Abstract
Genome-wide association studies (GWAS) allow the identification of associations between genetic variants and important phenotypes in domestic animals, including disease-resistance traits. Whole Genome Sequencing (WGS) data can help increase the resolution and statistical power of association mapping. Here, we conduced GWAS to asses he facultative intracellular bacterium Piscirickettsia salmonis, which affects farmed rainbow trout, Oncorhynchus mykiss, in Chile using imputed genotypes at the sequence level and searched for candidate genes located in genomic regions associated with the trait. A total of 2130 rainbow trout were intraperitoneally challenged with P. salmonis under controlled conditions and genotyped using a 57K single nucleotide polymorphism (SNP) panel. Genotype imputation was performed in all the genotyped animals using WGS data from 102 individuals. A total of 488,979 imputed WGS variants were available in the 2130 individuals after quality control. GWAS revealed genome-wide significant quantitative trait loci (QTL) in Omy02, Omy03, Omy25, Omy26 and Omy27 for time to death and in Omy26 for binary survival. Twenty-four (24) candidate genes associated with P. salmonis resistance were identified, which were mainly related to phagocytosis, innate immune response, inflammation, oxidative response, lipid metabolism and apoptotic process. Our results provide further knowledge on the genetic variants and genes associated with resistance to intracellular bacterial infection in rainbow trout.
1. Introduction
In 2010, the world production of rainbow trout was 752,400 tons, and in 2020, it increased to 960,600 tons; a very rapid growth of almost 27% in ten years [1]. In 2020, Chile harvested around 85,900 tons of rainbow trout [2]. The production of this species is highly affected by infectious diseases, such as piscirickettsiosis or Salmonid Rickettsial Syndrome (SRS) caused by P. salmonis, producing the greatest economic losses. In the first semester of 2022, 8.5% of the total mortality was attributed to SRS [3]. Prophylactic and control strategies against P. salmonis infection have been based mainly on vaccines and antibiotic administration during disease outbreaks. However, both approaches show low effectiveness in controlling SRS in field conditions [4,5]. During the first semester of 2022, 94.8% of the antibiotics administered to salmonids in Chile were for the treatment of P. salmonis infection [6]. The use of antibiotics might result in resistant bacterial strains, not only reducing the efficacy of treatments and affecting the success and sustainability of aquaculture but also representing an important threat to public health [7,8]. The limited efficacy of vaccines on reducing mortalities in sea-cages might be due to the variation in the host immune response between families and the complexity of the P. salmonis–host interaction [9,10,11,12,13,14,15]. An alternative strategy to reduce the detrimental effects of this disease in the long term, is the genetic selection of fish more resistant to P. salmonis infection [16,17,18]. In fact, the most important salmon and trout breeding companies in Chile currently offer SRS-resistant eggs within their products, which are generated from families which have shown superior survival in either experimental or field conditions.
With the advances in the genomics of aquatic organisms in recent years [18], it is now possible to include information from dense molecular markers (e.g., single nucleotide polymorphisms (SNPs), in aquaculture breeding programs [19]. Genomic information has two main applications for breeding programs: genomic selection and genome-wide association studies (GWAS). The genomic selection allows the estimation of genomic breeding values of selection candidates with greater accuracy than pedigree-based methods [20], which accelerates the genetic progress for the trait of interest. For example, the effect of genomic selection in increasing the accuracy on the estimation of the genetic merit for resistance to P. salmonis infection has been demonstrated in Atlantic Salmon [21], coho Salmon [22] and rainbow trout [18,23].
The GWAS aims to map quantitative trait loci (QTL) to understand the genetic architecture and find DNA variants associated with the trait of interest [24]. This approach is based on the evaluation of the association between genotypes and phenotypes, using different statistical methods [25]. Several factors may influence the accuracy of GWAS such as the heritability of the trait, genetic architecture, statistical method for the association, number of samples (n), and number of SNPs [26,27,28,29,30]. The inclusion of whole-genome sequence (WGS) genotypes might increase the accuracy of genomic prediction and QTL mapping compared to lower-density SNP arrays, especially if causal genetic variants are included in the analysis [31,32,33]. Despite the drastic reduction in WGS costs in recent years, it is still cheaper to genotype animals using SNP chips than re-sequencing individuals at the whole-genome level [34]. An interesting approach to reduce the cost of obtaining WGS individual data is the imputation of genotypes, which consists of predicting unknown genotypes in animals genotyped with a panel of SNPs of a lower density using animals genotyped with a panel of denser SNPs or whole-genome sequenced as reference [35]. The imputation of SNP genotypes to WGS has been performed in pigs [36,37], poultry [38,39], cattle [34,40,41], rabbits [42], rainbow trout [43] and Nile tilapia [44,45] as a strategy to increase the number of SNPs to boost statistical power and accuracy of association between markers and QTLs [33].
Mapping of QTLs through GWAS has been used to identify genomic regions related to disease resistance in different fish species [19]. GWAS for resistance against different diseases, including Flavobacterium psychrophilum [46,47,48], Aeromonas salmonicida subsp. salmonicida [49], Vibrio anguillarum [50], Ichthyophthirius multifiliis [51], IPN virus [52], Flavobacterium columnare [53] and Yersinia ruckeri [54], have been performed in rainbow trout using a medium density (57K) SNP chip. Potential QTLs associated with P. salmonis resistance have been found in rainbow trout and other salmonid species [21,22,23,55,56]. For instance, resistance to P. salmonis infection was determined to be a polygenic trait in rainbow trout, Atlantic salmon and coho salmon, with few QTL explaining a moderate proportion of the genetic variance for the trait [22,23,55]. All these studies were performed using medium-density SNP panels, including tens of thousands of markers, making it difficult to map genomic regions with high accuracy. This study aimed to perform GWAS for resistance against SRS using genotype data imputed to high-density SNP information (i.e., WGS) for increasing statistical power and resolution in the detection of genomic regions and putative genes controlling the trait.
2. Materials and Methods
2.1. Origin of Animals and Challenge to P. salmonis
The animals used in this study belonged to the 2011 year-class from the rainbow trout breeding nucleus owned by EFFIGEN S.A. (Puerto Montt, Chile). This breeding program has applied selection for different traits of interest (growth, carcass quality and appearance) since 2001. For the experimental challenge against P. salmonis, juveniles from 102 families, weighing approximately 7.0 ± 1.5 g were individually tagged with a PIT-tag, kept for seven months in a single tank, and then transferred to the Aquainnovo Research Station (X Region, Chile). The fish had an acclimatization period of 20 days before the challenge. Random sampling was performed to check for Flavobacterium spp., Infectious Pancreatic Necrosis virus (IPNV), infectious salmon anemia virus (ISAV), and Renibacterium salmoninarum using qRT-PCR. Then, 2130 juveniles (an average of 20 individuals per family, ranging from 12 to 27 fish) were inoculated intraperitoneally (IP) with 0.2 mL of the P. salmonis strain LF-89 at a lethal dose of 50, as described by [23]. After inoculation, the fish were distributed equally in three tanks keeping five to nine fish per family in each tank. The challenge lasted 32 days, in which daily mortality and final weight were recorded at death or at the end of the experiment if the fish survived. Survivor animals were euthanized, and their body weight were also recorded. Tissue samples were taken from the caudal fin to isolate the genomic DNA of all challenged fish and stored in 95% ethanol at −80 °C for subsequent genotyping.
Resistance against SRS was evaluated using the time of death (TD) with values ranging from day 10 to day 32 and binary survival (BS), where 1 indicates that the fish died and 0 that it survived challenge.
2.2. Genotyping of Challenged Animals
Genomic DNA was extracted from the 2130 sampled fins using a commercial kit (DNeasy Blood & Tissue Kit, Qiagen, China), following the manufacturer’s instructions. Individual genotypes were obtained using a commercial 57K SNP chip (Affymetrix Axiom) developed by the National Center for Cool and Cold Water Aquaculture of the USDA Agricultural Research Service [57]. The quality control (QC) of the genotypes was applied through the Plink v1.90 software [58], with a call rate for SNP and sample higher than 0.90, minor allele frequency (MAF) threshold of 0.01 and Hardy–Weinberg equilibrium (HWE) of p-value > .
2.3. Whole-Genome Sequence Data
Detailed information on the sequencing procedures is available in [43]. Briefly, genomic DNA was extracted from the fin-clips of 102 individuals using the DNeasy Blood & Tissue kit (Qiagen) according to the manufacturer’s instructions. The samples were sent to the Beijing Genomics Institute (BGI, China) for whole genome sequencing using DNBseq technology. The O. mykiss genome (GenBank Assembly Accession GCA_013265735.3 USDA_OmykA_1.1) was used to align re-sequencing data. This reference genome version is comprised of a final assembly length of ~2.33 Gbp, 1591 contigs with an N50 contig length of 9, 8 Mb, of which ~95% is anchored in 29 chromosomal sequences. The re-sequencing reads from each sample were mapped to the reference genome using the Burrows-Wheeler Aligner (BWA) analysis tool [59], resulting in a mapping rate and effective mapping depth between 97.51 and 98.16%, and 10.31× and 17.65×, respectively. A protocol implemented in the Genome Analysis Toolkit (GATK, https://www.broadinstitute.org/gatk/, accessed on 19 June 2021) was used for the SNP calling. The final VCF file consisted of 22.6 million non-redundant variants present in the 102 rainbow trout re-sequenced individuals. The quality control of genotypes was performed with Plink v1.90 [58] using the following criteria: SNP and sample call-rate higher than 0.80, MAF > 0.01 y HWE p-value > .
