Abstract
The SOX transcription factor family is pivotal in controlling aspects of development. To identify genotype–phenotype relationships of SOX proteins, we performed a non-biased study of SOX using 1890 open-reading frame and 6667 amino acid sequences in combination with structural dynamics to interpret 3999 gnomAD, 485 ClinVar, 1174 Geno2MP, and 4313 COSMIC human variants. We identified, within the HMG (High Mobility Group)- box, twenty-seven amino acids with changes in multiple SOX proteins annotated to clinical pathologies. These sites were screened through Geno2MP medical phenotypes, revealing novel SOX15 R104G associated with musculature abnormality and SOX8 R159G with intellectual disability. Within gnomAD, SOX18 E137K (rs201931544), found within the HMG box of ~0.8% of Latinx individuals, is associated with seizures and neurological complications, potentially through blood–brain barrier alterations. A total of 56 highly conserved variants were found at sites outside the HMG-box, including several within the SOX2 HMG-box-flanking region with neurological associations, several in the SOX9 dimerization region associated with Campomelic Dysplasia, SOX14 K88R (rs199932938) flanking the HMG box associated with cardiovascular complications within European populations, and SOX7 A379V (rs143587868) within an SOXF conserved far C-terminal domain heterozygous in 0.716% of African individuals with associated eye phenotypes. This SOX data compilation builds a robust genotype-to-phenotype association for a gene family through more robust ortholog data integration.
1. Introduction
SOX (SRY-related HMG box) genes are involved in organ development and cell fate decisions [1]. These molecular switches can be improperly activated or dysregulated in numerous disease states. The first SOX gene discovered, the testes-determining factor SRY, was mapped by identifying sex-reversal variants for XY females [2,3]. Since that initial discovery, 19 additional SOX genes have been identified in humans, resulting in 20 genes with orthologs found in bilaterians [4,5]. All SOX genes encode a highly conserved high mobility group (HMG) box that binds and bends DNA, serving as an architectural and recruitment domain essential to gene regulation [6,7,8,9]. While the functional roles of some domains and regions outside the HMG box have been determined, the bulk of the work on SOX proteins has been carried out on the shared HMG box. This is primarily because these proteins are intrinsically disordered and difficult to validate functionally. With the rapid increase in sequenced genomes for rare, monogenic disorders, an informatics analysis of this gene family is needed now more than ever to prioritize investment in robust, comprehensive SOX gene knowledge relevant to human diseases.
Pathogenic genomic variants have been identified in multiple SOX genes (Table 1 and Table 2). As indicated, SRY variants are associated with sex reversal [10]. SOX9 variants also result in sex reversal, with a well-established role in Campomelic Dysplasia [11], a severe skeletal dysplasia. Genetic variants within SOX2 are associated with anophthalmia and neural alterations [12,13], SOX3 with altered pituitary development [14], SOX10 with Waardenburg syndrome and Hirschsprung’s disease [15], SOX11 with Coffin–Siris syndrome [16], and SOX18 with Hypotrichosis–Lymphedema–Telangiectasia Syndrome (HLTRS) [17]. With the additive disease risks of variants in SOX genes, ongoing work has continued to define SOXopathies and summarize the phenotypic outcomes of modulating the gene family [18]. Recent work has suggested several cancer variants associated with gain-of-function variants in SOX17 based on the screening of cancer genomic databases, furthering the reach of SOX variants into somatic variant risks [19].
In this study, we systematically assessed vertebrate SOX sequences to identify amino acid variations and functional regions. We assessed genomic variants in multiple public human-sequencing databases, utilizing deep conservation analysis to rank the potential impact of identified variants. Initial analysis compiling disease variants onto the sequences of HMG proteins showed that most rare disease variants occurred at sites conserved across the gene family, at structurally essential regions of the HMG box [20]. Not only have SOX genes been linked to numerous rare diseases, but mutations contributing to altered protein function and gene regulation have been associated with multiple forms of cancer [21]. Building on those studies, we provide an analysis of the genomic landscape for each of the 20 SOX genes across vertebrate evolution to assess human variants.
2. Materials and Methods
2.1. Sequence and Structure Analysis
SOX gene/protein evolution was analyzed using our Sequence-to-Structure-to-Function analysis [22,23]. In short, open reading frame sequences (ORF) were isolated from NCBI so that only one sequence was used per species per gene. Sequences were aligned using the ClustalW [24] codon. Codon selection was called using a Muse–Gaut model of codon substitution [25] and dN/dS was called using HyPhy [26]. Any sequences with ambiguity or missing >9 nucleotides found in >90% of sequences for a gene were removed. All SOX sequences were aligned for the HMG box, and a phylogenetic tree was created using maximum likelihood with 1000 bootstrap analyses [27,28]. For each gene, the conservation score, a metric combining dN-dS and amino acid conservation [22], was placed on a 21-codon sliding window by adding the score of each position, with 10 upstream and 10 downstream. Domain annotations were identified using UniProt [29], and unknown domains were analyzed using ELM software [30]. Amino acid alignments were generated from NCBI gene orthologs [31] in November 2022, followed by alignment with COBALT [32]. The use of each amino acid across the 20 SOX proteins was called and colored onto the structure of SRY (PDB 1J46). Each protein was then modeled using 1J46 with YASARA homology modeling [33] and run for ten nanoseconds on molecular dynamic simulations (mds), as previously published in our extensive work on the HMG structures of SOX proteins [20,34]. The SOX18 E137K variant was assessed using PDB structure 4y60 [35] and processed for the wild type and variants over 20 nanoseconds of mds.
2.2. Genomic Variant Analysis
Genomic variants and phenotypic details were extracted from gnomAD [36], ClinVar [37], and Geno2MP [38] in November 2022. The Catalog of Somatic Mutations in Cancer (COSMIC) [39] was extracted in December 2022. Variants were called relative to the proximity to the HMG box, conservation in codon selection, conservation from amino acid sequence alignments, and frequency of the variant from each database. Further analysis of linkage disequilibrium using the phase 3 1000 genomes project was calculated using rAggr [40] with a specific focus on MXL and PEL populations. Additional disease-causing variants were curated from the literature (Table 1).
2.3. SOX18 Culture Experiments
For luciferase assays, HeLa cells were seeded in triplicate at 6.6 × 103 cells/cm2 in 24-well cassettes, cultured for 24 hr, and cotransfected using ViaFect. In each transfection reaction, we used 50 ng of effector plasmid (pHTC-Halo/SOX18, pHTC-Halo/SOX18-E137K, or empty pHTC-HaloTag vector), 500 ng of SOX-inducible luciferase reporter (as described previously [41]), and 1 ng of control vector (phRL-null Renilla). The cells were incubated for 30 hr and assessed using the Dual-Luciferase®® Reporter Assay System measured on a GloMax®® Luminometer. Each effector’s relative transcriptional activity was measured after normalizing Firefly luciferase to Renilla luciferase and then calculating the fold change relative to the empty effector plasmid.
The Amplified Luminescent Proximity Homogeneous Assay (Alpha) screening for SOX18 was performed as previously described [34,42,43]. A STRING [44] network map of SOX18, RBPJ, and MEF2C was created using no more than ten partners in both shells one and two of the network. GO enrichment was then assessed on the STRING tool.

Table 1.
SOX gene variants with potential functional outcomes extracted from the literature. Notes: a HMG box AA numbers are listed first based on UniProt and then listed based on Bowles/Koopman, where there is an offset of 2 bases in the annotations. References can be found in [10,17,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78].
Table 1.
SOX gene variants with potential functional outcomes extracted from the literature. Notes: a HMG box AA numbers are listed first based on UniProt and then listed based on Bowles/Koopman, where there is an offset of 2 bases in the annotations. References can be found in [10,17,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78].
3. Results
3.1. Sequence Evolution of SOX Genes/Proteins
A total of 1890 open reading frame (ORF) sequences of vertebrates were curated for 20 SOX genes, averaging just under 100 species per gene analyzed. In addition, we utilized more up-to-date (as of November 2022) amino acid multiple-species comparisons with 6667 sequences averaging 333 species for each SOX protein. The ORF alignments of the SOX genes allowed for the analysis of selection throughout each of the genes. Following codon and amino acid analysis for each of the 20 genes, a sliding window metric [22] was applied to identify conserved domains and linear motifs within each gene normalized such that the highest motif had a value of 1 (Figure 1). Only the HMG domain was conserved for SOX1, SOX2, SOX3, SOX15, SOX21, SOX30, and SRY (Figure 1, red boxes). The HMG domain numbers were based on UniProt, where additional SOX-centric numbering used by others [5] was shifted by two amino acids. In general, most of the 485 ClinVar variants were within HMG domains, while the 3999 gnomAD and 1174 Geno2MP variants were mostly found outside of the HMG domain (Figure 1, Table 2). Several SOX genes had conserved domains in addition to the HMG box annotated in databases such as UniProt (Figure 1, magenta boxes), and several regions were conserved without previous annotation (Figure 1, gray boxes). There were also ClinVar annotated variants within these regions that required further analyses.
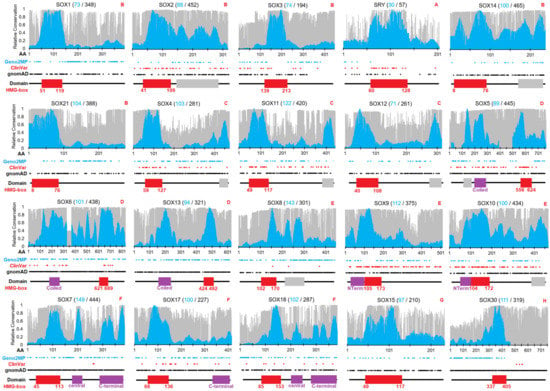
Figure 1.
Conservation and variants for each of the 20 SOX genes/proteins. In total, 1890 nucleic acid sequences (cyan) and 6667 amino acid sequences (gray) were analyzed for 20 SOX genes throughout vertebrate species. The evolutionary selection was mapped for each gene using the indicated number of sequences on a scale of 0 (weak conservation) to 1 (high conservation). Shown below the conservation data are the locations of human variants from Geno2MP (cyan), ClinVar (red), and gnomAD (black). UniProt-designated domains are shown below, each with the HMG box identified in red. HMG box human numbers are shown below the domain. Red letters at top right corner indicate the SOX subfamily annotation. The raw TIF file can be found at https://doi.org/10.6084/m9.figshare.21830343.v1 (accessed on 18 December 2022).

Table 2.
SOX genes with known disease associations and mapping data for genomic variants. In red at the bottom are averages or total numbers for various columns. AD—autosomal dominant; XL—X-linked; XLD—X-linked dominant; AR—autosomal recessive; NA—nucleic acid sequences; AA—amino acid sequences.
Table 2.
