Variation in Ovine DGAT1 and Its Association with Carcass Muscle Traits in Southdown Sheep
Abstract
1. Introduction
2. Materials and Methods
2.1. Sheep Investigated and Phenotypic Data Collection
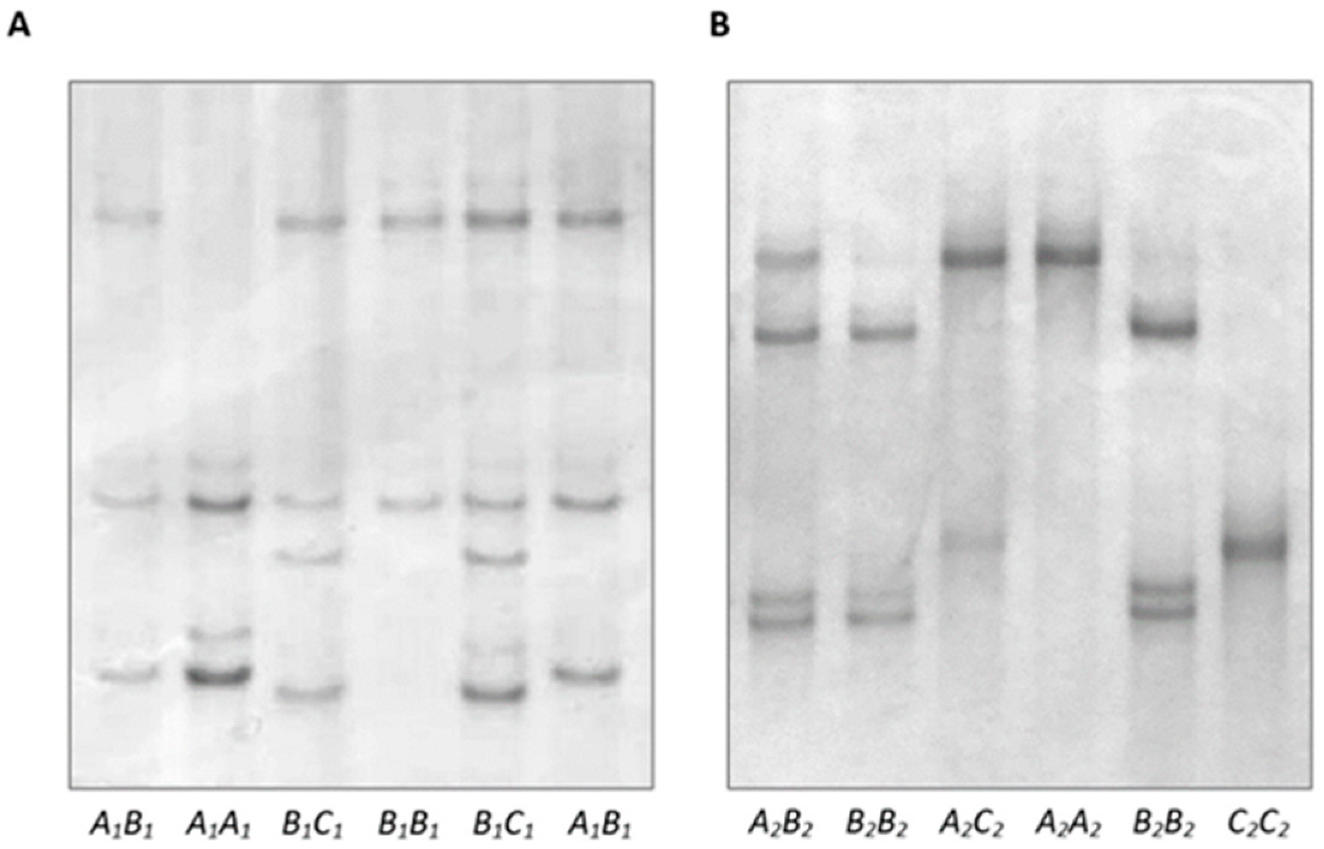
2.2. PCR-SSCP Analyses
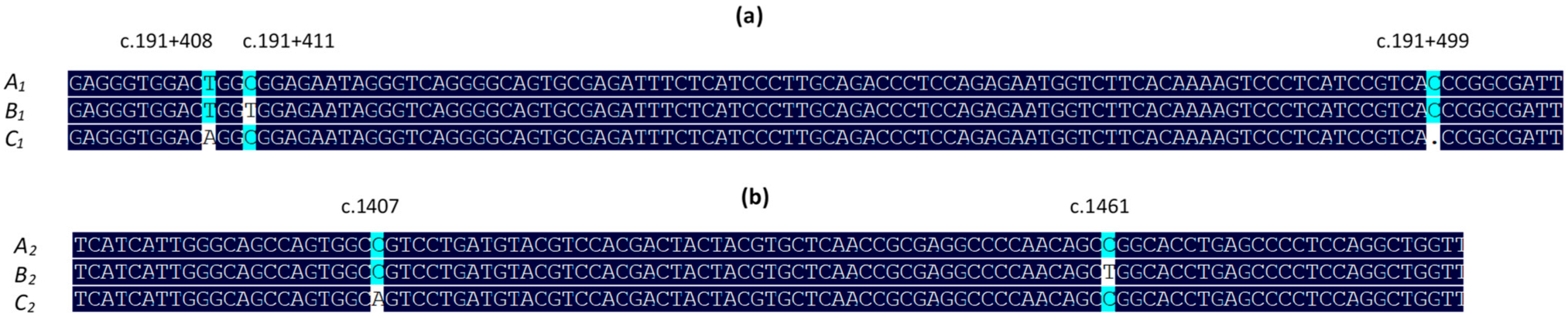
2.3. DNA Sequencing and Sequence Analyses
2.4. Statistical Analyses
3. Results
3.1. Variation in Ovine DGAT1
3.2. Association of DGAT1 Intron 1 Variation and Carcass Traits
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zong, G.; Li, Y.; Wanders, A.J.; Alssema, M.; Zock, P.L.; Willett, W.C.; Hu, F.B.; Sun, Q. Intake of individual saturated fatty acids and risk of coronary heart disease in US men and women: Two prospective longitudinal cohort studies. BMJ 2016, 355, i5796. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.; Enser, M.; Fisher, A.; Nute, G.; Sheard, P.; Richardson, R.; Hughes, S.; Whittington, F. 2008. Fat deposition, fatty acid composition and meat quality: A review. Meat Sci. 2008, 78, 343–358. [Google Scholar] [CrossRef]
- Lehner, R.; Kuksis, A. Biosynthesis of triacylglycerols. Prog. Lipid Res. 1996, 35, 169–201. [Google Scholar] [CrossRef]
- Grisart, B.; Coppieters, W.; Farnir, F.; Karim, L.; Ford, C.; Berzi, P.; Cambisano, N.; Mni, M.; Reid, S.; Simon, P. Positional candidate cloning of a QTL in dairy cattle: Identification of a missense mutation in the bovine DGAT1 gene with major effect on milk yield and composition. Genome Res. 2002, 12, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Schennink, A.; Stoop, W.M.; Visker, M.W.; Heck, J.M.; Bovenhuis, H.; van der Poel, J.J.; van Valenberg, H.J.; van Arendonk, J.A. DGAT1 underlies large genetic variation in milk-fat composition of dairy cows. Anim. Genet. 2007, 38, 467–473. [Google Scholar] [CrossRef]
- Thaller, G.; Kühn, C.; Winter, A.; Ewald, G.; Bellmann, O.; Wegner, J.; Zühlke, H.; Fries, R. DGAT1, a new positional and functional candidate gene for intramuscular fat deposition in cattle. Anim. Genet. 2003, 34, 354–357. [Google Scholar] [CrossRef]
- Winter, A.; Krämer, W.; Werner, F.A.; Kollers, S.; Kata, S.; Durstewitz, G.; Buitkamp, J.; Womack, J.E.; Thaller, G.; Fries, R. Association of a lysine-232/alanine polymorphism in a bovine gene encoding acyl-CoA:diacylglycerol acyltransferase (DGAT1) with variation at a quantitative trait locus for milk fat content. Proc. Natl. Acad. Sci. USA 2002, 99, 9300–9305. [Google Scholar] [CrossRef]