2.4. Imputation to Whole Genome Sequence Level
For the validation of the imputation, five cross-validation sets were formed using only the 102 parents divided into 20% validation (approximately 20 animals per group) and 80% reference (approximately 80 animals per group) sets. The validation animals had only the 57K SNPs in common with the genotyped animals (challenged animals)and the remaining SNPs were masked to perform genotype imputation. Reference animals had information at WGS level including only SNPs that passed by QC. FImpute v3.0 [60] was used to perform all genotype imputations. Accuracy was assessed using the r2 (Pearson’s squared correlation between observed and imputed genotypes) statistic, and SNPs with low imputation accuracy (lower than 0.8) were removed from the final set of imputable SNPs. Finally, the challenged animals genotyped with 57k SNPs were imputed using only validated SNPs with an accuracy above 0.8.
2.5. Genome-Wide Association Study (GWAS)
GWAS was performed for two trait definitions: time to death (TD) and binary survival (BS) using a linear mixed animal model implemented in the Genome Complex Trait Analysis (GCTA) software. This software uses a Restricted Maximum Likelihood (REML) analysis to estimate the proportion of additive genetic variation captured from all SNPs [58] through the mlma function. The linear mixed model applied was:
where Y is a vector of phenotypes (TD and BS), β is the vector of fixed effects (tank as fixed effect and body weight measured at the end of the experiment as covariate), is a vector of random additive polygenic genetic effects with distribution ∼N(0, ), where is the additive genetic variance and G is the genomic relationship matrix (GRM) calculated using the imputed genotypes [61], e is the vector of residuals with distribution ∼N(0, ), where I is the identity matrix, and is the residual variance, and X and Z are the incidence matrices for fixed and random effects, respectively. The allelic substitution effect and p-value for each SNP were also estimated using the GCTA.
For TD and BS, heritability () was estimated as follows:
where is the estimated additive genetic variance using the GRM, and is the residual variance.
2.6. Identification of Candidate Genes
The SNPs significantly associated with P. salmonis resistance were identified on the basis of the genomic significance threshold (0.05/number of SNPs) and the chromosomal significance threshold (0.05/number of SNPs), which represent α values corrected by Bonferroni. A systematic approach to search for candidate genes was performed using the Multi-marker Analysis of Genomic Annotation (MAGMA) software, which uses a multiple regression model to test the joint effect of multiple markers of a gene and then tests the association of the gene with the trait [62]. The Omyk_1.0 reference genome of O. mykiss (GenBank Assembly Accession GCA_002163495.1) was used in the gene search, and Bonferroni correction was also applied to obtain the significance threshold of the candidate genes.
3. Results
3.1. Summary Statistics
Similar mean values were observed for TD: 24.55 (SD = 7.60); 23.5 (SD = 7.82) and 22.73 (SD = 8.08) days for challenge tanks 1, 2, and 3, respectively. A Kaplan–Meier survival curve [63] was drawn for the best and worst families, as well as the average between families (Figure 1). Our results demonstrated significant (p > 0.05) phenotypic variation based on Kaplan–Meier survival analysis. For final weight (FW), values were: 177.5 (SD = 52.05); 167.3 (SD = 44.90) and 176.2 (SD = 58.62) grams for challenge tanks 1, 2 and 3, respectively (Figure 2).
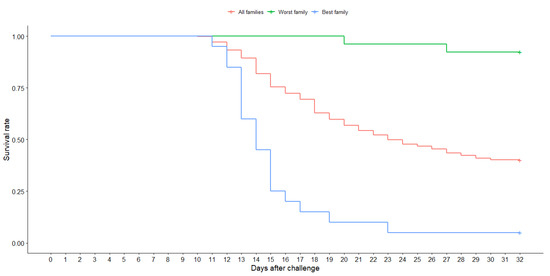
Figure 1.
Kaplan–Meier survival curves of the average of the 102 full-sib families, the best and the worst family after an experimental challenge with P. salmonis in a breeding rainbow trout (O. mykiss) population.
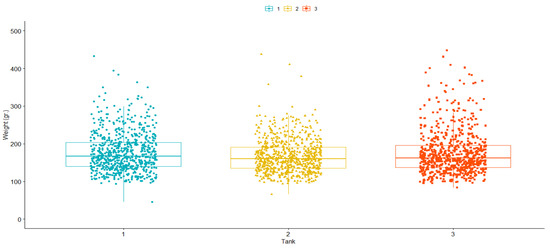
Figure 2.
Boxplot for the covariate final weight (FW), post-challenge against P. salmonis in a rainbow trout (O. mykiss) breeding population.
3.2. Quality Control of Genotypes and Imputation
A 57K SNP chip was used for genotyping 2130 challenged rainbow trout. Initial quality control (QC) of genotypes was applied with Affymetrix Axiom software according to default settings. Additional QC filters were sequentially applied, discarding 17.6% SNPs by call-rate, 2.7% SNPs by MAF and 17.14% by HWE, leaving a total of 31,275 SNPs available for downstream analyses. For the genotypes from WGS data, 22,649,022 SNPs from the 102 samples were obtained after SNP calling. The call-rate filter eliminated about 83.3% of the initial SNPs. MAF and HWE filters discarded 5.2% and 1% of SNPs, respectively, with a remaining list of 2,382,000 SNPs available for downstream analysis (a total of 10.5% out of the initial number of SNPs. After imputation validation, 24.3% of the SNPs had an r² value equal or greater than 0.80 and were used as the reference data set for the definitive imputation round. A post-imputation quality control (HWE < p-value > and MAF < 0.05) resulted in a total of 488,979 available imputed SNPs for the 2130 individuals reported as imputed WGS genotypes (Table 1).

Table 1.
Quality control results of genotyping of 57K SNP panel, whole genome sequence (WGS) and Imputed WGS data for the rainbow trout population.
3.3. Genome-Wide Association Studies
Genetic parameters were estimated using the 488,979 imputed genotypes. The estimated additive genetic variance was 13.59 and 0.045, and the heritability values were 0.27 (SD = 0.02) and 0.25 (SD = 0.02), for TD and BS, respectively (Table 2).

Table 2.
Estimates of variance components and heritability for P. salmonis resistance, defined as time to death (TD) and binary survival (BS), in a rainbow trout breeding population using whole-genome sequence imputed genotypes.
GWAS for resistance to P. salmonis measured as TD and BS was performed for each trait using the imputed WGS data (Figure 3). Five genomic regions were found to be genome-wide and significantly associated with TD and one region for BS. Genome-wide significant QTLs for TD were found in Omy2, Omy3, Omy25, Omy26 and Omy27, while for BS the only significant QTL was found in Omy26. In addition, chromosome-wide significant QTLs for TD were found in Omy3, Omy06, Omy10, Omy13, Omy14, Omy17 and Omy28 while for BS, only suggestive QTL were found in Omy3, Omy17, Omy26 and Omy27. All QTLs found here explained a low phenotypic variance for resistance against P. salmonis, reinforcing the fact that this trait is under polygenic control in rainbow trout (Supplementary Tables S1 and S2).
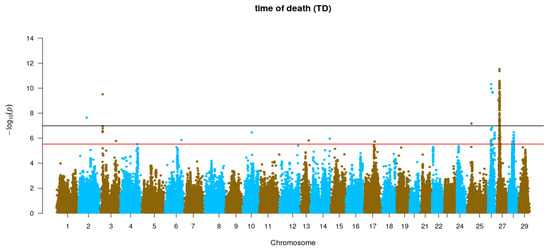
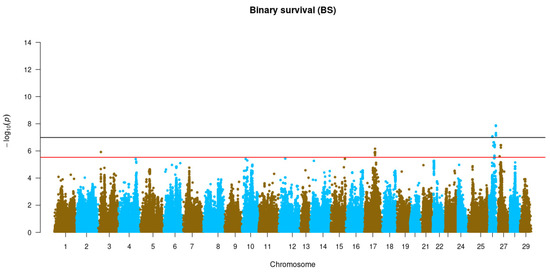
Figure 3.
Genome-wide association analysis for resistance to P. salmonis in rainbow trout (O. mykiss) using WGS-imputed data. The black and red lines represent the genome-wide and chromosome-wide significance thresholds, respectively.
3.4. Gene Identification
A total of 20,980 genes were available for a downstream gene search using MAGMA [62]. Table 3 shows the 24 candidate genes which were identified to be related to P. salmonis resistance, according to the Bonferroni-corrected significance threshold (0.05/20,980 = used for MAGMA analysis. Most of the identified genes are potentially involved in important biological functions related to the host response to P. salmonis infection, such as phagocytosis, innate immune response, inflammation, oxidative response, protein signaling pathway G, lipid metabolism, and the apoptotic process (Table 3).

Table 3.
Results of the gene-based analysis using MAGMA: Candidate genes related to resistance to P. salmonis in rainbow trout for time of death (TD) and binary survival (BS) traits.
4. Discussion
IP challenges have been extensively used to assess genetic variation for resistance to SRS given the high level of replication between experiments. In addition, the genetic correlation between IP and cohabitation challenges has been shown to be >0.8 [64,65] indicating that resistance to P. salmonis experimentally evaluated by IP or cohabitation is similar traits, from the genetic perspective. The estimated SNP-heritability for TD and BS using WGS-imputed genotypes were 0.27 ± 0.02 and 0.25 ± 0.02, respectively. These estimates were lower than those presented by [23], which were calculated in the same population using a different statistical approach implemented in BLUPF90 software, with heritability values of 0.48 ± 0.04 and 0.34 ± 0.04 for TD and BS, respectively. Here, we used a different density of SNPs compared with the previous study, which by imputation error may have had an impact on the estimation of genetic parameters. However, the results obtained are similar to those obtained when pedigree and genomic information was simultaneously used to estimate the genetic variance for resistance against P. salmonis in rainbow trout [18,23]. In general, our results confirm that there is a significant additive-genetic component involved in phenotypic variance for P. salmonis resistance.