SOX genes with known disease associations and mapping data for genomic variants. In red at the bottom are averages or total numbers for various columns. AD—autosomal dominant; XL—X-linked; XLD—X-linked dominant; AR—autosomal recessive; NA—nucleic acid sequences; AA—amino acid sequences.
| Gene | Group | Ensembl | NCBI | Missense Constraint | OMIM/ClinVar | Inheritance | AA | Species NA | Species AA | gnomAD | ClinVar | Geno2MP | COSMIC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SOX1 | B | ENST00000330949 | NP 005977 | 0.58 | - | - | 391 | 73 | 348 | 120 | 0 | 115 | 134 |
| SOX2 | B | ENST00000325404 | NP 003097 | 2.12 | Microphthalmia | AD | 317 | 88 | 452 | 119 | 70 | 66 | 203 |
| SOX3 | B | ENST00000370536 | NP 005625 | 2.21 | Panhypopituitarism | XL | 446 | 74 | 194 | 104 | 18 | 39 | 226 |
| SRY | A | ENST00000383070 | NP 003131 | −0.14 | sex reversal | XLD | 204 | 30 | 57 | 25 | 26 | 5 | 25 |
| SOX14 | B | ENST00000306087 | NP 004180 | 1.14 | - | - | 240 | 100 | 465 | 118 | 0 | 20 | 112 |
| SOX21 | B | ENST00000376945 | NP 009015 | 1.74 | - | - | 276 | 104 | 388 | 81 | 0 | 60 | 79 |
| SOX4 | C | ENST00000244745 | NP 003098 | 1.17 | Coffin-Siris syndrome | AD | 474 | 103 | 281 | 210 | 27 | 64 | 125 |
| SOX11 | C | ENST00000322002 | NP 003099 | 2.26 | Coffin-Siris syndrome | AD | 441 | 122 | 420 | 166 | 47 | 38 | 282 |
| SOX12 | C | ENST00000342665 | NP 008874 | 1.79 | - | - | 315 | 71 | 261 | 106 | 1 | 27 | 63 |
| SOX5 | D | ENST00000451604 | NP 008871 | 3.15 | Lamb-Shaffer syndrome | AD | 763 | 89 | 445 | 264 | 46 | 51 | 480 |
| SOX6 | D | ENST00000528429 | NP 001354802 | 1.94 | Tolchin-Le Caignec syndrome | AD | 828 | 101 | 438 | 365 | 17 | 86 | 380 |
| SOX13 | D | ENST00000367204 | NP 005677 | 1.74 | - | - | 622 | 94 | 321 | 296 | 0 | 91 | 199 |
| SOX8 | E | ENST00000293894 | NP 055402 | 0.89 | - | - | 446 | 143 | 301 | 254 | 3 | 98 | 183 |
| SOX9 | E | ENST00000245479 | NP 000337 | 1.63 | Campomelic dysplasia | AD | 509 | 112 | 375 | 259 | 97 | 55 | 524 |
| SOX10 | E | ENST00000396884 | NP 008872 | 2.77 | PCWH syndrome | AD | 466 | 100 | 434 | 176 | 100 | 44 | 198 |
| SOX7 | F | ENST0000030450 | NP 113627 | −0.74 | - | - | 388 | 149 | 444 | 420 | 2 | 81 | 204 |
| SOX17 | F | ENST00000297316 | NP 071899 | 0.77 | Vesicoureteral reflux | AD | 414 | 100 | 227 | 225 | 18 | 63 | 424 |
| SOX18 | F | ENST00000340356 | NP 060889 | 0.86 | HLTS | AD/AR | 384 | 102 | 287 | 151 | 9 | 75 | 93 |
| SOX15 | G | ENST00000250055 | NP 008873 | 0.62 | - | - | 233 | 97 | 210 | 118 | 0 | 29 | 83 |
| SOX30 | H | ENST00000265007 | NP 848511 | 0.78 | Male infertility | AD | 753 | 111 | 319 | 422 | 4 | 67 | 296 |
| Average | 1 | Total | 8910 | 1963 | 6667 | 3999 | 485 | 1174 | 4313 |
3.2. Mapping SOX Variants
Deep evolutionary analysis of the SOX genes has the potential to identify functional regions and amino acids, prioritizing variants of interest for future analyses. Advanced metrics to look at the general intolerance of mutations in genes across the entire genome, such as RVIS scores [79], suggest that SOX genes are generally tolerant to human variants, with a value around the 37th percentile of all genes (SOX2—45.64%; SOX4—31.22%; SOX5—5.30%; SOX6—4.93%; SOX7—22.47%; SOX8—76.58%; SOX9—9.40%; SOX10—27.70%; SOX11—33.83%; SOX13—61.79%; SOX14—40.18%; SOX15—48.78%; SOX17—34.82%; SOX30—73.37%). The RVIS (residual variation intolerance score) represents a value for how much a gene can tolerate variants, whereas most transcription factor scores suggest that they can tolerate changes. However, these metrics are biased based on the degree of domain and motif conservation relative to the size of the protein. The relative Z-scores of gnomAD variants (missense constraint, Table 2) computed based on the predicted number of variants based on all human genes also suggested a neutral level of SOX variants (average of 1). Therefore, a more systematic metric of SOX-gene-to-variant impact is needed.
We identified 3623 unique amino acids with germline variant sites within SOX proteins in population genomics (gnomAD), clinical genomes with matched phenotype descriptions (Geno2MP), and clinical genomes annotated within ClinVar (Figure 2A). The largest number of these amino acids had only gnomAD annotations, while Geno2MP and the overlap of gnomAD and Geno2MP were identified as the next-largest groups. As the conservation analysis highlighted, the HMG box had a critical function for all SOX members. Only within the ClinVar database were variants found more often within the HMG-box (red, Figure 2A), with all other databases having variants rarely falling within the domain.
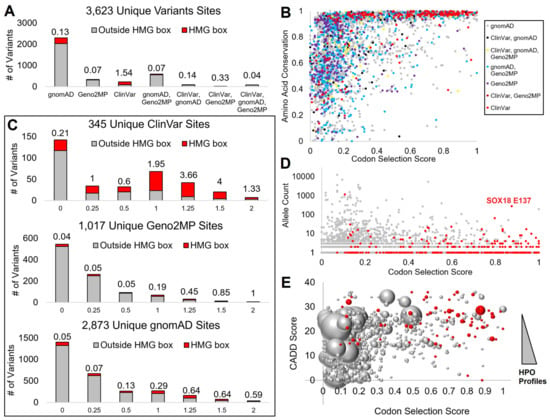
Figure 2.
Mapping SOX variants to conserved amino acids. (A) Assessment of each amino acid position and the number of variants at that site from each database. Those within the HMG box are in red and those outside the HMG box are in gray, with the value listed above each showing the ratio of variants within the HMG domain. If a variant, whether with the same or a different change, is found in multiple genomics databases for an amino acid, it is listed under the combination of databases. (B) Codon selection score (x-axis) relative to the amino acid conservation (y-axis) for each amino acid, colored as grouped in panel (A). (C) Codon selection score of unique sites for ClinVar, Geno2MP, and gnomAD, where the higher values indicate codon selection. Red represents variants within the HMG box and gray represents variants outside the HMG box. The number above each is the ratio of HMG to non-HMG variants. (D) Codon selection score (x-axis) relative to allele count (y-axis) of gnomAD variants. Those in red are within the HMG box. (E) Codon selection score (x-axis) relative to the functional variant CADD score of the highest annotation (y-axis). HMG box variants are in red and the bubble size represents the number of human phenotype profiles (HPO).
Our two metrics of conservation, the codon selection score generated by open reading frame alignments and amino acid conservation using a higher number of species protein sequences, showed that the codon selection scores provided a higher stratification of values. In contrast, amino acid conservation had more variants annotated at conserved sites (Figure 2B). Therefore, we focused on the codon selection scores to further annotate variants.
Within ClinVar, the HMG box variants had a higher codon selection score with high enrichment for the amino acids with evidence of selective pressure (ranking from 1 to 2, Figure 2C). Within Geno2MP and gnomAD, while HMG box variants were rare, when a variant fell within the HMG domain, they did fall at sites of higher conservation than those outside the HMG domain. Further, the amino acids with a high codon selection score within gnomAD had very low allele frequencies relative to sites with low scores (Figure 2D), suggesting that common variants of SOX proteins were at sites with limited selection. Additionally, many of the HMG box variants of gnomAD had low allele frequency, except for a SOX18 E137K variant. Similarly, Geno2MP variants had a lower number of HPO profiles when the codon selection scores were higher, while those rare variants often fell within the HMG domain with higher predicted CADD scores (Figure 2E). This suggests that a more detailed analysis of the HMG domain across the SOX members would benefit our ability to interpret SOX variants.
3.3. Interpreting HMG Box Variants
We began with a detailed analysis of amino acid alignments and structural insights for the HMG box between different SOX members, scaling insights into the orthologous domain. All 1890 ORF SOX HMG box sequences were aligned, and the evolutionary relationships were compared. Most sequences clustered well into the 20 genes, further clustering into the previously identified subgroups of SOX (https://doi.org/10.6084/m9.figshare.14544063, accessed on 18 December 2022, and https://doi.org/10.6084/m9.figshare.14544219, accessed on 18 December 2022). Using the alignment data, we mapped the number of amino acids used throughout the 20 human SOX proteins onto the solved HMG-box protein structure (PDB file 1J46, Figure 3). There was a high clustering of singly used amino acids (conserved throughout) in the core three-helix bundle of the HMG box, where conserved amino acids can be observed (red) in a dense core of the three-helix packing. Amino acid usage away from the HMG packing and DNA contacts could also be found (Figure 3A). The most amino acids used at any site of the HMG box was seven, occurring at five different sites (Figure 3B).
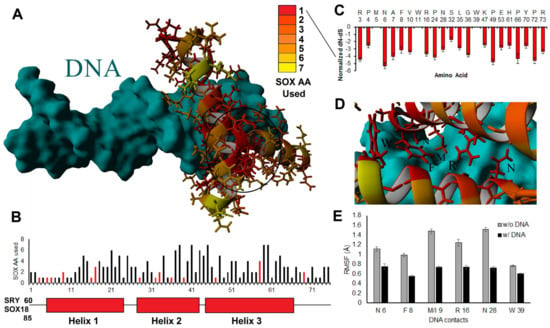
Figure 3.
SOX genes and conservation of DNA contacts. (A,B) Usage of different amino acids throughout the 1890 sequences shown on the structure of the HMG box (A) or the three-helix pictorial (B). In panel (A), we have circled the area of high conservation that drives the HMG box three-helix structure. In panel (B), we have included the amino acid in SRY or SOX18 that correspond to HMG box amino acid 1. The bars in red are amino acids that are in contact with the DNA. (C) The amino acids that are 100% conserved in all sequences analyzed show the relative selection at each site throughout evolution using a dN-dS metric. A dN-dS metric is the rate of nonsynonymous variants throughout evolution relative to synonymous variants. The more negative this value is, the more selective pressure there is to maintain the amino acid based on codon wobble. (D) Location of the 100% conserved sites relative to DNA binding on the structure. (E) Movement (root-mean-squared fluctuation, RMSF, in Å) of the six critical DNA contacts of SOX HMG proteins labeled in panel (D) following 20 nanoseconds of molecular dynamic simulations without DNA (gray) and with DNA (black) for all 20 structures of the SOX HMG domains. Error bars represent the standard error of the mean.