- Gautier, M.; Capitan, A.; Fritz, S.; Eggen, A.; Boichard, D.; Druet, T. Characterization of the DGAT1 K232A and variable number of tandem repeat polymorphisms in French dairy cattle. J. Dairy Sci. 2007, 90, 2980–2988. [Google Scholar] [CrossRef]
- Pareek, C.S.; Czarnik, U.; Zabolewicz, T.; Pareek, R.S.; Walawski, K. DGAT1 K232A quantitative trait nucleotide polymorphism in Polish Black-and-White cattle. J. Appl. Genet. 2005, 46, 85–87. [Google Scholar]
- Weller, J.; Golik, M.; Seroussi, E.; Ezra, E.; Ron, M. Population-wide analysis of a QTL affecting milk-fat production in the Israeli Holstein population. J. Dairy Sci. 2003, 86, 2219–2227. [Google Scholar] [CrossRef]
- Näslund, J.; Fikse, W.; Pielberg, G.; Lundén, A. Frequency and effect of the bovine acyl-CoA: Diacylglycerol acyltransferase 1 (DGAT1) K232A polymorphism in Swedish dairy cattle. J. Dairy Sci. 2008, 91, 2127–2134. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Jia, J.; Ma, Y.; Zhang, Y.; Wang, Y.; Yu, Y.; Zhang, Y. Effects of DGAT1 and GHR on milk yield and milk composition in the Chinese dairy population. Anim. Genet. 2009, 40, 997–1000. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, H.; Cheng, L.; Edwards, G.R.; Hickford, J.G.H. Effect of DGAT1 variant (K232A) on milk traits and milk fat composition in outdoor pasture-grazed dairy cattle. N. Z. J. Agric. Res. 2021, 64, 101–113. [Google Scholar] [CrossRef]
- Li, X.; Ekerljung, M.; Lundström, K.; Lundén, A. Association of polymorphisms at DGAT1, leptin, SCD1, CAPN1 and CAST genes with color, marbling and water holding capacity in meat from beef cattle populations in Sweden. Meat Sci. 2013, 94, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Pannier, L.; Mullen, A.; Hamill, R.; Stapleton, P.; Sweeney, T. Association analysis of single nucleotide polymorphisms in DGAT1, TG and FABP4 genes and intramuscular fat in crossbred Bos taurus cattle. Meat Sci. 2010, 85, 515–518. [Google Scholar] [CrossRef] [PubMed]
- Avilés, C.; Polvillo, O.; Peña, F.; Juárez, M.; Martinez, A.; Molina, A. Associations between DGAT1, FABP4, LEP, RORC, and SCD1 gene polymorphisms and fat deposition in Spanish commercial beef. J. Anim. Sci. 2013, 91, 4571–4577. [Google Scholar] [CrossRef] [PubMed]