Consistent with the results presented here, previous association-mapping studies in different salmonid species, indicate that resistance to P. salmonis is a polygenic trait [22,23,55]. Most of the previous works were performed through GWAS using low and medium-density SNPs [23,56,66]. Here, we found numerous SNPs significantly associated with P. salmonis resistance (Figure 1), most likely due to the polygenic architecture of this trait in rainbow trout, together with the reasonable sample size and increased SNP density. Although imputation or genotyping errors may generate false positives of association, the stringent filters used for the QC of true and WGS-imputed genotypes aimed at decreasing the probability of false positive detection. In addition, imputation results are in agreement with previous studies which performed genotype imputation to WGS in rainbow trout and other aquaculture species [43,44,45]. It is noteworthy that the most important QTL for TD, located in Omy27 and including several SNPs surpassing the genome-wide significance threshold, is in agreement with results from a previous study in which the same genomic region explained the highest proportion of the genetic variance for P. salmonis resistance [23]. The significant and suggestive QTLs identified for TD and BS in Omy3 and Omy 27, respectively, were also consistent with genomic regions explaining more than 1% of the genetic variance for P. salmonis resistance [23].
Host defense against infection by P. salmonis seems to depend on many biological features and processes. For instance, this bacterium enters mainly through the skin and gills, thus, physical barriers can be considered an important defense mechanism against the pathogen. In addition, phagocytosis is a key process in the life cycle of P. salmonis and it is considered the primary mode of pathogenesis. The bacterium is internalized by clathrin-dependent endocytosis in phagocytic cells, and once it enters the cell, the cytoskeleton is significantly reorganized by altering myosins, actins, among others [15], In addition, several other processes are involved in the interaction between P. salmonis and the host, including kinase and helicase activity, lipid metabolism, inflammation, GTP hydrolysis, and the innate immune response [56]. Several candidate genes were found flanking significant SNPs for TD Several candidate genes were found flanking significant SNPs for TD. In Omy03, we found the cation-dependent mannose 6-phosphate receptor (CD-MPR), which works as a pattern recognition receptor that interacts with invading bacteria and triggers the expression of antimicrobial peptides against pathogens [67]. This gene was described in rainbow trout [68] with participation in lysosome formation [69,70]. Ref. [71] described that a decrease in the lysosomal response may be related to a host immune evasion strategy by P. salmonis in Atlantic salmon (Salmo salar). In the same chromosome, we found the pleckstrin homology domain-containing, family G (with RhoGef domain) member 6 (PLEKHG6), also known as myosin interacting guanine nucleotide exchange factor (MyoGEF) that participates in the regulation of apoptosis with the appearance of blisters and cytokinesis [72,73], and possible suppressor of carcinogenic tumors [74]. Ref. [75] identified the MyoGEF gene as a mediator of the immune response, cell proliferation and response against the ectoparasite Neoparamoeba perurans in Atlantic salmon.
In Omy13, we found the specific erythropoietin receptor (EPOR), a fundamental signaling agent in erythropoiesis [76,77]. The EPO:EPOR junction activates the JAK/STAT signaling pathway, which plays an important role in modulating oxidative stress, innate immunity and inflammation [78,79,80]. This gene may also act in non-hematopoietic tissues under normal or infection conditions [78], such as wound healing and cardiovascular protection [76]. Due to skin lesions, such as petechial hemorrhages and the presence of ulcers, caused by P. salmonis in salmonids, the EPOR could be a candidate gene involved in defense mechanisms. In a study developed by [81], the authors identified that heme metabolism was affected by the infection of P. salmonis in Atlantic salmon by a lack of sufficient amounts of iron in the head kidney, resulting in a decrease in the expression of erythrocyte-specific EPOR genes.
In Omy26, we found the N-acetylglucosamine-6-sulfatase-like (GNSL) gene, which is related to phagocytosis [82]; the CBFA2T3 gene, also known as ETO2, which is a crucial regulator for oncogenesis [83,84], inflammation [85,86] and also a hematopoietic co-repressor [87]; and high-mobility group box family member 3 (TOX3), which is a regulator of innate lymphoid cells, in particular, CD8+ cytotoxic T lymphocytes, which in turn has been shown to be activated by the ectoparasite N.perurans in Atlantic salmon.
In Omy27, we found the cytoskeletal protein smoothelin-like protein 2 (SMTNL2) expressed in skeletal muscle, which is activated by c-Jun N-terminal kinase (JNK), in response to cellular environmental stress, including inflammatory stimuli and apoptosis [88,89]. Ref. [23] found the Smtnl2 gene associated with P. salmonis resistance in the same rainbow trout population. The leucine-rich repeat-containing protein 75A-like gene (LRRC75A), located on this same chromosome, belongs to the long non-coding RNAs (lncRNAs) that play multiple vital roles during inflammatory processes caused by pathogens [90,91]; possibly acting as a critical mediator of the inflammasome, an essential pro-inflammatory precursor [92]. Unconventional myosin-Ic (Myo1C), known to play an important role in bacterial diseases by encoding a member of the strange myosin family of proteins, was also found in Omy27 [93]. This gene is essential in lipid raft membrane recycling and proteins that regulate plasma membrane plasticity and pathogen entry [94]. An increase in Myo1c expression was identified upon infection with Aeromonas hydrophila, a Gram-negative bacterium, in Rohu carp (Labeo rohita) liver tissue [95]. The large neutral amino acid transporter small subunit 4 (LAT4) gene, also identified on chromosome 27, is essential in amino acid metabolism [96,97] and is involved in immune function [98,99]. In this same chromosome, we found the phosphatidylinositol transfer protein alpha (PITPNA) gene, essential for cell growth, organization of the cytoskeleton, metabolism, and apoptosis [100,101]. This gene was also previously identified to be associated with resistance to P. salmonis in rainbow trout [23]. The inositol polyphosphate 5-phosphatase K (INPP5K) gene also found in Omy27, participates in response to endoplasmic reticulum stress, organization of the cytoskeleton, renal osmoregulation, cell adhesion, and migration [102]. Loss of INPP5K may cause autophagy inhibition and lysosome depletion [103]. The INPP5K gene was identified to be involved in response to Aeromonas hydrophila infection in the European eel (Anguilla anguilla) [104]. F-box-only protein 39-like (FBXO39) participates in immune responses through the ubiquitin-proteasome system, promotes apoptosis of malignant cells [105], and has been described as an essential gene for animal behavior caused by stress in competition or fighting [106]. The X-linked inhibitor of apoptosis (XIAP)-associated factor 1 (XAF1), also found in Omy27, is a regulator of cytokinesis that considerably increases stress-induced apoptosis and decreases the invasive capacity of tumor cells [107,108]. XAF1 was identified as a gene involved in the primary responses to viruses with a pro-apoptotic function in Atlantic salmon [109,110], which could be considered a candidate gene for early response in microbial invasion in the host. In chromosome 27, we also found the bleomycin hydrolase gene (BLMH) that has a role in the regulation of inflammatory chemokines to initiate the immune response, wound healing [111], and skin integrity [112]. The expression of BLMH was found in the mucus of the skin of Greater amberjack (Seriola dumerili) infected with the ectoparasite Neobenedenia girallae [113]. In rainbow trout, BLMH expression has been identified as an innate response in the gill epithelial cell line to infection with ultraviolet-inactivated viral hemorrhagic septicemia virus [114]. The mediator of RNA polymerase II transcription subunit 19th gene (MED19), is an essential gene for the maintenance of white adipose tissue and adipogenesis. MED19 interacts with proliferator receptors activated peroxisome (PPARs) important in inflammatory processes [115]. The activation of PPARs decreases the expression of genes involved in oxidative stress, inflammation, migration, and cell growth [116]. Molecules that are not obviously related with processes which may be involved in host response against P. salmonis, were also identified in Omy27: Tektin 1 (TEKT1) has been shown to be related to sperm quality, cytoskeleton organization, and motility [117,118]; sodium-dependent serotonin transporter-like (SLC6A4) which is a gene that modulates many brain functions [119].
For BS, three essential candidate genes were found flanking the significant SNPs. In Omy03, we found the domain family protein 1 (TSC22D1) gene that participates as an early immune response molecule and that modulates the TGF-beta dependent signaling pathway [120], a tumor suppressor in cells [121,122], and induces cellular apoptosis and senescence [123]. These genes are from the TSC22 family, regulatory proteins with anti-inflammatory and immunosuppressive effects of interleukins and glucocorticoids [124]. In a transcriptome study in Atlantic salmon infected with P. salmonis, a decrease in the expression of TSC22D, from the TSC22 family, was shown in the head kidney [125]. In Omy17, we found the striatin-interacting protein one homolog (STRIP1) gene, which plays an important role in the migration and metastasis of cancer cells [126,127] and regulates the organization of the actin cytoskeleton [128]. In Omy26, we found a single gene called AP-2 complex subunit alpha-2 (AP2A2), involved in the activation of the leukocyte response, granulocytes, and myeloid cells involved in the immune response [129] and provides the first line of defense against a pathogen or virus [130].
Genes associated with both TD and BS were found in Omy26. Myotubularin-related protein 10 (MTMR10), which is associated with the protection of dendrites induced by oxidative stress or infection by pathogens and is present in several tissues was identified [131]. Nile tilapia exposed to ammonia showed a response in lipid metabolism-related to the MRM10 gene [132]. In Omy27, we found the protein Rap1 regulator of platelet activity [133,134]. It has been shown that (Rap1GAP2) is the only Rap1 GTPase activator protein in platelet segregation that is important in the immune system [135]. The expression of the immune-related gene Rap1GAP2 has been modulated in channel catfish (Ictalurus punctatus) after the infection with Flavobacterium columnare [136]. Ref. [137] observed high expression of this gene in both the spleen and kidney of giant grouper (Epinephelus lanceolatus) infected with spotted knifejaw iridovirus (SKIV). Slingshot phosphatases (SSH) are critical in controlling actin dynamics [138], especially the gene found on chromosome 27 called Slingshot phosphatase homolog 2 (SSH2), associated with inflammation. SSH2 is involved in the effective migration and invasion of tumor cells [139,140,141]. This gene could cause damage and autoinflammatory diseases due to excess neutrophils in healthy tissues associated with neutrophil chemotaxis [141].