The positions with only a single amino acid had high selective pressure (dN-dS metric, Figure 3C), except for M at position 5, W at 11, and W at 39. M/W used only a single codon and thus did not possess dN-dS selection metrics. In contrast, all other sites with a single amino acid were suggested based on dN-dS metrics to have high selective pressure throughout all 20 SOX genes. All the DNA contact positions had only a single amino acid used in all sequences (Figure 3D). They were significantly stabilized when DNA was added to all 20 proteins as determined by molecular dynamic simulations (Figure 3E). This suggests that all SOX HMG boxes recognized similar DNA sequences (such as AACAA) through the DNA binding domain. Any deviation in binding or target recognition was likely driven by protein-specific non-HMG flanking DNA interactions, protein–protein recruitment from regions outside DNA binding, dimerization, chromatin penetrance, or competition from other transcription factors within cellular environments, such as other HMG box or fork-head transcription factors.
Using the UniProt annotation of the HMG box, an analysis of relative HMG box positioning for all variants of ClinVar, Geno2MP, and gnomAD was conducted. This approach provides ortholog mapping insights where a clinical variant at an HMG box number could assist in further validating variants of uncertain biological impact. Flanking the HMG box (−2 to −10 and 76–100, where 1 is the first UniProt annotated amino acid of the HMG box), the amino acids had higher variability between SOX members (Figure 4A), a lower codon selection score (Figure 4B), and a lower amino acid conservation score (Figure 4C). Integrating ClinVar annotated variants and those within the literature (Table 1) identified many HMG box sites where multiple SOX genes had annotated pathologies connected to their variation (Figure 4D). Geno2MP (Figure 4E) and gnomAD variants (Figure 4F) were lower within the HMG box positions relative to those flanking the domain.
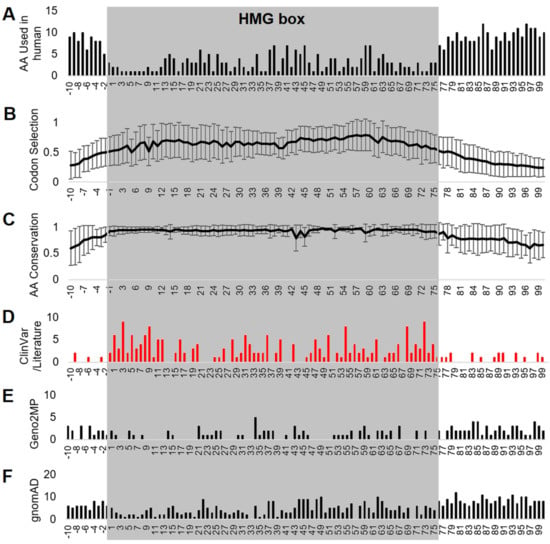
Figure 4.
HMG box alignment of all SOX proteins. Alignment of all HMG box sequences of the 20 SOX proteins showing the number of human amino acids used (A), codon selection scores (B), conservation (C), and genomic variants for different SOX proteins using ClinVar or the literature (D), Geno2MP (E), or gnomAD (F). Error bars for panels (B,C) represent plus and minus the standard error of the mean over all 20 genes/proteins. Amino acid 1 is the V/I annotated within UniProt as the first amino acid of the HMG box for each protein. Alignments were performed without any gaps.
Various resources provide different numbering of HMG amino acids. Using the UniProt numbering position −1 (2 within the Bowles/Koopman numbering) was the first to have an average of over 90% conserved (93 ± 9%) in the 6667 amino acid sequences analyzed. This position is always a polar basic (R/K/H). At the −2 UniProt numbering (1 within the Bowles/Koopman numbering), there was high variability in the amino acids used in SOX proteins (D/G/P/S/E), and only 86 ± 15% of the sequences were conserved with the human SOX member. In several of the SOX proteins, amino acids before this were conserved within the protein, but not across the family. On the other end of the HMG box, UniProt number 75 (77 of Bowles/Koopman numbering) could use a polar basic residue (R/K) in all, except for SOX30, and there was high conservation (93 ± 13%). At the next amino acid (76/78), there were seven different residues used in human SOX proteins (T/A/P/V/S/R/K) with only 87 ± 22% conservation. This would suggest that the best annotation of calling the overall HMG box was one amino acid shifted between the UniProt (−1 to 75) and the Bowles/Koopman numbering (2 to 77).
The additive variants at each HMG box position showed an overlap of SOX proteins with high conservation for ClinVar-connected human clinical sequencing (Table 3). The −1 position was the first HMG box to have a variant, where SOX4 H58P was found to have uncertain significance in ClinVar (Accession VCV001526184.1). Eight amino acids of the HMG box had multiple variant changes at a site within one protein (Table 3, 9/11—SOX10; 16/18—SOX2, SOX9, SOX11, and SOX5; 29/31—SOX10; 32/34—SOX11, SOX5, and SOX10; 36/38—SOX10; 39/41—SOX10; 58/60—SOX10; and 72/74—SOX2 and SOX9). Twenty-seven amino acids of the HMG box had more than one SOX protein with a known ClinVar variant (Table 3, 1/3, 2/4, 3/5, 5/7, 6/8, 7/9, 9/11, 11/13, 12/14, 16/18, 19/21, 20/22, 28/30, 31/33, 32/34, 34/36, 36/38, 39/41, 47/49, 50/52, 54/56, 56/58, 57/59, 58/60, 61/63, 68/70, and 72/74).

Table 3.
ClinVar variants within the HMG box using UniProt or Bowles/Koopman numbering. Rotating white to gray indicates different amino acids with multiple variants for some.
The power of this ClinVar ortholog knowledge is in the ability to aid in the screening and interpretation of additional HMG box variants of SOX proteins. From these HMG box positions with a ClinVar variant and high conservation, we further assessed if any of the Geno2MP variants fell on one of these sites and if that individual had a matching phenotype or newly identified phenotype in more than one individual (Table 4). Of these 26 Geno2MP variants with a CADD score of >20 and selection score of ≥1, 6 variants had interesting phenotypes noted. SOX11 G84S (HMG box location 36/38) was found in one affected individual with eye abnormality with globe alteration, in addition to head or neck abnormality. SOX11 was associated with Coffin–Siris syndrome 9, commonly with dysmorphic facial features [16].

Table 4.
Geno2MP variants within the HMG box using UniProt numbering. Those in red have matching phenotypes to the gene OMIM or in multiple Geno2MP individuals. Rotating white to gray indicates different SOX genes with multiple variants for some.
SOX14 R80Q (HMG box location 73/75) was found in two individuals with abnormality of the musculature, one with altered muscle physiology and another with muscular dystrophy. SOX14 did not have any OMIM annotated phenotypes. However, heterozygous global knockout of SOX14 has resulted in multiple altered mouse phenotypes, mostly morphological changes to various organs. but without any annotated changes to muscle phenotypes (https://www.mousephenotype.org/data/genes/MGI:98362, accessed on 18 December 2022). This suggests that the R to Q change may be conserved and that the phenotype is not connected to SOX14 variants within the individuals. SOX30 I367V (HMG box location 31/33) was absent from gnomAD and found in two individuals within Geno2MP with autism, intellectual disability, and mild cerebellar vermis hypoplasia. SOX30 variants have been associated with male infertility with testis-specific expression [80] and have never been associated with neurological traits. The conservation of the hydrophobic change suggests that this may not be the causal variant.
SOX15 R104G (HMG box location 56/58) was found in an individual with abnormality of the nervous system and musculature with noted fatigable weakness and progressive muscle weakness. Similar to SOX14, no current OMIM phenotypes have been annotated to SOX15, but SOX15 has been identified as critical for muscle differentiation and regeneration [81,82]. Position 56 was highly conserved as an R/K and found to have clinical variants in SOX2, SOX9, SOX5, and SOX6. The SOX 5 change was an R to G, matching that of the SOX15 Geno2MP variant, suggesting that it may be the individual’s causal pathogenic autosomal dominant variant.
SOX8 R159G (HMG box location 58/60) was absent in gnomAD and present in two individuals of Geno2MP with intellectual disability and microcephaly. SOX8 is not associated with any OMIM phenotypes, but it has been associated with developmental striatal projection neurons, glial cells, and the cerebellum [83,84]. The HMG box position 58 had ClinVar variants in the SOX10 homologous SOXE member linked to pathogenic annotation for Waardenburg syndrome type 2E, and SOX17 was also associated with disease due to changes at this site. None of the human SOX proteins utilized a G at this site, and R was highly selected in SOX8 evolutionary analysis, even with codon wobble occurring. This suggests that SOX8 R159G may be associated with altered neurological development. Thus, SOX15 with muscle disorders and SOX8 with neurological disorders may have supporting data within Geno2MP for novel genotype–phenotype associations.
3.4. HMG Box Variant SOX18 E137K
SOX18 E137K (HMG box location 53/55) was found within Geno2MP and gnomAD, while it was at a conserved site with known ClinVar variants in other SOX genes (Figure 5). The E137K of SOX18 fell on the third helix of the HMG box near the contacting sites with the first helix (Figure 5A). In the crystal structure of SOX18 (4y60), E137 formed a salt bridge with R140 and had hydrophobic packing with helix 1. E53 and R56 were highly conserved and selected in all 20 SOX proteins with multiple known ClinVar variants at several sites (yellow and magenta, Figure 5A). E137 was conserved in all 20 SOX genes in all 1890 sequences analyzed for the HMG box open reading frames. Over the HMG box sequences, the dN-dS value was −2.89. This amino acid was under greater than two standard deviations of selective pressure within SOX18, with a CADD score of 28.6 (near 0.1% of all deleterious variants) and a PolyPhen2 prediction of probably damaging. This suggests that E137K is of functional impact. Molecular dynamic simulations of the SOX18 protein showed that E137K resulted in elevated motion of the salt bridge (E/K137 with R140, https://youtu.be/CKg3dhkRHxY, accessed on 18 December 2022), which increased the availability of charges for potential protein interactions to increase, suggesting that it impacts SOX18’s function.
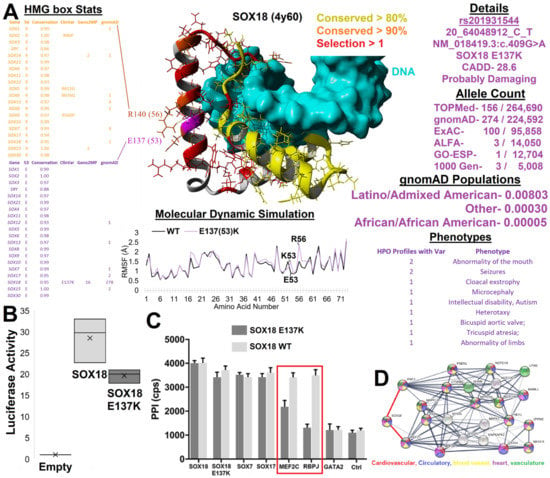
Figure 5.
