Support Interval for Two-Sample Summary Data-Based Mendelian Randomization
(This article belongs to the Section Bioinformatics)
Abstract
1. Introduction
- Relevance:
- It is associated with the exposure x (i.e., );
- Exclusion Restriction:
- It affects the outcome y only through its association with the exposure; and
- Exchangeability:
- It is not associated with any confounders of the exposure–outcome association, which implies .
2. Materials and Methods
2.1. One-Sample Individual-Level Data
2.2. Two Independent Samples with a Selected SNP
2.3. Support of Profile Likelihood
3. An Empirical Data Analysis
4. Discussion
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GWAS | genome-wide association study |
| IV | instrumental variable |
| MR | Mendelian randomization |
| SNP | single nucleotide polymorphism |
| TSLS | two-stage least-squares |
References
- Hemani, G.; Tilling, K.; Smith, G.D. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017, 13, e1007081. [Google Scholar]
- Zhu, Z.; Zhang, F.; Hu, H.; Bakshi, A.; Robinson, M.R.; Powell, J.E.; Montgomery, G.W.; Goddard, M.E.; Wray, N.R.; Visscher, P.M.; et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016, 48, 481. [Google Scholar] [CrossRef]
- Morrison, J.; Knoblauch, N.; Marcus, J.H.; Stephens, M.; He, X. Mendelian randomization accounting for correlated and uncorrelated pleiotropic effects using genome-wide summary statistics. Nat. Genet. 2020, 52, 740–747. [Google Scholar] [CrossRef]
- Davey Smith, G.; Ebrahim, S. ‘Mendelian randomization’: Can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 2003, 32, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Butterworth, A.; Thompson, S.G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 2013, 37, 658–665. [Google Scholar] [CrossRef] [PubMed]
- Bowden, J.; Davey Smith, G.; Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015, 44, 512–525. [Google Scholar] [CrossRef] [PubMed]
- Hemani, G.; Zheng, J.; Elsworth, B.; Wade, K.H.; Haberland, V.; Baird, D.; Laurin, C.; Burgess, S.; Bowden, J.; Langdon, R.; et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 2018, 7, e34408. [Google Scholar] [CrossRef]
- Zhao, Q.; Wang, J.; Hemani, G.; Bowden, J.; Small, D.S. Statistical inference in two-sample summary-data Mendelian randomization using robust adjusted profile score. Ann. Stat. 2020, 48, 1742–1769. [Google Scholar] [CrossRef]