We found 24 candidate genes that may be involved in P. salmonis resistance in rainbow trout, of which 14 have been identified in Omy27. Likewise, in [23], which used the same population, two genes were identified in common (PITPNA and SMTNL2) in Omy27 for TD. [56] used a genome-wide association study analysis applying a Bayesian approach to investigate the genetic architecture of resistance to P. salmonis in farmed populations of salmonids, including rainbow trout. The authors identified nine candidate genes in Omy27 (SMTNL2, LRRC75A, unconventional myosin-Ic, LAT4, PITPNA, INPP5K, TEKT1, BLMH, and SLC6A4). These same genes were identified in our study for the TD phenotype. These candidate genes may be considered strong candidates to explain P. salmonis resistance variation in rainbow trout and are potentially relevant to test further genetics- or pharmacological-based strategies for controlling this disease in rainbow trout.
5. Conclusions
Resistance to the infectious disease caused by P. salmonis (SRS) has a complex polygenic inheritance architecture in rainbow trout. Here, we use imputed whole genome sequence data to refine the GWAS results for P. salmonis resistance in rainbow trout. We found 24 putative functional genes related to phagocytosis, innate immune response, inflammation, oxidative response, G-protein signaling pathway, lipid metabolism, and apoptotic processes, which may be related to variation in the host response against the bacterium, which is mainly found in a significant QTL found on chromosome 27. Further studies are needed to validate the results obtained in this study, especially related to candidate genes located on Omy27.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes14010114/s1, Table S1: title phenotypic variance for resistance against P. salmonis, reinforcing the fact that this trait is under polygenic control in rainbow trout -BS and Table S2: title phenotypic variance for resistance against P. salmonis, reinforcing the fact that this trait is under polygenic control in rainbow trout -TD (Supplementary Tables S1 and S2).
Author Contributions
Conceptualization, J.M.Y.; methodology, C.S.-R., B.G. and J.G.-H.; formal analysis, C.S.-R., B.G. and J.G.-H.; writing—original draft preparation, C.S.-R., B.G. and J.G.-H.; writing—review and editing, J.M.Y.; supervision, J.M.Y. and B.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially funded by FONDECYT REGULAR (ANID, Chile), grant number 1171720.
Institutional Review Board Statement
Sampling procedures were approved by the Comité de Bioética Animal from the Facultad de Ciencias Veterinarias y Pecuarias, Universidad de Chile (Certificate No. 19270-VET-UCH).
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets generated and analyzed during the current study are available upon request.
Acknowledgments
We would like to acknowledge EFFIGEN S.A. (Puerto Montt, Chile), which provided the genotypic and phenotypic data used in this study. J.M.Y. is supported by Núcleo Milenio de Salmons Invasores Australes (INVASAL) funded by Chile’s government program, Iniciativa Cientifica Milenio from Ministerio de Economia, Fomento y Turismo.
Conflicts of Interest
The authors declared no conflict of interest.
References
- FAO. El Estado Mundial de la Pesca y la Acuicultura 2022. Hacia la Transformación Azul; FAO: Rome, Italy, 2022. [Google Scholar]
- Subsecretaria de Pesca y Acuicultura (SUBPESCA). Informe Sectorial de Pesca y Acuicultura Noviembre 2021; Subsecretaria de Pesca y Acuicultura (SUBPESCA): Valparaíso, Chile, 2021. [Google Scholar]
- Sernapesca. Informe Sanitario Con Información Sanitaria De Agua Dulce Y Mar 1° semestre Año 2022; Sernapesca: Santiago, Chile, 2022; pp. 1–54.
- Pérez-Valenzuela, J.; Mejías, M.; Ortiz, D.; Salgado, P.; Montt, L.; Chávez-Báez, I.; Vera-Tamargo, F.; Mandakovic, D.; Wacyk, J.; Pulgar, P. Increased dietary availability of selenium in rainbow trout (Oncorhynchus mykiss) improves its plasma antioxidant capacity and resistance to infection with Piscirickettsia salmonis. Veterinaty Res. 2021, 52, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, C.; Torrealba, D.; Morales-Lange, B.; Mercado, L.; Dixon, B.; Conejeros, P.; Silva, G.; Soto, C.; Gallardo, J.A. Commercial Vaccines Do Not Confer Protection against Two Genogroups of Piscirickettsia salmonis, LF-89 and EM-90, in Atlantic Salmon. Biology 2022, 11, 993. [Google Scholar] [CrossRef] [PubMed]
- Sernapesca. Informe Sobre Uso De Antimicrobianos En La Salmonicultura Nacional Año 2021; Sernapesca: Santiago, Chile, 2022.
- Kemper, N. Veterinary antibiotics in the aquatic and terrestrial environment. Ecol. Indic. 2008, 8, 1–13. [Google Scholar] [CrossRef]
- Schar, D.; Klein, E.; Laxminarayan, R.; Gilbert, M.; Van Boeckel, T. Global Trends in antimicrobial use in aquaculture. Sci. Rep. 2020, 10, 21878. [Google Scholar] [CrossRef]
- Maisey, K.; Montero; Christodoulides, M. Vaccines for piscirickettsiosis (salmonid rickettsial septicaemia, SRS): The Chile perspective. Expert Rev. Vaccines 2016, 16, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, C.; Veloso, P.; Espin, L.; Dixon, B.; Torrealba, D.; Said, I.; Afonso, J.; Soto, C.; Conejeros, P.; Gallardo, J. Host genetic variation explains reduced protection of commercial vaccines against Piscirickettsia salmonis in Atlantic salmon. Sci. Rep. 2020, 10, 18252. [Google Scholar] [CrossRef]
- Cabello, F.C.; Godfrey, H.P. Salmon aquaculture, Piscirickettsia salmonis virulence, and One Health: Dealing with harmful synergies between heavy antimicrobial use and piscine and human health comment on Avendaño-Herrera (2021). Aquaculture 2021, 537, 451–456. [Google Scholar] [CrossRef]
- Caruffo, M.; Vidal, S.; Santis, L.; Siel, D.; Pérez, O.; Huenchullan, P.; Sáenz, L. Effectiveness of a proteoliposome-based vaccine against salmonid rickettsial septicaemia in Oncorhynchus mykiss. Vet. Res. 2021, 52, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Rozas-Serri, M.; Ildefonso, R.; Peña, A.; Enríquez, R.; Barrientos, S.; Maldonado, L. Comparative pathogenesis of piscirickettsiosis in Atlantic salmon (Salmo salar L.) post-smolt experimentally challenged with LF-89-like and EM-90-like Piscirickettsia salmonis isolates. J. Fish Dis. 2017, 40, 1451–1472. [Google Scholar] [CrossRef]
- Rozas-Serri, M.; Peña, A.; Maldonado, L. Gene expression associated with immune response in Atlantic salmon head-kidney vaccinated with inactivated whole-cell bacterin of Piscirickettsia salmonis and pathogenic isolates. Fish Shellfish. Immunol. 2019, 93, 789–795. [Google Scholar] [CrossRef]
- Rozas-Serri, M. Why Does Piscirickettsia salmonis Break the Immunological Paradigm in Farmed Salmon? Biological Context to Understand the Relative Control of Piscirickettsiosis. Front. Immunol. 2022, 21, 856896. [Google Scholar] [CrossRef]
- Yáñez, J.; Martínez, V. Factores genéticos que inciden en la resistencia a enfermedades infecciosas en salmónidos y su aplicación en programas de mejoramiento. Arch. De Med. Vet. 2010, 42, 1–13. [Google Scholar] [CrossRef][Green Version]
- Yáñez, J.; Houston, R.; Newman, S. Genetics and genomics of disease resistance in salmonid species. Front. Genet. 2014, 5, 415. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, G.; Bangera, R.; Carvalheiro, R.; Correa, K.; Figueroa, R.; Lhorente, J.; Yáñez, J. Genomic prediction accuracy for resistance against Piscirickettsia salmonis in farmed rainbow trout. G3: Genes Genomes Genet. 2018, 8, 719–726. [Google Scholar] [CrossRef]
- Yáñez, J.; Peng, X.; Carvalheiro, R.; Hayes, B. Genomics applied to livestock and aquaculture breeding. Evol. Appl. 2022, 15, 517–522. [Google Scholar] [CrossRef]
- Georges, M.; Charlier, C.; and Hayes, B. Harnessing genomic information for livestock improvement. Nat. Rev. Genet. 2019, 20, 135–156. [Google Scholar] [CrossRef]