SOX18 E137K. (A) Integrated data and structure of SOX18 E137K. In the middle is the SOX18 structure (PDB file 4y60) bound to DNA (cyan). Amino acids are colored based on conservation and selection as labeled. To the left of the structure is the annotation of the HMG box compiled data for R56 (yellow) and E53 (magenta) amino acids. Below are 20 nanoseconds of mds for wild type (WT, black) or E137K (magenta), showing the root-mean-squared fluctuation for each amino acid. To the right are various data insights for E137K, including the multiple annotations, allele counts, and Geno2MP phenotypes. (B) Transcriptional activity of empty plasmid (empty normalized to 1), with SOX18 (gray) or SOX18 E137K (black) overexpressed in combination with an SOX response element driving luciferase production in HeLa cells. (C) ALPHAScreen assay to assess pairwise interactions between SOX18 WT or the SOX18 E137K variant with known protein partners. GATA2 is a known nonbinding control that provides a baseline ALPHAScreen signal similar to the control condition with buffer only (Ctrl). Error bars represent the standard error of the mean of three independent experiments. (D) STRING analysis of SOX18, MEF2C, and RBPJ with the first and second shells of the network with no more than ten interactions added in. Colors represent GO enrichments as follows: red—cardiovascular system development (GO:0072358; FDR—9.18 × 10−15); blue—circulatory system development (GO:0072359; FDR = 9.18 × 10−15); yellow—blood vessel development (GO:0001568; FDR = 2.67 × 10−13); magenta—heart development (GO:0007507; FDR = 2.67 × 10−13); green—vascular development (GO:0001944; FDR = 3.55 × 10−13). The raw TIF file can be found at https://doi.org/10.6084/m9.figshare.21830421.v1 (accessed on 18 December 2022).
SOX18 E137K (rs201931544, 20_64048912_C_T) was found in 156 TOPMed sequenced humans and 274 gnomAD genomes. Within gnomAD, the variant was enriched in those with a Latino/Admixed American population background. The phase3 1000 genomes of subpopulations showed an allele frequency of 0.8% in MXL (Mexican Ancestry from LA, USA) with 24 additional SNPs in LD (r2 of 1), an allele frequency of 0.6% in PEL (Peruvians from Lima, Peru) with 44 SNPs in LD (r2 of 1), and an allele frequency of 0 in all other subpopulations.
Rare variants in human SOX18 have been shown to result in Hypotrichosis–Lymphedema–Telangiectasia and renal syndrome (HLTRS), characterized by blood and lymphatic vascular symptoms and hair follicle defects [85]. In humans, HLTRS results from frameshift mutations that lead to the synthesis of a truncated version of the SOX18 protein, which acts as a dominant negative transcription factor, suppressing the endogenous functions of SOX7 and SOX17. In the case of SOX18 E137K, there was no frameshift in the ORF; however, it was not disregarded that amino acid variation in the third helix of the HMG-box interfered with protein partner recruitment.
Within Geno2MP, SOX18 E137K was found in 11 affected individuals. Two individuals were annotated with abnormality of the head and neck with the sub-phenotype term for abnormality of the mouth. Multiple individuals had a neurological phenotype, including two individuals with seizures, one with microcephaly, and one with intellectual disability with autism. Two individuals had an abnormality of the cardiovascular system due to altered value development, one with abnormality of limbs and one with cloacal exstrophy (external abdominal organs). The number of neurological variants is interesting, as the homozygous knockout of mice results in significant abnormal behavior and decreased thigmotaxis (touch stimulus changes, https://www.mousephenotype.org/data/genes/MGI:103559, accessed on 18 December 2022). As there were no significant changes within heterozygous mouse knockouts of IMPC, and SOX18 loss of function variants were autosomal recessive for HLTRS [17], it suggests that SOX18 E137K may be autosomal recessive.
Patients with SOX18 variants often suffer from capillary dysfunction, where mouse alterations of SOX18 do not modify the early vasculature in development, but rather the capillaries and lymphatics [86]. SOX18 has been shown to regulate the claudin-5 gene [87], which is critical for blood–brain barrier size selection [88]. The disruption of claudin-5 in mice results in spontaneous recurrent seizures and severe neuroinflammation [89]. Just recently, a patient with SOX18-associated HLTRS was observed to have idiosyncratic seizures following hyperbaric treatment [90]. This suggests that the neurological phenotypes of SOX18 E137K within Geno2MP may be environmentally regulated to contribute to changes in neuro-electrical control and, therefore, warrants additional characterization of functional outcomes for the missense variant.
SOX18 constructs were overexpressed in HeLa cells along with a SOX-regulated luciferase promoter, suggesting that the variant, E137K, decreased transcriptional regulation and was a loss-of-function (Figure 5B). SOX18 E137K additionally altered the interaction with known endothelial transcriptional regulators, such as MEF2C (adjP <0.0001) and RBPJ (adjP <0.0001), without disrupting dimerization to SOX7/17, as determined by the ALPHAScreen assay (Figure 5C), which measured pairwise protein–protein interactions. The analysis of a protein network built around a transcriptional regulatory node composed of SOX18, MEF2C, and RBPJ revealed a significant gene ontology (GO) enrichment for cardiovascular, circulatory, blood vessel, heart, and vasculature terms (Figure 5D). It should be noted that both MEF2C and RBPJ have been associated with intellectual disability and seizure disorders [91,92,93,94], matching the SOX18 E137K Geno2MP neurological phenotypes. This suggests a future need to study the role of SOX18 E137K in the blood–brain barrier and its role in neurological development and seizures.
3.5. Functional SOX Variants Outside the HMG Box
In screening variants, it was noted that multiple variants fell within SOX proteins at conserved motifs or domains outside the HMG box. Known dimerization regions of several SOX genes, including SOX9 (N-term region before the HMG box) [95] and SOX18 (central region) [96], were conserved within the dataset (Figure 1). Multiple SOX genes had regions under high conservation/selection that have not been curated within databases (Figure 1, gray boxes). SOX4 had a conserved C-terminal sequence (SOX4 440-SGSHFEFPDYCTPEVSEMISG, underlined residues with conservation score ≥1) consisting of multiple aromatic and charged residues that predicted potential GSK3 and ProDKin kinase sites with multiple docking motifs for proteins such as CKS1 and MAPK. SOX5 had a linear motif (SOX5 110-SLSSTALGTPERRKGSLADVVDTLKQRKMEELIKNEPEETPS) with multiple conserved charged residues that made up multiple potential phosphorylation sites. SOX8 had two regions (SOX8 220- QTHGPPTPPTTPKTE and SOX8 255-GRQNIDFSNVDISELSSEVMGT), with the first having a degradation control switch with a sumoylation site and the second harboring potential phosphorylation sites. SOX10/11/12/14 had conserved C-terminal domains. This compiled vertebrate SOX analysis revealed multiple putative functional domains that warrant further detailed mechanistic studies.
We identified 56 variants found within ClinVar, Geno2MP, or gnomAD at highly conserved amino acids with additional amino acids around the site conserved, suggesting that the variant potentially impacted domains or motifs (Table 5). Several of these variants had matching phenotypes for the gene. SOX2 had two neurological-associated variants (D123G and G130A) found within the region flanking the HMG box with multiple conserved charges (HMGbox-PRRKTKTLMKKDKYTLPGGLLAPGGNSMA, bold underlined letters are sites of variants). SOX6 had several non-HMG box variants connected to neurological alterations, including H209K, S310T, R485Q, R545Q, and E591. As noted, the SOX9 N-terminal region for dimerization was conserved and had multiple Campomelic Dysplasia-linked variants within the motif (I73T, A76E, L81V, and G83R). Flanking the HMG domain of SOX10 were multiple variants from ClinVar (K179N, G181R, H216Q, and T240P).

Table 5.
High-ranking SOX protein variants found outside the HMG box. Red text are those that have matching phenoptypes or are of high interest. Rotating white to gray indicates different SOX genes with multiple variants for some.
One of the most interesting variants outside of the HMG domain was that of SOX14 K88R (rs34393601). Within SOX14, the K was always conserved in all amino acid sequences of our alignment. The variant was predicted to be damaging in PolyPhen2 and had a CADD score of 25.8. The variant was found within 12 amino acids flanking the HMG box within a conserved motif with multiple prolines and charged residues (HMG box-PRRKPKNLLKKDRYVFPLPYLG). The most interesting observation is that, while the variant was present in gnomAD with an allele frequency of 0.018% of individuals (0.06% of north-western Europeans), it was present in five affected individuals with abnormality of the cardiovascular system, with an additional four with annotated thoracic aortic aneurysm. As noted above, SOX14 heterozygous knockout mice have been found to suffer from abnormal heart morphology (https://www.mousephenotype.org/data/genes/MGI:98362, accessed on 18 December 2022), suggesting some cardiovascular connections.
Finally, SOX7 A379V was identified at the far C-terminal end of the protein, with high conservation of amino acids around it. SOX7/17/18 had annotated C-terminal conserved regions with additional high levels of conservation/selection at the last ~20 amino acids in each of the three proteins that have not previously been noted in the literature. Segregation analysis of SOX18 relative to SOX7/17 revealed multiple conserved amino acids, but could also elucidate several conserved amino acids in SOX18 that differed in SOX7 and SOX17 (Figure 6). The far C-terminal region contained multiple highly conserved serine and threonine amino acids with the potential for phosphorylation and multiple hydrophobic amino acids, making this an ideal protein-interacting peptide. Moreover, the identification of ClinVar, Geno2MP, and gnomAD variants within this region suggests a need for SOX gene variant insights outside the HMG box.
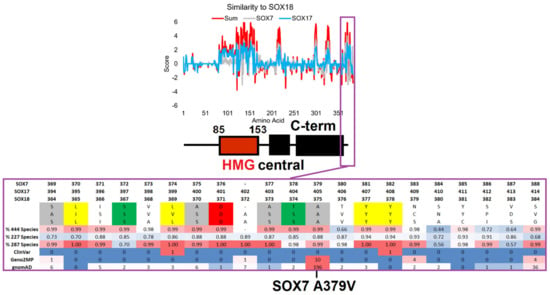
Figure 6.
Conservation of the SOX-F proteins highlighting the far C-terminal region. The top panel shows a comparative analysis of SOX18 relative to the SOX7 (gray) and SOX17 (cyan) conservation scores for each amino acid of SOX18, with the SOX18 domain annotation from Figure 1 shown below. When the amino acids are the same between SOX18 and the others, they have positive values; when they are different, the values are negative. The red line is the additive value of the SOX7 and SOX17 comparisons. In the bottom panel, the magenta call out shows the conservation of amino acids within the far C-terminal region of SOX-F members. The amino acid numbers of each protein are provided on top of the sequences. This is followed by the human amino acids, where those highlighted are conserved in each sequence (gray—flexible; yellow—hydrophobic; green—S or T; red—polar acidic). Then, the percentage of conservation for the amino acid sequence alignments is shown, with a value of 1 indicating 100% conservation. At the bottom is the number of variants seen in each genomics database. The conservation and variants are shown on a heatmap, with red representing the highest values and blue representing the lowest.
SOX7 A379V (rs143587868) was found to be heterozygous in 0.716% of African individuals from gnomAD, as confirmed in the TOPmed Bravo dataset (allele frequency of 0.2% total), and had a CADD score of 29.0 (near 0.1% of all deleterious variants). The variant was found on the most highly conserved motif of SOX7 (Figure 1), near multiple conserved Ser/Thr sites with potential phosphorylation, and was conserved on the C-terminal ends of SOX7, SOX17, and SOX18. We speculate that this region is critically involved in the phosphorylation control of SOXF genes; however, there is no information regarding the function of post-translational modification of these TFs. Within Geno2MP, SOX7 A379V was found in six affected individuals, with four having abnormal eye phenotypes (glaucoma or retinal degeneration). Glaucoma occurs five times more often in African American individuals [97], in which this variant is enriched. The SOXF genes have been identified to regulate the vascular development of the eye [98]. SOX7 A379V represents a novel potential for the future characterization of the C-terminal ends of SOXF genes and their roles in eye phenotypes.