- Gill, J.L.; Bishop, S.C.; McCorquodale, C.; Williams, J.L.; Wiener, P. Association of selected SNP with carcass and taste panel assessed meat quality traits in a commercial population of Aberdeen Angus-sired beef cattle. Genet. Sel. Evol. 2009, 41, 1–12. [Google Scholar] [CrossRef]
- Fortes, M.R.; Curi, R.A.; Chardulo, L.A.L.; Silveira, A.C.; Assumpção, M.E.; Visintin, J.A.; de Oliveira, H.N. Bovine gene polymorphisms related to fat deposition and meat tenderness. Genet. Mol. Biol. 2009, 32, 75–82. [Google Scholar] [CrossRef]
- Casas, E.; White, S.; Riley, D.; Smith, T.; Brenneman, R.; Olson, T.; Johnson, D.; Coleman, S.; Bennett, G.; Chase, C., Jr. Assessment of single nucleotide polymorphisms in genes residing on chromosomes 14 and 29 for association with carcass composition traits in Bos indicus cattle. J. Anim. Sci. 2005, 83, 13–19. [Google Scholar] [CrossRef]
- Armstrong, E.; Ciappesoni, G.; Iriarte, W.; Da Silva, C.; Macedo, F.; Navajas, E.; Brito, G.; San Julián, R.; Gimeno, D.; Postiglioni, A. Novel genetic polymorphisms associated with carcass traits in grazing Texel sheep. Meat Sci. 2018, 145, 202–208. [Google Scholar] [CrossRef]
- Scatà, M.; Napolitano, F.; Casu, S.; Carta, A.; De Matteis, G.; Signorelli, F.; Annicchiarico, G.; Catillo, G.; Moioli, B. Ovine acyl CoA: Diacylglycerol acyltransferase 1–molecular characterization, polymorphisms and association with milk traits. Anim. Genet. 2009, 40, 737–742. [Google Scholar] [CrossRef] [PubMed]
- Ala Noshahr, F.; Rafat, A. Polymorphism of DGAT1 gene and its relationship with carcass weight and dressing percentage in Moghani sheep breed. Iran. J. Appl. Anim. Sci. 2014, 4, 331–334. [Google Scholar]
- Mohammadi, H.; Shahrebabak, M.M.; Sadeghi, M. Association between single nucleotide polymorphism in the ovine DGAT1 gene and carcass traits in two Iranian sheep breeds. Anim. Biotechnol. 2013, 24, 159–167. [Google Scholar] [CrossRef]
- Xu, Q.L.; Chen, Y.L.; Ma, R.X.; Xue, P. Polymorphism of DGAT1 associated with intramuscular fat-mediated tenderness in sheep. J. Sci. Food Agric. 2009, 89, 232–237. [Google Scholar] [CrossRef]