- Bangera, R.; Correa, K.; Lhorente, J.; Figueroa, R.; Yáñez, J. Genomic predictions can accelerate selection for resistance against Piscirickettsia salmonis in Atlantic salmon (Salmo salar). BMC Genom. 2017, 18, 121. [Google Scholar] [CrossRef]
- Barría, A.; Christensen, K.; Yoshida, G.; Correa, K.; Jedlicki, A.; Lhorente, J.; Davidson, W.; Yáñez, J. Genomic predictions and genome-wide association study of resistance against Piscirickettsia salmonis in coho salmon (Oncorhynchus kisutch) using ddRAD sequencing. G3: Genes Genomes Genet. 2018, 8, 1183–1194. [Google Scholar] [CrossRef]
- Barría, A.; Marín-Nahuelpi, R.; Cáceres, P.; López, M.; Bassini, L.; Lhorente, J.; Yáñez, J. Single-step genome-wide association study for resistance to Piscirickettsia salmonis in rainbow trout (Oncorhynchus mykiss). G3: Genes Genomes Genet. 2019, 9, 3833–3841. [Google Scholar] [CrossRef]
- McCarthy, M.; Abecasis, G.; Cardon, L.; Goldstein, D.; Little, J.; Ioannidis, J.; Hirschhorn, J. Genome-wide association studies for complex traits: Consensus, uncertainty and challenges. Nat. Rev. Genet. 2008, 9, 356–369. [Google Scholar] [CrossRef]
- Goddard, M.; Hayes, B. Mapping genes for complex traits in domestic animals and their use in breeding programmes. Nat. Rev. Genet. 2009, 10, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Su, G.; Christensen, O.F.; Janss, L.; Lund, M.S. Comparison of genomic predictions using genomic relationship matrices built with different weighting factors to account for locus-specific variances. J. Dairy Sci. 2014, 10, 6547–6559. [Google Scholar] [CrossRef] [PubMed]
- Van Binsbergen, R.; Bink, M.; Calus, M.; van Eeuwijk, F.; Hayes, B.; Hulsegge, I.; Veerkamp, R. Accuracy of imputation to whole-genome sequence data in Holstein Friesian cattle. Genet. Sel. Evol. 2014, 41, 41. [Google Scholar] [CrossRef]
- Tan, C.H.; Wu, Z.; Ren, J.; Huang, Z.; Liu, D.; He, X.; Prakapenka, D.; Zhang, R.; Li, N.; Yang, D.; et al. Genome-wide association study and accuracy of genomic prediction for teat number in Duroc pigs using genotyping-by-sequencing. Genet. Sel. Evol. 2017, 49, 35. [Google Scholar] [CrossRef]
- Zhang, H.; Yin, L.; Wang, M.; Yuan, Y.; Liu, X. Factors affecting the accuracy of genomic selection for agricultural economic traits in maize, cattle, and pig populations. Front. Genet. 2019, 10, 189. [Google Scholar] [CrossRef] [PubMed]
- Uffelmann, E.; Huang, Q.; Munung, N.; Vries, J.; Okada, Y.; Martin, A.; Martin, H.; Lappalainen, T.; Posthuma, D. Genome-wide association studies. Nat. Rev. 2021, 1, 59. [Google Scholar] [CrossRef]
- Daetwyler, H.; Capitan, A.; Pausch, H.; Stothard, P.; Van Binsbergen, R.; Brøndum, R.; Liao, X.; Djari, A.; Rodriguez, S.; Grohs, C.; et al. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat. Genet. 2014, 46, 858–865. [Google Scholar] [CrossRef]
- Pérez-Enciso, M.; Rincón, J.; Legarra, A. Sequence- vs. chip-assisted genomic selection: Accurate biological information is advised. Genet. Sel. Evol. 2015, 47, 43. [Google Scholar] [CrossRef]
- MacLeod, I.; Bowman, P.; Vander, J.; Haile-Mariam, M.; Kemper, K.; Chamberlain, A.; Schrooten, C.; Hayes, B.; Goddard, E. Exploiting biological priors and sequence variants enhances QTL discovery and genomic prediction of complex traits. BMC Genom. 2016, 17, 144. [Google Scholar] [CrossRef]
- Fernandes Júnior, F.; Carvalheiro, R.; de Oliveira, H.; Sargolzaei, M.; Costilla, R.; Ventura, R.; Fonseca, L.; Neves, H.; de Hayes, B.; de Albuquerque, L.G. Imputation accuracy to whole-genome sequence in Nellore cattle. Genet. Sel. Evol. 2021, 53, 27. [Google Scholar] [CrossRef]
- Yoshida, G.; Carvalheiro, R.; Lhorente, J.; Correa, K.; Figueroa, R.; Houston, R.; Yáñez, J.M. Accuracy of genotype imputation and genomic predictions in a two-generation farmed Atlantic salmon population using high-density and low-density SNP panels. Aquaculture 2018, 491, 147–154. [Google Scholar] [CrossRef]
- Van den Berg, S.; Vandenplas, J.; van Eeuwijk, F.; Bouwman, A.; Lopes, M.; Veerkamp, R. Imputation to whole-genome sequence using multiple pig populations and its use in genome-wide association studies. Genet. Sel. Evol. 2019, 51, 2. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Duan, D.; Xue, X.; Han, X.; Wang, K.; Qiao, R.; Li, X.L.; Li, X.J. An association study on imputed whole-genome resequencing from high-throughput sequencing data for body traits in crossbred pig. Anim. Genet. 2022, 53, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.; Yuan, X.; Lin, X.; Gao, N.; Luo, Y.; Chen, Z.; Li, J.; Zhang, X.; Zhang, Z. Imputation from SNP chip to sequence: A case study in a Chinese indigenous chicken population. J. Anim. Sci. Biotechnol. 2018, 9, 30. [Google Scholar] [CrossRef] [PubMed]
- Teng, J.; Ye, S.; Gao, N.; Chen, Z.; Diao, S.; Li, X.; Yuan, X.; Zhang, H.; Li, J.; Zhang, X.; et al. Incorporating genomic annotation into single-step genomic prediction with imputed whole-genome sequence data. J. Integr. Agric. 2022, 4, 1126–1136. [Google Scholar] [CrossRef]
- Lopez, B.I.M.; An, N.; Srikanth, K.; Lee, S.; Oh, J.-D.; Park, W.; Chai, H.-H.; Park, J.-E.; Lim, D. Genomic Prediction Based on SNP Functional Annotation Using Imputed Whole-Genome Sequence Data in Korean Hanwoo Cattle. Front. Genet. 2021, 11, 603822. [Google Scholar] [CrossRef]
- Li, H.; Zhu, B.; Xu, L.; Wang, Z.; Xu, L.; Zhou, P.; Gao, H.; Guo, P.; Chen, Y.; Gao, X.; et al. Genomic Prediction Using LD-Based Haplotypes Inferred From High-Density Chip and Imputed Sequence Variants in Chinese Simmental Beef Cattle. Front. Genet. 2021, 12, 665382. [Google Scholar] [CrossRef]
- Mancin, E.; Sosa-Madrid, B.; Blasco, A.; Ibáñez-Escriche, N. Genotype imputation to improve the cost-efficiency of genomic selection in rabbits. Animals 2021, 11, 803. [Google Scholar] [CrossRef]
- Yoshida, G.; Yáñez, J. Increased accuracy of genomic predictions for growth under chronic thermal stress in rainbow trout by prioritizing variants from GWAS using imputed sequence data. Evol. Appl. 2021, 15, 537–552. [Google Scholar] [CrossRef]
- Garcia, B.; Yoshida, G.; Carvalheiro, R.; Yáñez, J. Accuracy of genotype imputation to whole genome sequencing level using different populations of Nile tilapia. Aquaculture 2022, 551, 737947. [Google Scholar] [CrossRef]
- Yoshida, G.; Yáñez, J. Multi-trait GWAS using imputed high-density genotypes from whole-genome sequencing identifies genes associated with body traits in Nile tilapia. BMC Genom. 2021, 22, 57. [Google Scholar] [CrossRef]
- Liu, S.; Vallejo, R.L.; Palti, Y.; Gao, G.; Marancik, D.P.; Hernandez, A.G.; Wiens, G.D. Identification of single nucleotide polymorphism markers associated with bacterial cold water disease resistance and spleen size in rainbow trout. Front. Genet. 2015, 6, 298. [Google Scholar] [CrossRef][Green Version]
- Vallejo, R.; Liu, S.; Gao, G.; Fragomeni, B.; Hernandez, A.; Leeds, T.; Parsons, J.; Martin, K.; Evenhuis, J.; Welch, T.; et al. Similar genetic architecture with shared and unique quantitative trait loci for bacterial cold water disease resistance in two rainbow trout breeding populations. Front. Genet. 2017, 8, 156. [Google Scholar] [CrossRef]
- Liu, S.; Vallejo, R.L.; Evenhuis, J.P.; Martin, K.E.; Hamilton, A.; Gao, G.; Leeds, T.D.; Wiens, G.D.; Palti, Y. Retrospective evaluation of marker-assisted selection for resistance to bacterial cold water disease in three generations of a commercial rainbow trout breeding population. Front. Genet. 2018, 9, 286. [Google Scholar] [CrossRef]
- Marana, M.; Karami, A.; Ødegård, J.; Zuo, S.; Jaafar, R.; Mathiessen, H.; Jørgensen, V.L.; Kania, P.; Dalsgaard, I.; Nielsen, T.; et al. Whole-genome association study searching for QTL for Aeromonas salmonicida resistance in rainbow trout. Sci. Rep. 2021, 11, 17857. [Google Scholar] [CrossRef]
- Karami, A.M.; Ødegård, J.; Marana, M.H.; Zuo, S.; Jaafar, R.; Mathiessen, H.; von Gersdorff, J.; Kania, P.W.; Dalsgaard, I.; Nielsen, T.; et al. A Major QTL for Resistance to Vibrio anguillarum in Rainbow Trout. Front. Genet. 2020, 11, 607558. [Google Scholar] [CrossRef]