3.6. Somatic SOX Variants in Cancer
Our final genomics analysis of SOX proteins was the analysis of somatic variants within cancer using the COSMIC database. As of December 2022, there were 4313 SOX-based COSMIC variants. Similar to ClinVar, where variants were enriched within the HMG box of several proteins, cancer somatic variants also showed an HMG box enrichment for SOX11, SOX9, SOX4, SOX10, SOX1, SRY, SOX18, SOX21, SOX3, SOX12, SOX14, SOX17, SOX2, and SOX13 (cyan, Figure 7A). Nearly all of the SOX proteins had a significant de-enrichment for variants within the HMG box for gnomAD and Geno2MP. This suggests that functional SOX somatic variants might be elevated within cancer samples. Most of the SOX HMG box variants fell on conserved sites (Figure 7B), with multiple sites of the HMG domain annotated for ClinVar or the literature’s phenotype connections (Figure 7C). These HMG box variants were primarily found in Adenocarcinoma and large intestine samples (Figure 7D, 117 samples), with many of the SOX proteins having high-risk variants for the pathology. Of the top-ten cancer types, SOX9 had the leading high-risk variants for large intestine adenocarcinoma and stomach non-specified (NS) cancer. SOX17 was the leading protein for skin NS cancer, stomach adenocarcinoma, endometrium endometrioid carcinoma, lung adenocarcinoma, and urinary tract NS cancer. SOX13 accounted for 22% of the high-risk variants in lung squamous cell carcinoma, SOX 2 for 33% of pancreas ductal carcinoma, and SOX7 for 40% of thyroid NS cancer. This suggests that further analysis of somatic variants within SOX proteins may be critical and that the gene family has functionality outside early development.
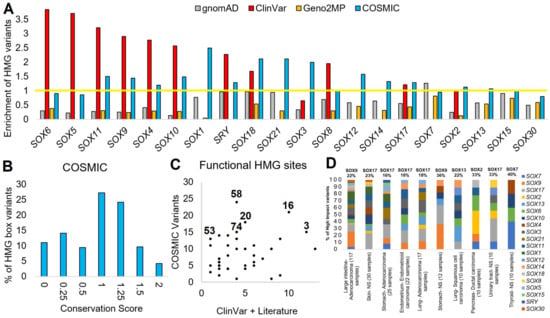
Figure 7.
COSMIC variants of SOX genes. (A) Enrichment of variants within the HMG box for each SOX protein for gnomAD (gray), ClinVar (red), Geno2MP (orange), or COSMIC (cyan). Genes are ranked based on the highest enrichment for any database. The enrichment was calculated by normalizing the HMG box size relative to the total protein such that a value of 1 (yellow line) is the random probability of a variant falling within the HMG box. (B) Codon selection score of variants from COSMIC that fall with the HMG domain. (C) Amino acids found with ClinVar or annotations in the literature (x-axis) associated with disease for the HMG box that have COSMIC variants (y-axis) (D) Annotation of HMG box variants from panel (C) showing tissue type and histology annotations for each protein of the top-10 tumor types. The number of COSMIC samples with each cancer type with high-impact variants is shown on the x-axis labels. The top SOX gene for the percent of variants is labeled on top of each bar.
4. Discussion
Since 1990, with the discovery of SRY variants in sex reversal [57], SOX genes have had known disease implications [18,99]. Our group characterized the rat chromosome-Y Sry duplications and point mutations involved in blood pressure regulation [41,100]. One of the duplicated Sry genes, Sry2, found on the rat Y-chromosome, was inserted into the intron of Kdm5d, driving a ubiquitous expression profile [41]. We hypothesized that this ubiquitous expression profile of the Sry2 gene altered the phenotype of an ancient rat, such that a loss-of-function mutation (R21H, HMG box location 17) occurred and was selected to compensate for the ubiquitous expression of Sry2. This insight led us to explore additional SRY and SOX gene variants that might impact phenotypes while attempting to understand the molecular mechanisms of the variants. While this initially appeared to be an easy task, we were surprised to find a lack of functional domain/motif mapping in SOX genes outside the HMG box. This has recently been identified as an emerging need within SOX gene knowledge [18].
Therefore, we developed a deep evolutionary assessment of SOX ORF sequences to map functional sites (Figure 1), followed by an assessment of human variants in gnomAD, COSMIC, the published literature, and ClinVar. Future work could be focused outside vertebrate SOX genes, focusing on comparative evolutionary analysis of invertebrate species, which was not performed here. Further work on domain annotations could also be performed. Conservation analysis can be challenging [101]. Utilizing the codon selection strategy described in this work [22,23], a few divergent sequences decreased their conservation scores. For example, the loss of conservation in several gene members shown in Figure 1 was likely the result of a few divergent species sequences. This is why amino acid alignments were performed with more species, yielding insights over all species that were not biased by a few divergent sequences.
Before the 2000s, most variant insights were published in the literature [102], and therefore we developed a list of common SOX genetic variant papers in Table 1. This table is not meant to be all-inclusive, but captures the most notable SOX genetic variant papers. Following the establishment of ClinVar and other databases, it became more typical for variants to be listed in both publications and within the database. Thus, combining the early variant manuscripts (Table 1) with the most common variant databases makes a larger map of the genomics landscape of SOX genes possible.
Using multiple data sources, this study has reaffirmed many published observations while making multiple novel discoveries: (1) uncharacterized, but highly conserved, motifs found in SOX2, SOX14, SOX4, SOX11, SOX12, SOX5, SOX8, and SOX10; (2) 100% conservation of all HMG box DNA-specific contact amino acids; (3) enrichment of variants in the HMG boxes of multiple SOX proteins; (4) potential high-impact human variants in SOX18 and SOX7; (5) potential novel syndromes for SOX15 in musculature abnormalities and SOX8 with intellectual disabilities; and (6) the role of functional variants outside the HMG box, including those of SOX2, SOX9, SOX14, and SOX7.
In our COSMIC analysis, we can confirm the recent findings of the Jauch lab [19] that a few cancer variants are found at critical sites of the SOX2/17-OCT4 interaction residues [9,103] (HMG boxes numbered 44, 48, 51, 55, 62, and 69). Most notable is the role of amino acid changes to SOX9 at position 62 (deletion of residue K), seen multiple times in cancer. These amino acids represent one of the fascinating parts of the evolution of SOX genes, specifically HMG box sites 44, 55, and 62 (Bowles annotated sites 46, 57, and 64), where our evolutionary conservation shows high intra-protein multiple-species conservation while variation across SOX proteins.
Understanding the clinical impact of variants between 0.01 and 1% is incredibly challenging [23]. This can be seen for our identified SOX18 and SOX7 human variants. SOX18 E137K (rs201931544) was not found on most SNP-Chips used in genotyping patients, thus making phenotypic association rather challenging. This indicates that the variant and all of the LD SNPs of Latino individuals have been included in limited published genome-wide association studies (GWAS). Genotyped individuals in large biobanks cannot have phenome-wide association studies (PheWAS) performed for this site due to the low allele frequency, underpowering discovery. The lower limits of detectable allele frequencies for significance within GWAS and PheWAS are greater than 1–5% of the global population [104], while SOX18 E137K is estimated to have a frequency between 0.1% and 0.05% globally. This highlights the ethnic/racial bias currently present in genomic knowledge that needs to be actively addressed [105].
To compensate for this lack of information, we took an alternative approach based on a preliminary molecular assessment of the functional impact of SOX18 E137K. This revealed a change in transcriptional activity and affinity for protein partners, likely yielding a change in transcription factor biology. Our preliminary laboratory experiments on SOX18 E137K suggest a loss of function by the variant. Additional molecular and biochemical experiments, including co-immunoprecipitations, nuclear localization, DNA binding, and structural changes, are required to further elucidate the changes to SOX18 biology. Most notably, based on the Geno2MP annotated phenotypes, we suggest that it is critical to characterize the gene within the endothelial cells of the blood–brain barrier. As whole-genome sequencing is being implemented in more studies, particularly for Latino populations, the variant will be identified more often and could be linked to phenotypes. This highlights the increasing importance of extensive data integrations into genomic medicine, an undertaking within the All of Us precision medicine initiative [106] that will make the systematic assessment of gene variants more interpretable.
As sequencing increases in multiple species and millions of humans, systematic assessments of gene families are needed. This paper uses bioinformatics to assess the genotype-to-phenotype associations of each of the 20 human SOX gene family members. This work highlights the utility of studying gene families with a more systematic approach, which should be applied to more transcription factor families to segregate genotype-to-phenotype associations further, which is not possible when studying only a single gene family member. These strategies will ultimately help when prioritizing understudied, yet fruitful, avenues of genetic research within gene families in the age of data integration.
5. Conclusions
This study is the first to systematically analyze the evolutionary conservation within thousands of sequences of SOX genes/proteins with multiple database integrations of human variants linked to phenotypes. From discovering novel domains outside the HMG box, linking several SOX genes to novel phenotypes, and identifying several inherited variants linked to phenotypic traits, we show the promise of new genomic discoveries within a large transcription factor family. While we continue to advance our knowledge of SOX members, this work also highlights the importance of using paralog mapping to understand variants better, especially when occurring in a paralog family member that is less studied with new knowledge to be gained. Overall, this shows that, even in 2023, there is much genomic knowledge to be learned, and bioinformatics holds much promise in advancing our genomic insights.
