. Biol. Evol. 2012, 29, 1695–1701. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Tang, P.; Zhu, J.C.; Zheng, B.Y.; Wei, S.J.; Sharkey, M.; Chen, X.X.; Vogler, A.P. Mitochondrial phylogenomics of the Hymenoptera. Mol. Phylogenet. Evol. 2019, 131, 8–18. [Google Scholar] [CrossRef]
- Cameron, S.L.; Dowton, M.; Castro, L.R.; Ruberu, K.; Whiting, M.F.; Austin, A.D.; Diement, K.; Stevens, J. Mitochondrial genome organization and phylogeny of two vespid wasps. Genome 2008, 51, 800–808. [Google Scholar] [CrossRef] [PubMed]
- Cha, S.Y.; Yoon, H.J.; Lee, E.M.; Yoon, M.H.; Hwang, J.S.; Jin, B.R.; Han, Y.S.; Kim, I. The complete nucleotide sequence and gene organization of the mitochondrial genome of the bumblebee, Bombus ignitus (Hymenoptera: Apidae). Gene 2007, 392, 206–220. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.J.; Shi, M.; He, J.H.; Sharkey, M.; Chen, X.X. The complete mitochondrial genome of Diadegma semiclausum (Hymenoptera: Ichneumonidae) indicates extensive independent evolutionary events. Genome 2009, 52, 308–319. [Google Scholar] [CrossRef] [PubMed]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Ruparel, H.; Bi, L.R.; Li, Z.M.; Bai, X.P.; Kim, D.H.; Turro, N.J.; Ju, J.Y. Design and synthesis of a 3 ‘-O-allyl photocleavable fluorescent nucleotide as a reversible terminator for DNA sequencing by synthesis. Proc. Natl. Acad. Sci. USA 2005, 102, 5932–5937. [Google Scholar] [CrossRef]
- Oliveira, D.; Raychoudhury, R.; Lavrov, D.V.; Werren, J.H. Rapidly evolving mitochondrial genome and directional selection in mitochondrial genes in the parasitic wasp Nasonia (Hymenoptera: Pteromalidae). Mol. Biol. Evol. 2008, 25, 2167–2180. [Google Scholar] [CrossRef]
- Wei, S.J.; Li, Q.; van Achterberg, K.; Chen, X.X. Two mitochondrial genomes from the families Bethylidae and Mutillidae: Independent rearrangement of protein-coding genes and higher-level phylogeny of the Hymenoptera. Mol. Phylogenet. Evol. 2014, 77, 1–10. [Google Scholar] [CrossRef]
- Ma, Y.; Zheng, B.Y.; Zhu, J.C.; van Achterberg, C.; Tang, P.; Chen, X.X. The first two mitochondrial genomes of wood wasps (Hymenoptera: Symphyta): Novel gene rearrangements and higher-level phylogeny of the basal hymenopterans. Int. J. Biol. Macromol. 2019, 123, 1189–1196. [Google Scholar] [CrossRef]
- Zhu, J.C.; Tang, P.; Zheng, B.Y.; Wu, Q.; Wei, S.J.; Chen, X.X. The first two mitochondrial genomes of the family Aphelinidae with novel gene orders and phylogenetic implications. Int. J. Biol. Macromol. 2018, 118, 386–396. [Google Scholar] [CrossRef] [PubMed]
- Mason, W.R.M. The polyphyletic nature of Apanteles Förster (Hymenoptera: Braconidae): A phylogeny and reclassification of Microgastrinae. Mem. Entomol. Soc. Can. 1981, 113, 1–147. [Google Scholar] [CrossRef]
- Dowton, M.; Austin, A.D. Phylogenetic Relationships among the Microgastroid Wasps (Hymenoptera: Braconidae): Combined Analysis of 16S and 28S rDNA Genes and Morphological Data. Mol. Phylogenet. Evol. 1998, 10, 354. [Google Scholar] [CrossRef] [PubMed]

| Species | Whole Genome | Protein-Coding Genes | ||||
|---|---|---|---|---|---|---|
| Length (bp) | A + T (%) | Length (bp) | A + T (%) | AT-Skew | GC-Skew | |
| Pa. hypsidrae | 17,063 | 86.15 | 11,311 | 85.12 | −0.0696 | 0.1266 |
| Pr. immunis | 16,397 | 86.56 | 11,088 | 85.52 | −0.1040 | 0.0847 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, D.; Wang, Z.; Zhu, J.; Zhou, X.; Tang, P.; Chen, X. The Mitochondrial Genomes of Two Parasitoid Wasps Protapanteles immunis and Parapanteles hyposidrae (Hymenoptera: Braconidae) with Phylogenetic Implications and Novel Gene Rearrangements. Genes 2023, 14, 230. https://doi.org/10.3390/genes14010230
Xiao D, Wang Z, Zhu J, Zhou X, Tang P, Chen X. The Mitochondrial Genomes of Two Parasitoid Wasps Protapanteles immunis and Parapanteles hyposidrae (Hymenoptera: Braconidae) with Phylogenetic Implications and Novel Gene Rearrangements. Genes. 2023; 14(1):230. https://doi.org/10.3390/genes14010230
Chicago/Turabian StyleXiao, Dandan, Ziqi Wang, Jiachen Zhu, Xiaogui Zhou, Pu Tang, and Xuexin Chen. 2023. "The Mitochondrial Genomes of Two Parasitoid Wasps Protapanteles immunis and Parapanteles hyposidrae (Hymenoptera: Braconidae) with Phylogenetic Implications and Novel Gene Rearrangements" Genes 14, no. 1: 230. https://doi.org/10.3390/genes14010230
APA StyleXiao, D., Wang, Z., Zhu, J., Zhou, X., Tang, P., & Chen, X. (2023). The Mitochondrial Genomes of Two Parasitoid Wasps Protapanteles immunis and Parapanteles hyposidrae (Hymenoptera: Braconidae) with Phylogenetic Implications and Novel Gene Rearrangements. Genes, 14(1), 230. https://doi.org/10.3390/genes14010230