- Jaafar, R.; Ødegård, J.; Mathiessen, H.; Karami, A.; Marana, A.; von Gersdorff, L.J.; Zuo, S.; Nielsen, T.; Kania, P.; Buchmann, K. Quantitative trait loci (QTL) associated with resistance of rainbow trout Oncorhynchus mykiss against the parasitic ciliate Ichthyophthirius multifiliis. J. Fish Dis. 2020, 43, 1591–1602. [Google Scholar] [CrossRef]
- Rodríguez, F.; Flores-Mara, R.; Yoshida, G.; Barría, A.; Jedlicki, A.; Lhorente, J.; Reyes-López, F.; Yáñez, J. Genome-Wide Association Analysis for Resistance to Infectious Pancreatic Necrosis Virus Identifies Candidate Genes Involved in Viral Replication and Immune Response in Rainbow Trout (Oncorhynchus mykiss). G3: Genes Genomes Genet. 2019, 9, 2897–2904. [Google Scholar] [CrossRef]
- Silva, R.; Evenhuis, J.; Vallejo, R.; Gao, G.; Martin, K.; Leeds, T.; Palti, Y.; Lourenco, D.R. Whole-genome mapping of quantitative trait loci and accuracy of genomic predictions for resistance to columnaris disease in two rainbow trout breeding populations. Genet. Sel. Evol. 2019, 51, 42. [Google Scholar] [CrossRef]
- Zuo, S.; Karami, A.M.; Ødegård, J.; Mathiessen, H.; Marana, M.H.; Jaafar, R.M.; von Gersdorff, J.L.; Abdu, M.; Kania, P.W.; Dalsgaard, I.; et al. Immune gene expression and genome-wide association analysis in rainbow trout with different resistance to Yersinia ruckeri infection. Fish Shellfish. Immunol. 2020, 106, 441–450. [Google Scholar] [CrossRef]
- Correa, K.; Lhorente, J.; López, L.; Bassini, L.; Naswa, S.; Deeb, N.; Di Genova, A.; Maass, A.; Davidson, W.; Yáñez, J. Genome-wide association analysis reveals loci associated with resistance against Piscirickettsia salmonis in two Atlantic salmon (Salmo salar L.) chromosomes. BMC Genom. 2015, 16, 854. [Google Scholar] [CrossRef]
- Yáñez, J.; Yoshida, G.; Parra, A.; Correa, K.; Barría, A.; Bassini, L.; Christensen, K.; López, L.; Carvalheiro, R.; Lhorente, J.; et al. Comparative genomic analysis of three salmonid species identifies functional candidate genes involved in resistance to the intracellular bacterium Piscirickettsia salmonis. Front. Genet. 2019, 10, 00665. [Google Scholar] [CrossRef]
- Palti, Y.; Vallejo, R.; Gao, G.; Liu, S.; Hernandez, A.; Rexroad III, C.; Wiens, G. Detection and Validation of QTL AffectingBacterial Cold Water Disease Resistance inRainbow Trout Using Restriction-SiteAssociated DNA Sequencing. PLoS ONE 2015, 10, e0138435. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 2010, 5, 559–575. [Google Scholar] [CrossRef]
- Sargolzaei, M.; Chesnais, J.P.; Schenkel, F.S. A new approach for efficient genotype imputation using information from relatives. BMC Genom. 2014, 15, 478. [Google Scholar] [CrossRef]
- Yang., J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- de Leeuw, C.; Mooij, J.; Heskes, T.; Posthuma, D. MAGMA:Generalized Gene-Set Analysis of GWAS Data. PLoS Computational. Biol. 2015, 11, 11–19. [Google Scholar] [CrossRef]
- Kaplan, E.; Meier, L. Nonparametric Estimation from Incomplete Observations. J. Am. Stat. Assoc. 1985, 53, 457–481. [Google Scholar] [CrossRef]
- Martinez, V. Genomic Selection Applied to Piscirickettsia salmonis Resistance in Chilean Atlantic Salmon. In Proceedings of the International Plant & Animal Genome XXII, San Diego, CA, USA, 11–15 January 2014. [Google Scholar]
- Dettleff, P.; Bravo, C.; Patel, A.; Martinez, V. Patterns of Piscirickettsia salmonis load in susceptible and resistant families of Salmo salar. Fish Shellfish. Immunol. 2015, 45, 67–71. [Google Scholar] [CrossRef]
- Barría, A.; Trịnh, Q.; Mahmuddin, M.; Peñaloza, C.; Papadopoulou, A.; Gervais, O.; Chadag, V.; Benzie, J.; Houston, R. A major quantitative trait locus affecting resistance to Tilapia lake virus in farmed Nile tilapia (Oreochromis niloticus). Heredity 2021, 127, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.Y.; Yuan, W.J.; Xu, J.D.; Wang, J.X. Cation-dependent mannose-6-phosphate receptor functions as a pattern recognition receptor in anti-bacterial immunity of Marsupenaeus japonicus. Dev. Comp. Immunol. 2018, 89, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Nadimpalli, S.; Yerramalla, U.; Hille-Rehfeld, A.; Figura, K. Mannose 6-phosphate receptors (MPR 300 and MPR 46) from a teleostean fish (trout). Physiol.—B Biochem. Mol. Biol. 1999, 23, 261–265. [Google Scholar] [CrossRef]
- Dahms, N.M.; Hancock, M.K. P-type lectins. Biochim. Et Biophys. Acta—Gen. Subjects 2002, 1572, 317–340. [Google Scholar] [CrossRef]
- Nolan, C.; McCarthy, M.; Eivers, E.; Jirtle, R.; Byrnes, L. Mannose 6-phosphate receptors in an ancient vertebrate, zebrafish. Dev. Genes Evol. 2006, 216, 144–151. [Google Scholar] [CrossRef]
- Pérez-Stuardo, D.; Morales-Reyes, J.; Tapia, S.; Ahumada, D.; Espinoza, A.; Soto-Herrera, V.; Brianson, B.; Ibaceta, V.; Sandino, A.; Spencer, E.; et al. Non-lysosomal activation in macrophages of atlantic salmon (Salmo salar) after infection with Piscirickettsia salmonis. Front. Immunol. 2019, 10, 434. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Asiedu, M.; Adelstein, R.; Wei, Q. A novel guanine nucleotide exchange factor MyoGEF is required for cytokinesis. Cell Cycle 2006, 5, 1234–1239. [Google Scholar] [CrossRef]
- Jiao, M.; Wu, D.; Wei, Q. Myosin II-interacting guanine nucleotide exchange factor promotes bleb retraction via stimulating cortex reassembly at the bleb membran. Mol. Biol. Cell 2018, 29, 643–656. [Google Scholar] [CrossRef]
- Ke, J.; Tian, J.; Mei, S.; Ying, P.; Yang, N.; Wang, X.; Zou, D.; Peng, X.; Yang, Y.; Zhu, Y.; et al. Genetic Predisposition to Colon and Rectal Adenocarcinoma Is Mediated by a Super-enhancer Polymorphism Coactivating CD9 and PLEKHG6. Cancer Epidemiol. Biomark. Prev. 2020, 29, 850–859. [Google Scholar] [CrossRef]
- Botwright, N.A.; Mohamed, A.R.; Slinger, J.; Lima, P.C.; Wynne, J.W. Host-Parasite Interaction of Atlantic salmon (Salmo salar) and the Ectoparasite Neoparamoeba perurans in Amoebic Gill Disease. Front. Immunol. 2021, 12, 672700. [Google Scholar] [CrossRef]
- Katakura, F.; Katzenback, B.A.; Belosevic, M. Molecular and functional characterization of erythropoietin receptor of the goldfish (Carassius auratus L.). Dev. Comp. Immunol. 2014, 45, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Malik, M.S.; Bjørgen, H.; Dhamotharan, K.; Wessel, Ø.; Koppang, E.O.; Di Cicco, E.; Hansen, E.F.; Dahle, M.K.; Rimstad, E. Erythroid Progenitor Cells in Atlantic Salmon (Salmo salar) May Be Persistently and Productively Infected with Piscine Orthoreovirus (PRV). Viruses 2019, 11, 824. [Google Scholar] [CrossRef]
- Chu, C.-Y.; Cheng, C.-H.; Yang, C.-H.; Huang, C.-J. Erythropoietins from teleosts. Cell. Mol. Life Sci. 2008, 65, 3545–3552. [Google Scholar] [CrossRef] [PubMed]
- Ostrowski, D.; Heinrich, R. Alternative Erythropoietin Receptors in the Nervous System. J. Clin. Med. 2018, 7, 24. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Chen, X.; Gao, R.; Wang, K.; Zheng, W.; Liu, H. A cytokine receptor domeless promotes white spot syndrome virus infection via JAK/STAT signaling pathway in red claw crayfish Cherax quadricarinatus. Dev. Comp. Immunol. 2020, 111, 103749. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Caballero-Solares, A.; Hall, J.R.; Umasuthan, N.; Kumar, S.; Jakob, E.; Skugor, S.; Hawes, C.; Santander, J.; Taylor, R.G.; et al. Transcriptome Profiling of Atlantic Salmon (Salmo salar) Parr With Higher and Lower Pathogen Loads Following Piscirickettsia salmonis Infection. Front. Immunol. 2021, 12, 789465. [Google Scholar] [CrossRef]
- Yang, Q.; Zhang, X.; Lu, Z.; Huang, R.; Tuan, N.; Wu, J.; Yang, F.; Ge, H.; Zhong, C.; Sun, Q.; et al. Transcriptome and Metabolome Analyses of Sea Cucumbers Apostichopus japonicus in Southern China During the Summer Aestivation Period. J. Ocean. Univ. China 2021, 20, 198–212. [Google Scholar] [CrossRef]
- Kumar, R.; Cheney, K.; McKirdy, R.; Neilsen, P.; Schulz, R.; Lee, J.; Cohen, J.; Booker, G.; Callen, D. CBFA2T3-ZNF652 corepressor complex regulates transcription of the E-box gene HEB. J. Biol. Chem. 2008, 283, 19026–19038. [Google Scholar] [CrossRef]