- Hopkins, D.L.; Safari, E.; Thompson, J.M.; Smith, C.R. Video image analysis in the Australian meat industry—Precision and accuracy of predicting lean meat yield in lamb carcasses. Meat Sci. 2004, 67, 269–274. [Google Scholar] [CrossRef]
- Zhou, H.; Hickford, J.G.H.; Fang, Q. A two-step procedure for extracting genomic DNA from dried blood spots on filter paper for polymerase chain reaction amplification. Anal. Biochem. 2006, 354, 159–161. [Google Scholar] [CrossRef] [PubMed]
- Byun, S.O.; Fang, Q.; Zhou, H.; Hickford, J.G.H. An effective method for silver-staining DNA in large numbers of polyacrylamide gels. Anal. Biochem. 2009, 385, 174–175. [Google Scholar] [CrossRef]
- Kijas, J.W.; Townley, D.; Dalrymple, B.P.; Heaton, M.P.; Maddox, J.F.; McGrath, A.; Wilson, P.; Ingersoll, R.G.; McCulloch, R.; McWilliam, S. A genome wide survey of SNP variation reveals the genetic structure of sheep breeds. PLoS ONE 2009, 4, e4668. [Google Scholar] [CrossRef]
- Gong, H.; Zhou, H.; Hickford, J.G.H. Diversity of the glycine/tyrosine-rich keratin-associated protein 6 gene (KAP6) family in sheep. Mol. Biol. Rep. 2011, 38, 31–35. [Google Scholar] [CrossRef]
- Jo, B.S.; Choi, S.S. Introns: The functional benefits of introns in genomes. Genom. Inform. 2015, 13, 112. [Google Scholar] [CrossRef]
- Park, S.G.; Hannenhalli, S.; Choi, S.S. Conservation in first introns is positively associated with the number of exons within genes and the presence of regulatory epigenetic signals. BMC Genom. 2014, 15, 526. [Google Scholar] [CrossRef] [PubMed]
| Region Amplified | Primer Sequence (5′-3′) | Expected Size (bp) | SSCP Condition |
|---|---|---|---|
| Intron 1 | GTGATCCCACCACGCACAG | 271 | 10% gel containing 1% glycerol, 150 V, 24 °C and 17 h. |
| CACTTTGACCCTAGAGCAG | |||
| Exon 17 | TGATGGCACAGGTGAGCAG | 310 | 12% gel, 250 V, 22 °C and 17 h. |
| CAGGCTCCAGTACAGCAGC |
| Trait | Variant Assessed | Other Variant Fitted | Mean ± SE | p1 | |
|---|---|---|---|---|---|
| Present | Absent | ||||
| HCW (kg) | A1 | None | 19.9 ± 0.37 (n = 78) | 19.6 ± 0.94 (n = 9) | 0.766 |
| B1 | None | 20.0 ± 0.46 (n = 48) | 19.8 ± 0.48 (n = 39) | 0.662 | |
| C1 | None | 19.6 ± 0.78 (n = 16) | 20.0 ± 0.39 (n = 71) | 0.639 | |
| Loin yield (%) | A1 | None | 15.5 ± 0.09 (n = 78) | 15.8 ± 0.24 (n = 9) | 0.196 |
| B1 | None | 15.8 ± 0.11 (n = 48) | 15.3 ± 0.11 (n = 39) | 0.001 | |