- Wang, K.; Han, S. Effect of selection bias on two sample summary data based Mendelian randomization. Sci. Rep. 2021, 11, 7585. [Google Scholar] [CrossRef]
- Ye, T.; Shao, J.; Kang, H. Debiased inverse-variance weighted estimator in two-sample summary-data Mendelian randomization. Ann. Stat. 2021, 49, 2079–2100. [Google Scholar] [CrossRef]
- Bigdeli, T.B.; Lee, D.; Webb, B.T.; Riley, B.P.; Vladimirov, V.I.; Fanous, A.H.; Kendler, K.S.; Bacanu, S.A. A simple yet accurate correction for winner’s curse can predict signals discovered in much larger genome scans. Bioinformatics 2016, 32, 2598–2603. [Google Scholar] [CrossRef]
- Jiang, T.; Gill, D.; Butterworth, A.S.; Burgess, S. An empirical investigation into the impact of winner’s curse on estimates from Mendelian randomization. medRxiv 2022. [Google Scholar] [CrossRef]
- Forde, A.; Hemani, G.; Ferguson, J. Review and further developments in statistical corrections for Winner’s Curse in genetic association studies. bioRxiv 2022. [Google Scholar] [CrossRef]
- Zhao, Q.; Chen, Y.; Wang, J.; Small, D.S. Powerful three-sample genome-wide design and robust statistical inference in summary-data Mendelian randomization. Int. J. Epidemiol. 2019, 48, 1478–1492. [Google Scholar] [CrossRef]
- Jo, E.J.; Han, S.; Wang, K. Estimation of Causal Effect of Age at Menarche on Pubertal Height Growth Using Mendelian Randomization. Genes 2022, 13, 710. [Google Scholar] [CrossRef]
- Hannon, E.; Gorrie-Stone, T.J.; Smart, M.C.; Burrage, J.; Hughes, A.; Bao, Y.; Kumari, M.; Schalkwyk, L.C.; Mill, J. Leveraging DNA-methylation quantitative-trait loci to characterize the relationship between methylomic variation, gene expression, and complex traits. Am. J. Hum. Genet. 2018, 103, 654–665. [Google Scholar] [CrossRef]
- Lee, B.; Yao, X.; Shen, L. Integrative analysis of summary data from GWAS and eQTL studies implicates genes differentially expressed in Alzheimer’s disease. BMC Genom. 2022, 23, 414. [Google Scholar] [CrossRef]
- Porcu, E.; Rüeger, S.; Lepik, K.; Santoni, F.A.; Reymond, A.; Kutalik, Z. Mendelian randomization integrating GWAS and eQTL data reveals genetic determinants of complex and clinical traits. Nat. Commun. 2019, 10, 3300. [Google Scholar] [CrossRef]