- Brown, R.; Jacobse, J.; Anant, S.; Blunt, K.; Chen, B.; Vega, P.; Jones, C.; Pilat, J.; Revetta, R.; Gorby, A.; et al. MTG16 (CBFA2T3) regulates colonic epithelial differentiation, colitis, and tumorigenesis by repressing E protein transcription factors. bioRXiv 2022, 11, 1–41. [Google Scholar] [CrossRef]
- Li, R.; Li, C.; Chen, H.; Li, R.; Chong, Q.; Xiao, H.; Chen, S. Genome-wide scan of selection signatures in Dehong humped cattle for heat tolerance and disease resistance. Anim. Genet. 2019, 51, 292–299. [Google Scholar] [CrossRef]
- Alshawi, A.; Essa, A.; Al-Bayatti, S.; Hanotte, O. Genome Analysis Reveals Genetic Admixture and Signature of Selection for Productivity and Environmental Traits in Iraqi Cattle. Front. Genet. 2019, 10, 609. [Google Scholar] [CrossRef]
- Steinauer, N.; Guo, C.; Zhang, J. The transcriptional corepressor CBFA2T3 inhibits all- trans-retinoic acid-induced myeloid gene expression and differentiation in acute myeloid leukemia. J. Biol. Chem. 2020, 295, 8887–8900. [Google Scholar] [CrossRef]
- Kasuya, Y.; Kim, J.-D.; Hatano, M.; Tatsumi, K.; Matsuda, S. Pathophysiological Roles of Stress-Activated Protein Kinases in Pulmonary Fibrosis. Int. J. Mol. Sci. 2021, 11, 6041. [Google Scholar] [CrossRef]
- Gordon, E.A.; Whisenan, T.C.; Zeller, M.; Kaake, R.M.; Gordon, W.M.; Krotee, P.; Patel, V.; Huang, L.; Baldi, P.; Bardwell, L. Combining docking site and phosphosite predictions to find new substrates: Identification of smoothelin-like-2 (SMTNL2) as a c-Jun N-terminal kinase (JNK) substrate. Cell. Signal. 2013, 25, 2518–2529. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, H.; Zhang, R.; Li, D.; Gao, M.Q. LRRC75A antisense lncRNA1 knockout attenuates inflammatory responses of bovine mammary epithelial cells. Int. J. Biol. Sci. 2020, 16, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, J.; Huang, Z.; Jing, H.; Yin, B.; Guo, S.; Deng, G.; Guo, M. Exosomal lnc-AFTR as a novel translation regulator of FAS ameliorates Staphylococcus aureus-induced mastitis. BioFactors 2021, 48, 148–163. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.K.; Wen, H.; Ting, J.P.Y. The Inflammasome NLRs in immunity, inflammation, and associated diseases. Annu. Rev. Immunol. 2011, 29, 707–735. [Google Scholar] [CrossRef]
- Lin, S.; Ke, C.; Liu, L.; Gao, Y.; Xu, L.; Han, B.; Zhao, Y.; Zhang, S.; Sun, D. Genome-wide association studies for immunoglobulin concentrations in colostrum and serum in Chinese Holstein. BMC Genom. 2022, 23, 41. [Google Scholar] [CrossRef]
- Brandstaetter, H.; Kendrick-Jones, J.; Buss, F. Myo1c regulates lipid raft recycling to control cell spreading, migration and Salmonella invasion. J. Cell Sci. 2012, 125, 1991–2003. [Google Scholar] [CrossRef] [PubMed]
- Nissa, M.U.; Pinto, N.; Ghosh, B.; Singh, U.; Goswami, M.; Srivastava, S. Proteomic analysis of liver tissue reveals Aeromonas hydrophila infection mediated modulation of host metabolic pathways in Labeo rohita. bioRXiv 2021. [Google Scholar] [CrossRef]
- Gomes, F.; Watanabe, L.; Vianez, J.; Nunes, M.; Cardoso, J.; Lima, C.; Schneider, H.; Sampaio, I. Comparative analysis of the transcriptome of the Amazonian fish species Colossoma macropomum (tambaqui) and hybrid tambacu by next generation sequencing. PLoS Genet. 2019, 14, e0212755. [Google Scholar] [CrossRef]
- To, V.P.T.H.; Masagounder, K.; Loewen, M.E. SLC transporters ASCT2, B0AT1-like, y+LAT1, and LAT4-like associate with methionine electrogenic and radio-isotope flux kinetics in rainbow trout intestine. Physiol. Rep. 2019, 7, e14274. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Ding, M.; Liang, Q.; Yang, Y.; Chen, M.; Wei, X.; Wang, A.; Liao, S.; Ye, J. The key differentially expressed genes and proteins related to immune response in the spleen of pufferfish (Takifugu obscurus) infected by Aeromonas hydrophila. Fish Shellfish. Immunol. 2019, 91, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bertucci, E.M.; Mason, M.W.; Camus, A.C.; Rhodes, O.E.; Parrott, B.B. Chronic low dose irradiation alters hepatic transcriptional profiles, but not global DNA methylation in medaka (Oryzias latipes). Sci. Total Environ. 2020, 729, 138680. [Google Scholar] [CrossRef] [PubMed]
- Cockcroft, S.; Garner, K. Function of the phosphatidylinositol transfer protein gene family: Is phosphatidylinositol transfer the mechanism of action? Crit. Rev. Biochem. Mol. Biol. 2011, 46, 89–117. [Google Scholar] [CrossRef] [PubMed]
- Vieira, N.M.; Spinazzola, J.M.; Alexander, M.S.; Moreira, Y.B.; Kawahara, G.; Gibbs, D.E.; Mead, L.C.; Verjovski-Almeida, S.; Zatz, M.; Kunkel, L.M. Repression of phosphatidylinositol transfer protein α ameliorates the pathology of Duchenne muscular dystrophy. Proc. Natl. Acad. Sci. USA 2017, 114, 6080–6085. [Google Scholar] [CrossRef] [PubMed]
- Schurmans, S.; Vande Catsyne, C.A.; Desmet, C.; Moës, B. The phosphoinositide 5-phosphatase INPP5K: From gene structure to in vivo functions. Adv. Biol. Regul. 2021, 79, 100760. [Google Scholar] [CrossRef]
- McGrath, M.; Eramo, E.; Gurung, R.; Sriratana, A.; Gehrig, S.; Lynch, G.; Lourdes, S.; Koentgen, F.; Feeney, S.; Lazarou, L.; et al. Defective lysosome reformation during autophagy causes skeletal muscle disease. J. Clin. Investig. 2021, 4, 131. [Google Scholar] [CrossRef]
- Xiong, F.; Xiong, J.; Wu, Y.F.; Cao, L.; Huang, W.S.; Chang, M.X. Time-resolved RNA-seq provided a new understanding of intestinal immune response of European eel (Anguilla anguilla) following infection with Aeromonas hydrophila. Fish Shellfish. Immunol. 2020, 105, 297–309. [Google Scholar] [CrossRef]
- Zheng, J.; You, W.; Zheng, C.; Wan, P.; Chen, J.; Jiang, X.; Zhu, Z.; Zhang, Z.; Gong, A.; Li, W.; et al. Knockdown of fbxo39 inhibits proliferation and promotes apoptosis of human osteosarcoma u-2os cells. Oncol. Lett. 2018, 16, 1849–1854. [Google Scholar] [CrossRef]
- Ponsuksili, S.; Zebunke, M.; Murani, E.; Trakooljul, N.; Krieter, J.; Puppe, B.; Schwerin, M.; Wimmers, K. Integrated Genome-wide association and hypothalamus eQTL studies indicate a link between the circadian rhythm-related gene PER1 and coping behavior. Sci. Rep. 2015, 5, 16264. [Google Scholar] [CrossRef] [PubMed]
- Zou, B.; Chim, C.; Pang, R.; Zeng, H.; Dai, Y.; Zhang, R.; Lam, C.S.C.; Tan, V.; Hung, I.F.N.; Lan, H.; et al. XIAP-associated factor 1 (XAF1), a novel target of p53, enhances p53-mediated apoptosis via post-translational modification. Mol. Carcinog. 2012, 51, 422–432. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.; Kim, J.; Ko, K.; Ryu, B.; Lee, M.; Kim, H.; Chi, S. XAF1 forms a positive feedback loop with IRF-1 to drive apoptotic stress response and suppress tumorigenesis. Cell Death Dis. 2018, 9, 806. [Google Scholar] [CrossRef] [PubMed]
- Krasnov, A.; Timmerhaus, G.; Schiøtz, B.L.; Torgersen, J.; Afanasyev, S.; Iliev, D.; Jørgensen, J.; Takle, H.; Jørgensen, S.M. Genomic survey of early responses to viruses in Atlantic salmon, Salmo salar L. Mol. Immunol. 2011, 49, 163–174. [Google Scholar] [CrossRef]
- Xu, C.; Evensen, Ø.; Munang’andu, H.M. De novo assembly and transcriptome analysis of Atlantic salmon macrophage/dendritic-like TO cells following type I IFN treatment and Salmonid alphavirus subtype-3 infection. BMC Genom. 2015, 16, 96. [Google Scholar] [CrossRef]
- Riise, R.; Odqvist, L.; Mattsson, J.; Monkley, S.; Abdillahi, S.; Tyrchan, C.; Muthas, D.; Yrlid, L. Bleomycin hydrolase regulates the release of chemokines important for inflammation and wound healing by keratinocytes. Sci. Rep. 2019, 9, 20407. [Google Scholar] [CrossRef]
- Schwartz, D.R.; Homanics, G.E.; Hoyt, D.G.; Klein, E.; Abernethy, J.; Lazo, J.S. The neutral cysteine protease bleomycin hydrolase is essential for epidermal integrity and bleomycin resistance. Proc. Natl. Acad. Sci. USA 1999, 96, 4680–4685. [Google Scholar] [CrossRef]