Author Contributions
Conceptualization, A.U., T.M.F., A.M., R.J., T.J.T.J., C.M.K., C.P.B., S.R., M.F. and J.W.P.; methodology, A.U., D.T.R., D.H., M.M., Y.G., E.S., F.F., M.F. and J.W.P.; formal analysis, A.U., D.T.R., D.H., J.M., J.K.Z., J.M., N.E.A., T.W.C., M.M., Y.G., E.S., S.V., A.S.D., W.C., O.S., X.S., K.B., M.A., M.G., S.H., A.M.W., D.Q., M.F. and J.W.P.; data curation, A.U., D.T.R., J.K.Z., A.M.W., T.M.F., D.Q., A.M., R.J. and J.W.P.; writing—original draft preparation, A.U., M.F. and J.W.P.; writing—review and editing, A.U., D.T.R., D.H., J.T.M., J.K.Z., J.M., N.E.A., T.W.C., M.M., Y.G., E.S., F.F., S.V., A.S.D., W.C., O.S., X.S., K.B., M.A., M.G., S.H., A.M.W., T.M.F., D.Q., A.M., R.J., T.J.T.J., C.M.K., C.P.B., S.R., M.F. and J.W.P.; supervision, A.U., T.M.F., A.M., C.P.B., S.R., M.F. and J.W.P.; funding acquisition, T.J.T.J., C.M.K., C.P.B., S.R. and J.W.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Gerber Foundation (to C.P.B., S.R., J.W.P.), National Institutes of Health (K01ES025435 to J.W.P.; R01AI171984 to T.J.T.J., C.M.K. and J.W.P.), Michigan State University, and the National Health and Medical Research Council (APP1107643 and APP1111169 to M.F.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The compiled dataset for SOX codons/amino acids and variants can be found at https://doi.org/10.6084/m9.figshare.21761000 (accessed on 18 December 2022). The file contains three tabs: All AAs—lists each of the SOX genes and every codon/amino acid of each along with data of conservation and variants; HMG box data—Aligned data over all SOX genes for HMG box positions; and All var AA—a list of all amino acids with variants. The molecular dynamic simulation data can be found at https://doi.org/10.6084/m9.figshare.14544339 (accessed on 18 December 2022). The SOX HMG box alignments can be found at https://doi.org/10.6084/m9.figshare.14544063 (accessed on 18 December 2022) and the phylogenetic tree at https://doi.org/10.6084/m9.figshare.14544219 (accessed on 18 December 2022).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pevny, L.H.; Lovell-Badge, R. Sox genes find their feet. Curr. Opin. Genet. Dev. 1997, 7, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, A.H.; Berta, P.; Palmer, M.S.; Hawkins, J.R.; Griffiths, B.L.; Smith, M.J.; Foster, J.W.; Frischauf, A.M.; Lovell-Badge, R.; Goodfellow, P.N. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 1990, 346, 240–244. [Google Scholar] [CrossRef]
- Goodfellow, P.N.; Lovell-Badge, R. SRY and sex determination in mammals. Annu. Rev. Genet. 1993, 27, 71–92. [Google Scholar] [CrossRef] [PubMed]
- Kiefer, J.C. Back to basics: Sox genes. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2007, 236, 2356–2366. [Google Scholar] [CrossRef]
- Bowles, J.; Schepers, G.; Koopman, P. Phylogeny of the SOX Family of Developmental Transcription Factors Based on Sequence and Structural Indicators. Dev. Biol. 2000, 227, 239–255. [Google Scholar] [CrossRef] [PubMed]
- Pontiggia, A.; Whitfield, S.; Goodfellow, P.N.; Lovell-Badge, R.; Bianchi, M.E. Evolutionary conservation in the DNA-binding and -bending properties of HMG-boxes from SRY proteins of primates. Gene 1995, 154, 277–280. [Google Scholar] [CrossRef]
- Read, C.M.; Cary, P.D.; Preston, N.S.; Lnenicek-Allen, M.; Crane-Robinson, C. The DNA sequence specificity of HMG boxes lies in the minor wing of the structure. EMBO J. 1994, 13, 5639–5646. [Google Scholar] [CrossRef]
- Murphy, E.C.; Zhurkin, V.B.; Louis, J.M.; Cornilescu, G.; Clore, G.M. Structural basis for SRY-dependent 46-X,Y sex reversal: Modulation of DNA bending by a naturally occurring point mutation. J. Mol. Biol. 2001, 312, 481–499. [Google Scholar] [CrossRef]
- Williams, D.C., Jr.; Cai, M.; Clore, G.M. Molecular basis for synergistic transcriptional activation by Oct1 and Sox2 revealed from the solution structure of the 42-kDa Oct1.Sox2.Hoxb1-DNA ternary transcription factor complex. J. Biol. Chem. 2004, 279, 1449–1457. [Google Scholar] [CrossRef]
- Hawkins, J.R.; Taylor, A.; Berta, P.; Levilliers, J.; Van der Auwera, B.; Goodfellow, P.N. Mutational analysis of SRY: Nonsense and missense mutations in XY sex reversal. Hum. Genet. 1992, 88, 471–474. [Google Scholar] [CrossRef] [PubMed]
- Wagner, T.; Wirth, J.; Meyer, J.; Zabel, B.; Held, M.; Zimmer, J.; Pasantes, J.; Bricarelli, F.D.; Keutel, J.; Hustert, E.; et al. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell 1994, 79, 1111–1120. [Google Scholar] [CrossRef] [PubMed]
- Fantes, J.; Ragge, N.K.; Lynch, S.-A.; McGill, N.I.; Collin, J.R.O.; Howard-Peebles, P.N.; Hayward, C.; Vivian, A.J.; Williamson, K.; van Heyningen, V.; et al. Mutations in SOX2 cause anophthalmia. Nat. Genet. 2003, 33, 461–463. [Google Scholar] [CrossRef] [PubMed]
- Hagstrom, S.A.; Pauer, G.J.T.; Reid, J.; Simpson, E.; Crowe, S.; Maumenee, I.H.; Traboulsi, E.I. SOX2 mutation causes anophthalmia, hearing loss, and brain anomalies. Am. J. Med. Genet. A 2005, 138A, 95–98. [Google Scholar] [CrossRef] [PubMed]
- Woods, K.S.; Cundall, M.; Turton, J.; Rizotti, K.; Mehta, A.; Palmer, R.; Wong, J.; Chong, W.K.; Al-Zyoud, M.; El-Ali, M.; et al. Over- and underdosage of SOX3 is associated with infundibular hypoplasia and hypopituitarism. Am. J. Hum. Genet. 2005, 76, 833–849. [Google Scholar] [CrossRef]
- Pingault, V.; Bondurand, N.; Kuhlbrodt, K.; Goerich, D.E.; Préhu, M.O.; Puliti, A.; Herbarth, B.; Hermans-Borgmeyer, I.; Legius, E.; Matthijs, G.; et al. SOX10 mutations in patients with Waardenburg-Hirschsprung disease. Nat. Genet. 1998, 18, 171–173. [Google Scholar] [CrossRef]
- Tsurusaki, Y.; Koshimizu, E.; Ohashi, H.; Phadke, S.; Kou, I.; Shiina, M.; Suzuki, T.; Okamoto, N.; Imamura, S.; Yamashita, M.; et al. De novo SOX11 mutations cause Coffin-Siris syndrome. Nat. Commun. 2014, 5, 4011. [Google Scholar] [CrossRef]
- Irrthum, A.; Devriendt, K.; Chitayat, D.; Matthijs, G.; Glade, C.; Steijlen, P.M.; Fryns, J.-P.; Van Steensel, M.A.M.; Vikkula, M. Mutations in the transcription factor gene SOX18 underlie recessive and dominant forms of hypotrichosis-lymphedema-telangiectasia. Am. J. Hum. Genet. 2003, 72, 1470–1478. [Google Scholar] [CrossRef]
- Angelozzi, M.; Lefebvre, V. SOXopathies: Growing Family of Developmental Disorders Due to SOX Mutations. Trends Genet. TIG 2019, 35, 658–671. [Google Scholar] [CrossRef]
- Srivastava, Y.; Tan, D.S.; Malik, V.; Weng, M.; Javed, A.; Cojocaru, V.; Wu, G.; Veerapandian, V.; Cheung, L.W.T.; Jauch, R. Cancer-associated missense mutations enhance the pluripotency reprogramming activity of OCT4 and SOX17. FEBS J. 2020, 287, 122–144. [Google Scholar] [CrossRef]
- Prokop, J.W.; Leeper, T.C.; Duan, Z.-H.; Milsted, A. Amino acid function and docking site prediction through combining disease variants, structure alignments, sequence alignments, and molecular dynamics: A study of the HMG domain. BMC Bioinform. 2012, 13 (Suppl. S2), S3. [Google Scholar] [CrossRef]
- Dong, C.; Wilhelm, D.; Koopman, P. Sox genes and cancer. Cytogenet. Genome Res. 2004, 105, 442–447. [Google Scholar] [CrossRef] [PubMed]
- Prokop, J.W.; Lazar, J.; Crapitto, G.; Smith, D.C.; Worthey, E.A.; Jacob, H.J. Molecular modeling in the age of clinical genomics, the enterprise of the next generation. J. Mol. Model. 2017, 23, 75. [Google Scholar] [CrossRef]
- Prokop, J.W.; Jdanov, V.; Savage, L.; Morris, M.; Lamb, N.; VanSickle, E.; Stenger, C.L.; Rajasekaran, S.; Bupp, C.P. Computational and Experimental Analysis of Genetic Variants. Compr. Physiol. 2022, 12, 3303–3336. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Muse, S.V.; Gaut, B.S. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol. Biol. Evol. 1994, 11, 715–724. [Google Scholar] [PubMed]
- Pond, S.L.K.; Frost, S.D.W.; Muse, S.V. HyPhy: Hypothesis testing using phylogenies. Bioinform. Oxf. Engl. 2005, 21, 676–679. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783. [Google Scholar] [CrossRef]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. UniProt: The Universal Protein knowledgebase. Nucleic Acids Res. 2004, 32, D115–D119. [Google Scholar] [CrossRef]
- Dinkel, H.; Michael, S.; Weatheritt, R.J.; Davey, N.E.; Van Roey, K.; Altenberg, B.; Toedt, G.; Uyar, B.; Seiler, M.; Budd, A.; et al. ELM—The database of eukaryotic linear motifs. Nucleic Acids Res. 2012, 40, D242–D251. [Google Scholar] [CrossRef]
- Brown, G.R.; Hem, V.; Katz, K.S.; Ovetsky, M.; Wallin, C.; Ermolaeva, O.; Tolstoy, I.; Tatusova, T.; Pruitt, K.D.; Maglott, D.R.; et al. Gene: A gene-centered information resource at NCBI. Nucleic Acids Res. 2015, 43, D36–D42. [Google Scholar] [CrossRef]
- Papadopoulos, J.S.; Agarwala, R. COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics 2007, 23, 1073–1079. [Google Scholar] [CrossRef]
- Krieger, E.; Joo, K.; Lee, J.; Lee, J.; Raman, S.; Thompson, J.; Tyka, M.; Baker, D.; Karplus, K. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins 2009, 77 (Suppl. S9), 114–122. [Google Scholar] [CrossRef]
- Fontaine, F.R.; Goodall, S.; Prokop, J.W.; Howard, C.B.; Moustaqil, M.; Kumble, S.; Rasicci, D.T.; Osborne, G.W.; Gambin, Y.; Sierecki, E.; et al. Functional domain analysis of SOX18 transcription factor using a single-chain variable fragment-based approach. mAbs 2018, 10, 596–606. [Google Scholar] [CrossRef] [PubMed]