- Porcu, E.; Sadler, M.C.; Lepik, K.; Auwerx, C.; Wood, A.R.; Weihs, A.; Sleiman, M.S.B.; Ribeiro, D.M.; Bandinelli, S.; Tanaka, T.; et al. Differentially expressed genes reflect disease-induced rather than disease-causing changes in the transcriptome. Nat. Commun. 2021, 12, 5647. [Google Scholar] [CrossRef]
- Zhu, Z.; Zheng, Z.; Zhang, F.; Wu, Y.; Trzaskowski, M.; Maier, R.; Robinson, M.R.; McGrath, J.J.; Visscher, P.M.; Wray, N.R.; et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat. Commun. 2018, 9, 224. [Google Scholar] [CrossRef]
- Jin, C.; Lee, B.; Shen, L.; Long, Q.; Alzheimer’s Disease Neuroimaging Initiative; Alzheimer’s Disease Metabolomics Consortium. Integrating multi-omics summary data using a Mendelian randomization framework. Brief. Bioinform. 2022, 23, bbac376. [Google Scholar] [CrossRef] [PubMed]
- Pustejovsky, J.E. 2SLS Standard Errors and the Delta-Method. 2017. Available online: https://www.jepusto.com/delta-method-and-2sls-ses/ (accessed on 11 November 2022).
- Greene, W.H. Econometric Analysis, 6th ed.; Pearson-Prentice Hall: New York, NY, USA, 2008. [Google Scholar]
- Zhao, Q.; Wang, J.; Spiller, W.; Bowden, J.; Small, D.S. Two-sample instrumental variable analyses using heterogeneous samples. Stat. Sci. 2019, 34, 317–333. [Google Scholar] [CrossRef]
- Burgess, S.; Dudbridge, F.; Thompson, S.G. Combining information on multiple instrumental variables in Mendelian randomization: Comparison of allele score and summarized data methods. Stat. Med. 2016, 35, 1880–1906. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Zou, F.; Wright, F.A. Estimating odds ratios in genome scans: An approximate conditional likelihood approach. Am. J. Hum. Genet. 2008, 82, 1064–1074. [Google Scholar] [CrossRef]
- Edwards, A.W.F. Likelihood; CUP Archive: New York, NY, USA, 1984. [Google Scholar]
- Perry, J.R.; Day, F.; Elks, C.E.; Sulem, P.; Thompson, D.J.; Ferreira, T.; He, C.; Chasman, D.I.; Esko, T.; Thorleifsson, G.; et al. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature 2014, 514, 92–97. [Google Scholar] [CrossRef]
- Cousminer, D.L.; Berry, D.J.; Timpson, N.J.; Ang, W.; Thiering, E.; Byrne, E.M.; Taal, H.R.; Huikari, V.; Bradfield, J.P.; Kerkhof, M.; et al. Genome-wide association and longitudinal analyses reveal genetic loci linking pubertal height growth, pubertal timing and childhood adiposity. Hum. Mol. Genet. 2013, 22, 2735–2747. [Google Scholar] [CrossRef]