- Fernández-Montero, Á.; Torrecillas, S.; Montero, D.; Acosta, F.; Prieto-Álamo, M.; Abril, N.; Jurado, J. Proteomic profile and protease activity in the skin mucus of greater amberjack (Seriola dumerili) infected with the ectoparasite Neobenedenia girellae—An immunological approach. Fish Shellfish. Immunol. 2021, 110, 100–115. [Google Scholar] [CrossRef]
- Misk, E.; Huber, P.; MacInnes, J.I.; Sherif, S.M.; Abo-Ismail, M.; Lumsden, J.S. Innate response of rainbow trout gill epithelial (RTgill-W1) cell line to ultraviolet-inactivated VHSV and FliC and rhabdovirus infection. Fish Shellfish. Immunol. Rep. 2022, 3, 100043. [Google Scholar] [CrossRef]
- Dean, J.M.; He, A.; Tan, M.; Wang, J.; Lu, D.; Razani, B.; Lodhi, I. MED19 Regulates Adipogenesis and Maintenance of White Adipose Tissue Mass by Mediating PPARγ-Dependent Gene Expression. Cell Rep. 2020, 33, 108228. [Google Scholar] [CrossRef]
- Zelechower, H.; Elbert, A.E. PPARs—Receptores activados por proliferadores peroxisomales. Rev. Nefrol. Dial. Traspl. 2009, 29, 74–83. [Google Scholar]
- Chen, J.J.; Xia, X.H.; Wang, L.F.; Jia, Y.F.; Nan, P.; Li, L.; Chang, Z.J. Identification and comparison of gonadal transcripts of testis and ovary of adult common carp Cyprinus carpio using suppression subtractive hybridizatio. Theriogenology 2015, 83, 1416–1427. [Google Scholar] [CrossRef] [PubMed]
- Judycka, S.; Nynca, J.; Hliwa, P.; Ciereszko, A. Characteristics and Cryopreservation of Semen of Sex-Reversed Females of Salmonid Fish. Int. J. Mol. Sci. 2021, 22, 964. [Google Scholar] [CrossRef] [PubMed]
- Murphy, D.L.; Fox, M.A.; Timpano, K.R.; Moya, P.R.; Ren-Patterson, R.; Andrews, A.M.; Holmes, A.; Lesch, K.P.; Wendland, J.R. How the serotonin story is being rewritten by new gene-based discoveries principally related to SLC6A4, the serotonin transporter gene, which functions to influence all cellular serotonin systems. Neuropharmacology 2008, 55, 932–960. [Google Scholar] [CrossRef]
- Salem, M.; Kenney, P.B.; Rexroad, C.E.; Yao, J. Development of a 37 k high-density oligonucleotide microarray: A new tool for functional genome research in rainbow trout. J. Fish Biol. 2008, 72, 2187–2206. [Google Scholar] [CrossRef]
- Wu, X.; Yamada-Mabuchi, M.; Morris, E.J.; Tanwar, P.S.; Dobens, L.; Gluderer, S.; Khan, S.; Cao, J.; Stocker, H.; Hafen, E.; et al. The Drosophila homolog of human tumor suppressor TSC-22 promotes cellular growth, proliferation, and survival. Proc. Natl. Acad. Sci. USA 2008, 14, 5414–5419. [Google Scholar] [CrossRef]
- Dragotto, J.; Canterini, S.; Del Porto, P.; Bevilacqua, A.; Fiorenza, M.T. The interplay between TGF-β-stimulated TSC22 domain family proteins regulates cell-cycle dynamics in medulloblastoma cells. J. Cell. Physiol. 2019, 234, 18349–18360. [Google Scholar] [CrossRef]
- Kamimura, R.; Uchida, D.; Kanno, S.-I.; Shiraishi, R.; Hyodo, T.; Sawatani, Y.; Shimura, M.; Hasegawa, T.; Tsubura-Okubo, M.; Yaguchi, E.; et al. Identification of Binding Proteins for TSC22D1 Family Proteins Using Mass Spectrometry. Int. J. Mol. Sci. 2021, 22, 10913. [Google Scholar] [CrossRef]
- Vogel, P.; Hans-Jfirgen, M.; Cieslak, A.; Adermann, K.; Forssmann, W. hDIP—A potential transcriptional regulator related to murine TSC-22 and Drosophila shortsighted (shs)—Is expressed in a large number of human tissues. Biochim. Et Biophys. Acta (BBA)-Gene Struct. Expr. 1996, 1309, 200–204. [Google Scholar] [CrossRef]
- Tacchi, L.; Bron, J.E.; Taggart, J.B.; Secombes, C.J.; Bickerdike, R.; Adler, M.A.; Takle, H.; Martin, S.A. Multiple tissue transcriptomic responses to Piscirickettsia salmonis in Atlantic salmon (Salmo salar). Physiol. Genom. 2011, 43, 1241–1254. [Google Scholar] [CrossRef]
- Rodriguez-Cupello, C.; Dam, M.; Serini, L.; Wang, S.; Lindgren, D.; Englund, E.; Kjellman, P.; Axelson, H.; García-Mariscal, A.; Madsen, C.D. The STRIPAK Complex Regulates Response to Chemotherapy Through p21 and p27. Front. Cell Dev. Biol. 2020, 8, 146. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Dong, Y.; Qiang, R.; Zhang, Y.; Zhang, X.; Chen, Y.; Jiang, P.; Ma, X.; Wu, L.; Ai, J.; et al. Characterization of Strip1 Expression in Mouse Cochlear Hair Cells. Front. Genet. 2021, 12, 625867. [Google Scholar] [CrossRef] [PubMed]
- Kück, U.; Radchenko, D.; Teichert, I. STRIPAK, a highly conserved signaling complex, controls multiple eukaryotic cellular and developmental processes and is linked with human diseases. Biol. Chem. 2019, 400, 1005–1022. [Google Scholar] [CrossRef] [PubMed]
- Puente-Marin, S.; Nombela, I.; Ciordia, S.; Mena, M.C.; Chico, V.; Coll, J.; Ortega-Villaizan, M.D.M. In Silico Functional Networks Identified in Fish Nucleated Red Blood Cells by Means of Transcriptomic and Proteomic Profiling. Genes 2018, 9, 202. [Google Scholar] [CrossRef] [PubMed]
- Rowland, K.; Saelao, P.; Wang, Y.; Fulton, J.E.; Liebe, G.N.; McCarron, A.M.; Wolc, A.; Gallardo, R.A.; Kelly, T.; Zhou, H.; et al. Association of Candidate Genes with Response to Heat and Newcastle Disease Virus. Genes 2018, 9, 560. [Google Scholar] [CrossRef]
- Kaur, S.; Sang, Y.; Aballay, A. Myotubularin-related protein protects against neuronal degeneration mediated by oxidative stress or infection. J. Biol. Chem. 2022, 298, 101614. [Google Scholar] [CrossRef]
- Zhu, Z.X.; Lin, Y.L.; Qin, H.; Xiong, Y.Y.; Jiang, D.L.; Lin, H.R.; Yu, Z.L.; Xia, J.L. Identifying a genome-wide QTL interval controlling for ammonia-nitrogen tolerance on chrLG1 of Nile tilapia. Aquaculture 2021, 543, 736946. [Google Scholar] [CrossRef]
- Stefanini, L.; Boulaftali, Y.; Ouellette, T.; Holinstat, M.; Désiré, L.; Leblond, B.; Andre, P.; Conley, P.; Bergmeier, W. Rap1-Rac1 Circuits Potentiate Platelet Activation. Arterioscler. Thromb. Vasc. Biol. 2011, 32, 434–441. [Google Scholar] [CrossRef]
- Nagy, Z.; Smolenski, A. Cyclic nucleotide-dependent inhibitory signaling interweaves with activating pathways to determine platelet responses. Res. Pract. Trombos. Haemost. 2018, 2, 558–571. [Google Scholar] [CrossRef]
- Neumüller, O.; Hoffmeister, M.; Babica, J.; Prelle, C.; Gegenbauer, K.; Smolenski, A.P. Synaptotagmin-like protein 1 interacts with the GTPase-activating protein Rap1GAP2 and regulates dense granule secretion in platelets. Blood. 2009, 114, 1396–1404. [Google Scholar] [CrossRef]
- Tan, S.; Wang, W.; Tian, C.; Niu, D.; Zhou, T.; Yang, Y.; Gao, D.; Liu, Z. Post-transcriptional regulation through alternative splicing after infection with Flavobacterium columnare in channel catfish (Ictalurus punctatus). Fish Shellfish. Immunol. 2019, 91, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Zhou, Q.; Li, K.; Xu, W.; Wang, L.; Hu, G.; Chen, S. Transcriptome analysis of Giant grouper (Epinephelus lanceolatus) kidney and spleen in response to spotted knifejaw iridovirus (SKIV) infection. Aquac. Res. 2020, 52, 1954–1964. [Google Scholar] [CrossRef]
- Niwa, V.; Nagata-Ohashi, K.; Takeichi, M.; Mizuno, K.; Uemura, T. Control of actin reorganization by slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell 2002, 108, 233–246. [Google Scholar] [CrossRef]
- Sun, B.; Fang, Y.T.; Jin, D.J.; Chen, Z.Y.; Li, Z.Y.; Gu, X.D.; Xiang, J.B. miR-194 Inhibits the Proliferation of SW620 Colon Cancer Stem Cells Through Downregulation of SSH2 Expression. Cancer Manag. Res. 2019, 11, 10229–10238. [Google Scholar] [CrossRef]
- Zheng, C.; Li, R.; Zheng, S.; Fang, H.; Xu, M.; Zhong, L. LINC00174 Facilitates Cell Proliferation, Cell Migration and Tumor Growth of Osteosarcoma via Regulating the TGF-β/SMAD Signaling Pathway and Upregulating SSH2 Expression. Front. Mol. Biosci. 2021, 8, 697773. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Kim, K.T.; Kim, C.H.; Lee, E.Y.; Lee, S.G.; Seo, M.E.; Kim, J.H.; Chung, C.K. Unveiling the genetic variation of severe continuous/mixed-type ossification of the posterior longitudinal ligament by whole-exome sequencing and bioinformatic analysis. Spine J. 2021, 21, 1847–1856. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).