- Klaus, M.; Prokoph, N.; Girbig, M.; Wang, X.; Huang, Y.-H.; Srivastava, Y.; Hou, L.; Narasimhan, K.; Kolatkar, P.R.; Francois, M.; et al. Structure and decoy-mediated inhibition of the SOX18/Prox1-DNA interaction. Nucleic Acids Res. 2016, 44, 3922–3935. [Google Scholar] [CrossRef] [PubMed]
- Lek, M.; Karczewski, K.J.; Minikel, E.V.; Samocha, K.E.; Banks, E.; Fennell, T.; O’Donnell-Luria, A.H.; Ware, J.S.; Hill, A.J.; Cummings, B.B.; et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016, 536, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Hoover, J.; et al. ClinVar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016, 44, D862–D868. [Google Scholar] [CrossRef]
- Wang, J.; Al-Ouran, R.; Hu, Y.; Kim, S.-Y.; Wan, Y.-W.; Wangler, M.F.; Yamamoto, S.; Chao, H.-T.; Comjean, A.; Mohr, S.E.; et al. MARRVEL: Integration of Human and Model Organism Genetic Resources to Facilitate Functional Annotation of the Human Genome. Am. J. Hum. Genet. 2017, 100, 843–853. [Google Scholar] [CrossRef]
- Forbes, S.A.; Bindal, N.; Bamford, S.; Cole, C.; Kok, C.Y.; Beare, D.; Jia, M.; Shepherd, R.; Leung, K.; Menzies, A.; et al. COSMIC: Mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011, 39, D945–D950. [Google Scholar] [CrossRef]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef]
- Prokop, J.W.; Tsaih, S.-W.; Faber, A.B.; Boehme, S.; Underwood, A.C.; Troyer, S.; Playl, L.; Milsted, A.; Turner, M.E.; Ely, D.; et al. The phenotypic impact of the male-specific region of chromosome-Y in inbred mating: The role of genetic variants and gene duplications in multiple inbred rat strains. Biol. Sex Differ. 2016, 7, 10. [Google Scholar] [CrossRef] [PubMed]
- Sierecki, E.; Stevers, L.M.; Giles, N.; Polinkovsky, M.E.; Moustaqil, M.; Mureev, S.; Johnston, W.A.; Dahmer-Heath, M.; Skalamera, D.; Gonda, T.J.; et al. Rapid mapping of interactions between Human SNX-BAR proteins measured in vitro by AlphaScreen and single-molecule spectroscopy. Mol. Cell. Proteom. 2014, 13, 2233–2245. [Google Scholar] [CrossRef] [PubMed]
- Sierecki, E.; Giles, N.; Polinkovsky, M.; Moustaqil, M.; Alexandrov, K.; Gambin, Y. A cell-free approach to accelerate the study of protein-protein interactions in vitro. Interface Focus 2013, 3, 20130018. [Google Scholar] [CrossRef] [PubMed]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; von Mering, C.; et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013, 41, D808–D815. [Google Scholar] [CrossRef]
- Gimelli, G.; Gimelli, S.; Dimasi, N.; Bocciardi, R.; Di Battista, E.; Pramparo, T.; Zuffardi, O. Identification and molecular modelling of a novel familial mutation in the SRY gene implicated in the pure gonadal dysgenesis. Eur. J. Hum. Genet. 2007, 15, 76–80. [Google Scholar] [CrossRef]
- Domenice, S.; Yumie Nishi, M.; Correia Billerbeck, A.E.; Latronico, A.C.; Aparecida Medeiros, M.; Russell, A.J.; Vass, K.; Marino Carvalho, F.; Costa Frade, E.M.; Prado Arnhold, I.J.; et al. A novel missense mutation (S18N) in the 5’ non-HMG box region of the SRY gene in a patient with partial gonadal dysgenesis and his normal male relatives. Hum. Genet. 1998, 102, 213–215. [Google Scholar] [CrossRef]
- Canto, P.; de la Chesnaye, E.; López, M.; Cervantes, A.; Chávez, B.; Vilchis, F.; Reyes, E.; Ulloa-Aguirre, A.; Kofman-Alfaro, S.; Méndez, J.P. A mutation in the 5’ non-high mobility group box region of the SRY gene in patients with Turner syndrome and Y mosaicism. J. Clin. Endocrinol. Metab. 2000, 85, 1908–1911. [Google Scholar] [CrossRef]
- Goji, K.; Nishijima, E.; Tsugawa, C.; Nishio, H.; Pokharel, R.K.; Matsuo, M. Novel missense mutation in the HMG box of SOX9 gene in a Japanese XY male resulted in campomelic dysplasia and severe defect in masculinization. Hum. Mutat. 1998, 11 (Suppl. S1), S114–S116. [Google Scholar] [CrossRef]
- Kwok, C.; Weller, P.A.; Guioli, S.; Foster, J.W.; Mansour, S.; Zuffardi, O.; Punnett, H.H.; Dominguez-Steglich, M.A.; Brook, J.D.; Young, I.D. Mutations in SOX9, the gene responsible for Campomelic dysplasia and autosomal sex reversal. Am. J. Hum. Genet. 1995, 57, 1028–1036. [Google Scholar]
- Scherer, G.; Held, M.; Erdel, M.; Meschede, D.; Horst, J.; Lesniewicz, R.; Midro, A.T. Three novel SRY mutations in XY gonadal dysgenesis and the enigma of XY gonadal dysgenesis cases without SRY mutations. Cytogenet. Cell Genet. 1998, 80, 188–192. [Google Scholar] [CrossRef]
- Meyer, J.; Südbeck, P.; Held, M.; Wagner, T.; Schmitz, M.L.; Bricarelli, F.D.; Eggermont, E.; Friedrich, U.; Haas, O.A.; Kobelt, A.; et al. Mutational analysis of the SOX9 gene in campomelic dysplasia and autosomal sex reversal: Lack of genotype/phenotype correlations. Hum. Mol. Genet. 1997, 6, 91–98. [Google Scholar] [CrossRef]
- Braun, A.; Kammerer, S.; Cleve, H.; Löhrs, U.; Schwarz, H.P.; Kuhnle, U. True hermaphroditism in a 46,XY individual, caused by a postzygotic somatic point mutation in the male gonadal sex-determining locus (SRY): Molecular genetics and histological findings in a sporadic case. Am. J. Hum. Genet. 1993, 52, 578–585. [Google Scholar] [PubMed]
- Staffler, A.; Hammel, M.; Wahlbuhl, M.; Bidlingmaier, C.; Flemmer, A.W.; Pagel, P.; Nicolai, T.; Wegner, M.; Holzinger, A. Heterozygous SOX9 mutations allowing for residual DNA-binding and transcriptional activation lead to the acampomelic variant of campomelic dysplasia. Hum. Mutat. 2010, 31, E1436–E1444. [Google Scholar] [CrossRef] [PubMed]
- Affara, N.A.; Chalmers, I.J.; Ferguson-Smith, M.A. Analysis of the SRY gene in 22 sex-reversed XY females identifies four new point mutations in the conserved DNA binding domain. Hum. Mol. Genet. 1993, 2, 785–789. [Google Scholar] [CrossRef] [PubMed]
- Chaoui, A.; Watanabe, Y.; Touraine, R.; Baral, V.; Goossens, M.; Pingault, V.; Bondurand, N. Identification and functional analysis of SOX10 missense mutations in different subtypes of Waardenburg syndrome. Hum. Mutat. 2011, 32, 1436–1449. [Google Scholar] [CrossRef]
- Haqq, C.M.; King, C.Y.; Ukiyama, E.; Falsafi, S.; Haqq, T.N.; Donahoe, P.K.; Weiss, M.A. Molecular basis of mammalian sexual determination: Activation of Müllerian inhibiting substance gene expression by SRY. Science 1994, 266, 1494–1500. [Google Scholar] [CrossRef]
- Berta, P.; Hawkins, J.R.; Sinclair, A.H.; Taylor, A.; Griffiths, B.L.; Goodfellow, P.N.; Fellous, M. Genetic evidence equating SRY and the testis-determining factor. Nature 1990, 348, 448–450. [Google Scholar] [CrossRef]
- Zeng, Y.T.; Ren, Z.R.; Zhang, M.L.; Huang, Y.; Zeng, F.Y.; Huang, S.Z. A new de novo mutation (A113T) in HMG box of the SRY gene leads to XY gonadal dysgenesis. J. Med. Genet. 1993, 30, 655–657. [Google Scholar] [CrossRef]
- Schmitt-Ney, M.; Thiele, H.; Kaltwasser, P.; Bardoni, B.; Cisternino, M.; Scherer, G. Two novel SRY missense mutations reducing DNA binding identified in XY females and their mosaic fathers. Am. J. Hum. Genet. 1995, 56, 862–869. [Google Scholar]
- Lundberg, Y.; Ritzén, M.; Harlin, J.; Wedell, A. Novel missense mutation (P131R) in the HMG box of SRY in XY sex reversal. Hum. Mutat. 1998, 11, S328–S329. [Google Scholar] [CrossRef]
- Fernandez, R.; Marchal, J.A.; Sanchez, A.; Pasaro, E. A point mutation, R59G, within the HMG-SRY box in a female 45,X/46,X, psu dic(Y)(pter-->q11::q11-->pter). Hum. Genet. 2002, 111, 242–246. [Google Scholar] [CrossRef] [PubMed]
- Vilain, E.; McElreavey, K.; Jaubert, F.; Raymond, J.P.; Richaud, F.; Fellous, M. Familial case with sequence variant in the testis-determining region associated with two sex phenotypes. Am. J. Hum. Genet. 1992, 50, 1008–1011. [Google Scholar] [PubMed]
- Hiort, O.; Gramss, B.; Klauber, G.T. True hermaphroditism with 46,XY karyotype and a point mutation in the SRY gene. J. Pediatr. 1995, 126, 1022. [Google Scholar] [CrossRef] [PubMed]
- Wada, Y.; Nishimura, G.; Nagai, T.; Sawai, H.; Yoshikata, M.; Miyagawa, S.; Hanita, T.; Sato, S.; Hasegawa, T.; Ishikawa, S.; et al. Mutation analysis of SOX9 and single copy number variant analysis of the upstream region in eight patients with campomelic dysplasia and acampomelic campomelic dysplasia. Am. J. Med. Genet. A 2009, 149A, 2882–2885. [Google Scholar] [CrossRef]
- Imai, A.; Takagi, A.; Tamaya, T. A novel sex-determining region on Y (SRY) missense mutation identified in a 46,XY female and also in the father. Endocr. J. 1999, 46, 735–739. [Google Scholar] [CrossRef] [PubMed]
- Okuhara, K.; Tajima, T.; Nakae, J.; Fujieda, K. A novel missense mutation in the HMG box region of the SRY gene in a Japanese patient with an XY sex reversal. J. Hum. Genet. 2000, 45, 112–114. [Google Scholar] [CrossRef]
- Cunha, J.L.; Soardi, F.C.; Bernardi, R.D.; Oliveira, L.E.C.; Benedetti, C.E.; Guerra-Junior, G.; Maciel-Guerra, A.T.; de Mello, M.P. The novel p.E89K mutation in the SRY gene inhibits DNA binding and causes the 46,XY disorder of sex development. Braz. J. Med. Biol. Res. 2011, 44, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, J.R.; Taylor, A.; Goodfellow, P.N.; Migeon, C.J.; Smith, K.D.; Berkovitz, G.D. Evidence for increased prevalence of SRY mutations in XY females with complete rather than partial gonadal dysgenesis. Am. J. Hum. Genet. 1992, 51, 979–984. [Google Scholar]
- Dörk, T.; Stuhrmann, M.; Miller, K.; Schmidtke, J. Independent observation of SRY mutation I90M in a patient with complete gonadal dysgenesis. Hum. Mutat. 1998, 11, 90–91. [Google Scholar] [CrossRef]