- Staiger, D.O.; Stock, J.H. Instrumental Variables Regression with Weak Instruments; Cowles Foundation Discussion Papers: London, UK, 1994. [Google Scholar]
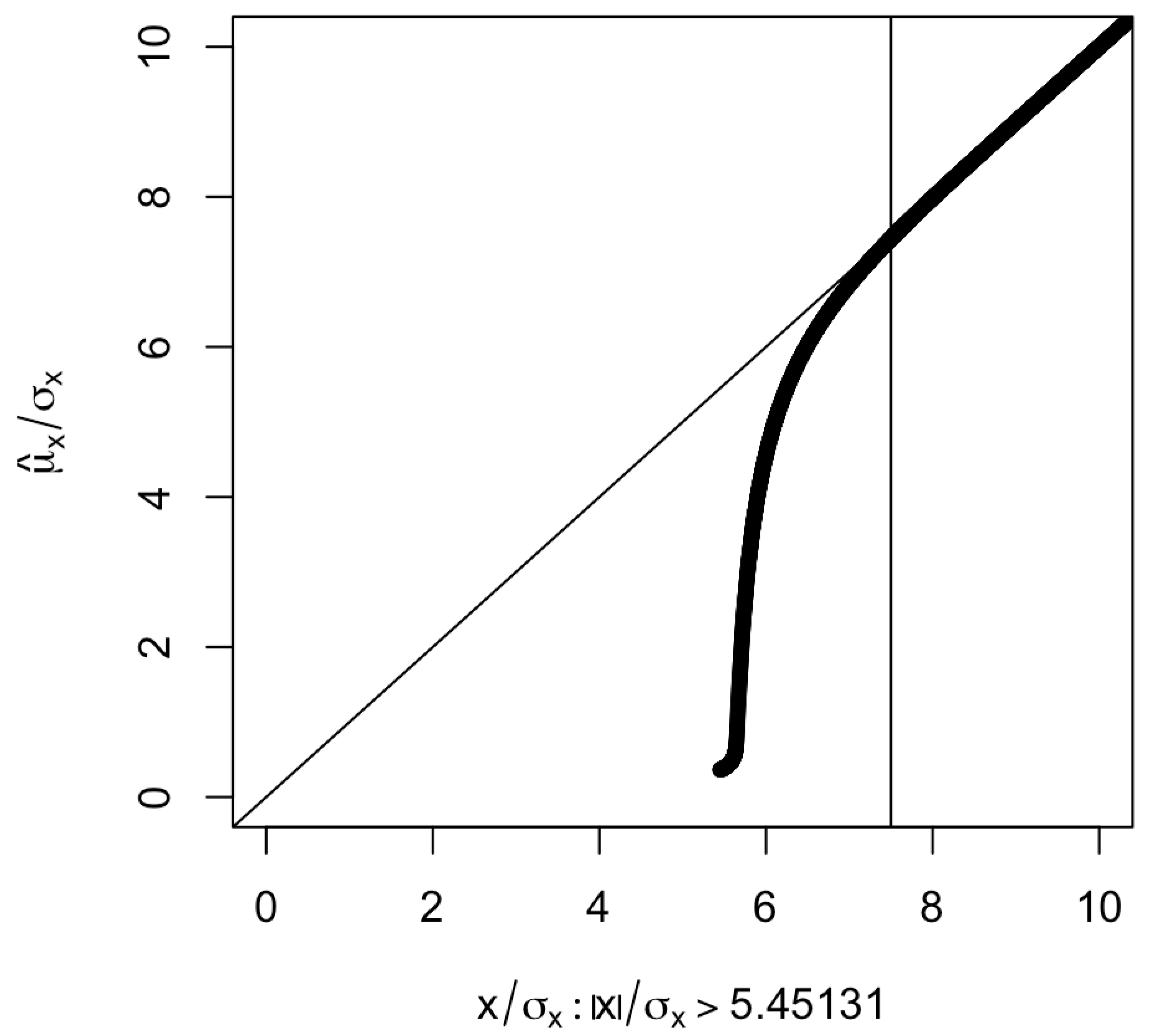
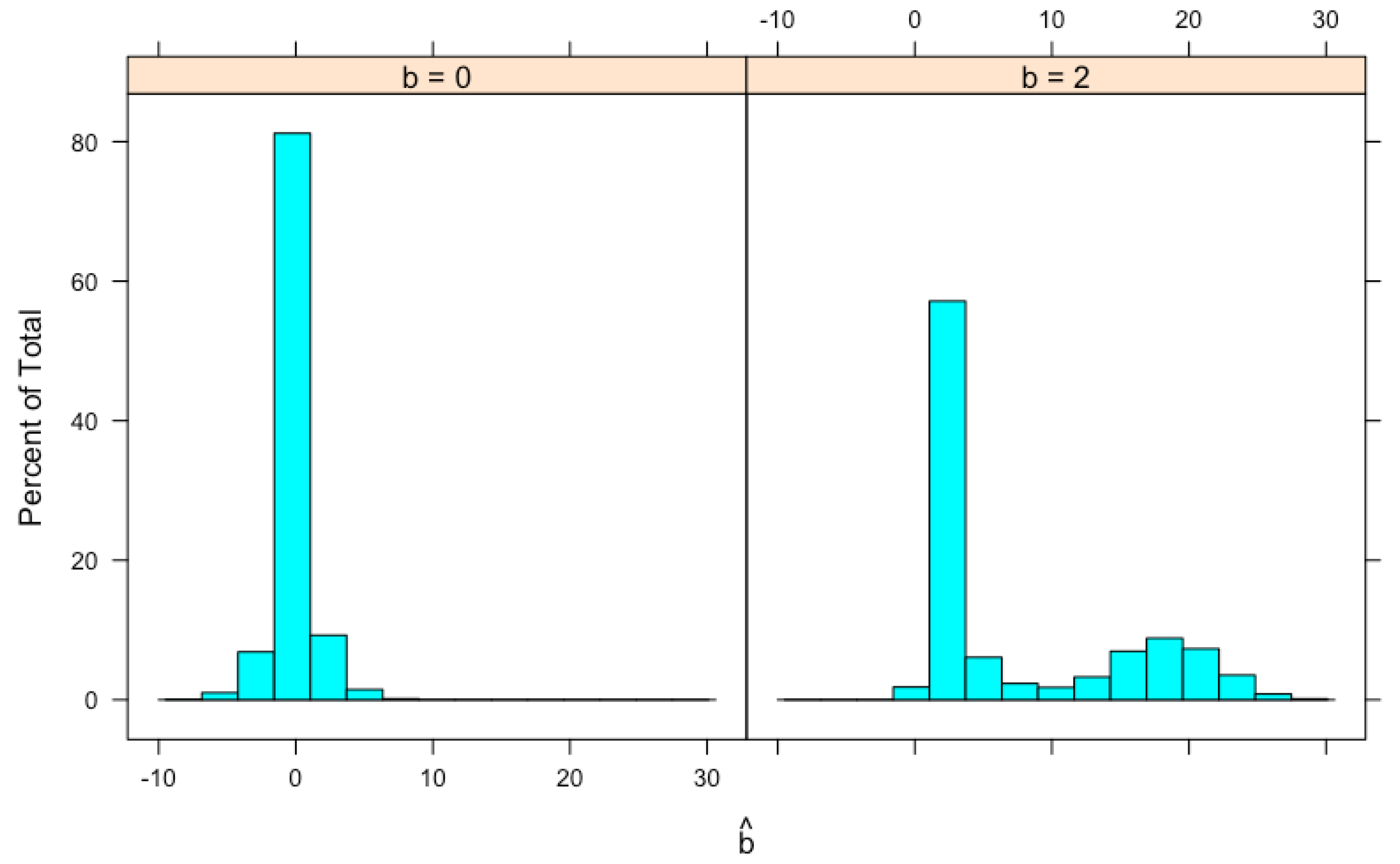

| b | |||||
|---|---|---|---|---|---|
| Method | 0 | 0.5 | 1 | 1.5 | 2 |
| Winner’s-curse-corrected | |||||
| Mean of | 0.0073 | 1.8743 | 3.7414 | 5.6084 | 7.4755 |
| Median of | 0.0022 | 0.6843 | 1.3091 | 1.9327 | 2.5700 |
| Coverage of 2-unit support | 0.9587 | 0.9725 | 0.9803 | 0.9816 | 0.9811 |
| Power of T for testing | 0.0471 | 0.5217 | 0.9807 | 1.0000 | 1.0000 |
| SMR | |||||
| Mean of | 0.0019 | 0.3424 | 0.6829 | 1.0234 | 1.3639 |
| Median of | −0.3310 | 0.3405 | 0.6795 | 1.0199 | 1.3615 |
| Coverage of 95% CI | 0.9648 | 0.8524 | 0.6511 | 0.4966 | 0.3958 |
| Power for testing | 0.0353 | 0.4721 | 0.9726 | 1.0000 | 1.0000 |
| Winner’s-Curse-Corrected Method | |||
|---|---|---|---|
| SNP | Gene Name | (5.9-Unit Support) | p-Value |
| Total pubertal height growth | |||
| rs7514705 | TNNI3K | 2.048 (0.889, 3.807) | |
| rs7642134 | POU1F1 | 2.474 (1.264, 4.433) | |
| Late pubertal height growth | |||
| rs7514705 | TNNI3K | 1.822 (0.057, 5.091) | |
| rs7759938 | LIN28B | 0.931 (0.335, 1.571) | |
| SMR Method | |||
| SNP | Gene Name | (99.94% CI) | p-Value |
| Total pubertal height growth | |||
| rs7514705 | TNNI3K | 2.042 (0.330, 3.754) | |
| rs7642134 | POU1F1 | 2.466 (0.647, 4.284) | |
| Late pubertal height growth | |||
| rs7759938 | LIN28B | 0.931 (0.330, 1.533) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, K. Support Interval for Two-Sample Summary Data-Based Mendelian Randomization. Genes 2023, 14, 211. https://doi.org/10.3390/genes14010211
Wang K. Support Interval for Two-Sample Summary Data-Based Mendelian Randomization. Genes. 2023; 14(1):211. https://doi.org/10.3390/genes14010211
Chicago/Turabian StyleWang, Kai. 2023. "Support Interval for Two-Sample Summary Data-Based Mendelian Randomization" Genes 14, no. 1: 211. https://doi.org/10.3390/genes14010211
APA StyleWang, K. (2023). Support Interval for Two-Sample Summary Data-Based Mendelian Randomization. Genes, 14(1), 211. https://doi.org/10.3390/genes14010211