- Maier, E.M.; Leitner, C.; Löhrs, U.; Kuhnle, U. True hermaphroditism in an XY individual due to a familial point mutation of the SRY gene. J. Pediatr. Endocrinol. Metab. 2003, 16, 575–580. [Google Scholar] [CrossRef]
- Schäffler, A.; Barth, N.; Winkler, K.; Zietz, B.; Rümmele, P.; Knüchel, R.; Schölmerich, J.; Palitzsch, K.D. Identification of a new missense mutation (Gly95Glu) in a highly conserved codon within the high-mobility group box of the sex-determining region Y gene: Report on a 46,XY female with gonadal dysgenesis and yolk-sac tumor. J. Clin. Endocrinol. Metab. 2000, 85, 2287–2292. [Google Scholar] [CrossRef] [PubMed]
- Jakubiczka, S.; Bettecken, T.; Stumm, M.; Neulen, J.; Wieacker, P. Another mutation within the HMG-box of the SRY gene associated with Swyer syndrome. Hum. Mutat. 1999, 13, 85. [Google Scholar] [CrossRef]
- Jäger, R.J.; Harley, V.R.; Pfeiffer, R.A.; Goodfellow, P.N.; Scherer, G. A familial mutation in the testis-determining gene SRY shared by both sexes. Hum. Genet. 1992, 90, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Preiss, S.; Argentaro, A.; Clayton, A.; John, A.; Jans, D.A.; Ogata, T.; Nagai, T.; Barroso, I.; Schafer, A.J.; Harley, V.R. Compound effects of point mutations causing campomelic dysplasia/autosomal sex reversal upon SOX9 structure, nuclear transport, DNA binding, and transcriptional activation. J. Biol. Chem. 2001, 276, 27864–27872. [Google Scholar] [CrossRef] [PubMed]
- McDowall, S.; Argentaro, A.; Ranganathan, S.; Weller, P.; Mertin, S.; Mansour, S.; Tolmie, J.; Harley, V. Functional and structural studies of wild type SOX9 and mutations causing campomelic dysplasia. J. Biol. Chem. 1999, 274, 24023–24030. [Google Scholar] [CrossRef] [PubMed]
- Poulat, F.; Soullier, S.; Gozé, C.; Heitz, F.; Calas, B.; Berta, P. Description and functional implications of a novel mutation in the sex-determining gene SRY. Hum. Mutat. 1994, 3, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Jordan, B.K.; Jain, M.; Natarajan, S.; Frasier, S.D.; Vilain, E. Familial mutation in the testis-determining gene SRY shared by an XY female and her normal father. J. Clin. Endocrinol. Metab. 2002, 87, 3428–3432. [Google Scholar] [CrossRef]
- Thong, M.K.; Scherer, G.; Kozlowski, K.; Haan, E.; Morris, L. Acampomelic campomelic dysplasia with SOX9 mutation. Am. J. Med. Genet. 2000, 93, 421–425. [Google Scholar] [CrossRef] [PubMed]
- Petrovski, S.; Wang, Q.; Heinzen, E.L.; Allen, A.S.; Goldstein, D.B. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013, 9, e1003709. [Google Scholar] [CrossRef]
- Feng, C.-W.A.; Spiller, C.; Merriner, D.J.; O’Bryan, M.K.; Bowles, J.; Koopman, P. SOX30 is required for male fertility in mice. Sci. Rep. 2017, 7, 17619. [Google Scholar] [CrossRef]
- Béranger, F.; Méjean, C.; Moniot, B.; Berta, P.; Vandromme, M. Muscle differentiation is antagonized by SOX15, a new member of the SOX protein family. J. Biol. Chem. 2000, 275, 16103–16109. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-J.; Göring, W.; Ochs, M.; Mühlfeld, C.; Steding, G.; Paprotta, I.; Engel, W.; Adham, I.M. Sox15 is required for skeletal muscle regeneration. Mol. Cell. Biol. 2004, 24, 8428–8436. [Google Scholar] [CrossRef] [PubMed]
- Merchan-Sala, P.; Nardini, D.; Waclaw, R.R.; Campbell, K. Selective neuronal expression of the SoxE factor, Sox8, in direct pathway striatal projection neurons of the developing mouse brain. J. Comp. Neurol. 2017, 525, 2805–2819. [Google Scholar] [CrossRef]
- Cheng, Y.C.; Lee, C.J.; Badge, R.M.; Orme, A.T.; Scotting, P.J. Sox8 gene expression identifies immature glial cells in developing cerebellum and cerebellar tumours. Brain Res. Mol. Brain Res. 2001, 92, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, I.; Fernández-Alvarez, P.; Plaja, A.; Ariceta, G.; Sabaté-Rotés, A.; García-Arumí, E.; Vendrell, T.; Tizzano, E. Further delineation of the SOX18-related Hypotrichosis, Lymphedema, Telangiectasia syndrome (HTLS). Eur. J. Med. Genet. 2018, 61, 269–272. [Google Scholar] [CrossRef]
- Downes, M.; François, M.; Ferguson, C.; Parton, R.G.; Koopman, P. Vascular defects in a mouse model of hypotrichosis-lymphedema-telangiectasia syndrome indicate a role for SOX18 in blood vessel maturation. Hum. Mol. Genet. 2009, 18, 2839–2850. [Google Scholar] [CrossRef] [PubMed]
- Fontijn, R.D.; Volger, O.L.; Fledderus, J.O.; Reijerkerk, A.; de Vries, H.E.; Horrevoets, A.J.G. SOX-18 controls endothelial-specific claudin-5 gene expression and barrier function. Am. J. Physiol. Heart Circ. Physiol. 2008, 294, H891–H900. [Google Scholar] [CrossRef]
- Nitta, T.; Hata, M.; Gotoh, S.; Seo, Y.; Sasaki, H.; Hashimoto, N.; Furuse, M.; Tsukita, S. Size-selective loosening of the blood-brain barrier in claudin-5-deficient mice. J. Cell Biol. 2003, 161, 653–660. [Google Scholar] [CrossRef]
- Greene, C.; Hanley, N.; Reschke, C.R.; Reddy, A.; Mäe, M.A.; Connolly, R.; Behan, C.; O’Keeffe, E.; Bolger, I.; Hudson, N.; et al. Microvascular stabilization via blood-brain barrier regulation prevents seizure activity. Nat. Commun. 2022, 13, 2003. [Google Scholar] [CrossRef]
- Dailey, C.; Oshodi, R.B.; Boull, C.; Aggarwal, A. Expanding the clinical spectrum of SOX18-related Hypotrichosis-lymphedema-telangiectasia-renal defect syndrome. Eur. J. Med. Genet. 2022, 65, 104607. [Google Scholar] [CrossRef]
- Nowakowska, B.A.; Obersztyn, E.; Szymańska, K.; Bekiesińska-Figatowska, M.; Xia, Z.; Ricks, C.B.; Bocian, E.; Stockton, D.W.; Szczałuba, K.; Nawara, M.; et al. Severe mental retardation, seizures, and hypotonia due to deletions of MEF2C. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2010, 153B, 1042–1051. [Google Scholar] [CrossRef]
- Vrečar, I.; Innes, J.; Jones, E.A.; Kingston, H.; Reardon, W.; Kerr, B.; Clayton-Smith, J.; Douzgou, S. Further Clinical Delineation of the MEF2C Haploinsufficiency Syndrome: Report on New Cases and Literature Review of Severe Neurodevelopmental Disorders Presenting with Seizures, Absent Speech, and Involuntary Movements. J. Pediatr. Genet. 2017, 6, 129–141. [Google Scholar] [CrossRef]
- Borlot, F.; Whitney, R.; Cohn, R.D.; Weiss, S.K. MEF2C-related epilepsy: Delineating the phenotypic spectrum from a novel mutation and literature review. Seizure 2019, 67, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, T.; Saitsu, H.; Endo, W.; Kikuchi, A.; Uematsu, M.; Haginoya, K.; Hino-fukuyo, N.; Kobayashi, T.; Iwasaki, M.; Tominaga, T.; et al. RBPJ is disrupted in a case of proximal 4p deletion syndrome with epilepsy. Brain Dev. 2014, 36, 532–536. [Google Scholar] [CrossRef] [PubMed]
- Bernard, P.; Tang, P.; Liu, S.; Dewing, P.; Harley, V.R.; Vilain, E. Dimerization of SOX9 is required for chondrogenesis, but not for sex determination. Hum. Mol. Genet. 2003, 12, 1755–1765. [Google Scholar] [CrossRef]
- Overman, J.; Fontaine, F.; Moustaqil, M.; Mittal, D.; Sierecki, E.; Sacilotto, N.; Zuegg, J.; Robertson, A.A.; Holmes, K.; Salim, A.A.; et al. Pharmacological targeting of the transcription factor SOX18 delays breast cancer in mice. eLife 2017, 6, e21221. [Google Scholar] [CrossRef]
- Restrepo, N.A.; Cooke Bailey, J.N. Primary Open-Angle Glaucoma Genetics in African Americans. Curr. Genet. Med. Rep. 2017, 5, 167–174. [Google Scholar] [CrossRef]
- Zhou, Y.; Williams, J.; Smallwood, P.M.; Nathans, J. Sox7, Sox17, and Sox18 Cooperatively Regulate Vascular Development in the Mouse Retina. PloS ONE 2015, 10, e0143650. [Google Scholar] [CrossRef]
- Sreenivasan, R.; Gonen, N.; Sinclair, A. SOX Genes and Their Role in Disorders of Sex Development. Sex. Dev. 2022, 16, 80–91. [Google Scholar] [CrossRef]
- Prokop, J.W.; Deschepper, C.F. Chromosome Y genetic variants: Impact in animal models and on human disease. Physiol. Genom. 2015, 47, 525–537. [Google Scholar] [CrossRef]
- Kemena, C.; Notredame, C. Upcoming challenges for multiple sequence alignment methods in the high-throughput era. Bioinformatics 2009, 25, 2455–2465. [Google Scholar] [CrossRef] [PubMed]
- Brookes, A.J.; Robinson, P.N. Human genotype-phenotype databases: Aims, challenges and opportunities. Nat. Rev. Genet. 2015, 16, 702–715. [Google Scholar] [CrossRef] [PubMed]
- Merino, F.; Ng, C.K.L.; Veerapandian, V.; Schöler, H.R.; Jauch, R.; Cojocaru, V. Structural basis for the SOX-dependent genomic redistribution of OCT4 in stem cell differentiation. Structure 2014, 22, 1274–1286. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Clayton, D.G.; Concannon, P.; Akolkar, B.; Cooper, J.D.; Erlich, H.A.; Julier, C.; Morahan, G.; Nerup, J.; Nierras, C.; et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 2009, 41, 703–707. [Google Scholar] [CrossRef] [PubMed]
- Peterson, R.E.; Kuchenbaecker, K.; Walters, R.K.; Chen, C.-Y.; Popejoy, A.B.; Periyasamy, S.; Lam, M.; Iyegbe, C.; Strawbridge, R.J.; Brick, L.; et al. Genome-wide Association Studies in Ancestrally Diverse Populations: Opportunities, Methods, Pitfalls, and Recommendations. Cell 2019, 179, 589–603. [Google Scholar] [CrossRef]
- The All of Us Research Program Investigators; Denny, J.C.; Rutter, J.L.; Goldstein, D.B.; Philippakis, A.; Smoller, J.W.; Jenkins, G.; Dishman, E. The “All of Us” Research Program. N. Engl. J. Med. 2019, 381, 668–676. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).