| C1 | None | 15.3 ± 0.19 (n = 16) | 15.6 ± 0.10 (n = 71) | 0.200 | |
| B1 | A1 | 15.8 ± 0.14 (n = 48) | 15.4 ± 0.16 (n = 39) | 0.003 | |
| Shoulder yield (%) | A1 | None | 16.9 ± 0.11 (n = 78) | 17.1 ± 0.29 (n = 9) | 0.424 |
| B1 | None | 17.1 ± 0.14 (n = 48) | 16.7 ± 0.14 (n = 39) | 0.014 | |
| C1 | None | 16.5 ± 0.23 (n = 16) | 17.0 ± 0.12 (n = 71) | 0.080 | |
| B1 | C1 | 17.0 ± 0.20 (n = 48) | 16.7 ± 0.15 (n = 39) | 0.065 | |
| C1 | B1 | 16.7 ± 0.25 (n = 16) | 16.9 ± 0.12 (n = 71) | 0.501 | |
| Leg yield (%) | A1 | None | 22.3 ± 0.13 (n = 78) | 22.8 ± 0.34 (n = 9) | 0.168 |
| B1 | None | 22.6 ± 0.16 (n = 48) | 22.1 ± 0.17 (n = 39) | 0.037 | |
| C1 | None | 22.2 ± 0.28 (n = 16) | 22.4 ± 0.14 (n = 71) | 0.460 | |
| B1 | A1 | 22.7 ± 0.20 (n = 48) | 22.3 ± 0.24 (n = 39) | 0.073 | |
| Eye muscle width (mm) | A1 | None | 68.7 ± 0.63 (n = 78) | 67.6 ± 1.58 (n = 9) | 0.485 |
| B1 | None | 68.8 ± 0.78 (n = 48) | 68.4 ± 0.81 (n = 39) | 0.734 | |
| C1 | None | 68.4 ± 1.31 (n = 16) | 68.6 ± 0.65 (n = 71) | 0.850 | |
| Eye muscle depth (mm) | A1 | None | 29.2 ± 0.29 (n = 78) | 29.1 ± 0.74 (n = 9) | 0.878 |
| B1 | None | 29.3 ± 0.36 (n = 48) | 29.2 ± 0.38 (n = 39) | 0.817 | |
| C1 | None | 29.5 ± 0.61 (n = 16) | 29.2 ± 0.30 (n = 71) | 0.661 | |
| IMF (%) | A1 | None | 3.32 ± 0.135 (n = 78) | 2.94 ± 0.330 (n = 9) | 0.252 |
| B1 | None | 3.35 ± 0.172 (n = 48) | 3.23 ± 0.168 (n = 39) | 0.554 | |
| C1 | None | 3.18 ± 0.280 (n = 16) | 3.31 ± 0.141 (n = 71) | 0.664 | |
| Subcutaneous fat depth (mm) | A1 | None | 4.12 ± 0.193 (n = 78) | 4.25 ± 0.557 (n = 9) | 0.830 |
| B1 | None | 4.18 ± 0.240 (n = 48) | 4.08 ± 0.255 (n = 39) | 0.767 | |
| C1 | None | 3.79 ± 0.438 (n = 16) | 4.21 ± 0.206 (n = 71) | 0.397 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, R.; Zhou, H.; Fang, Q.; Zhou, P.; Yang, Y.; Jiang, S.; Hickford, J.G.H. Variation in Ovine DGAT1 and Its Association with Carcass Muscle Traits in Southdown Sheep. Genes 2022, 13, 1670. https://doi.org/10.3390/genes13091670
Dai R, Zhou H, Fang Q, Zhou P, Yang Y, Jiang S, Hickford JGH. Variation in Ovine DGAT1 and Its Association with Carcass Muscle Traits in Southdown Sheep. Genes. 2022; 13(9):1670. https://doi.org/10.3390/genes13091670
Chicago/Turabian StyleDai, Rong, Huitong Zhou, Qian Fang, Ping Zhou, Yang Yang, Shuang Jiang, and Jonathan G. H. Hickford. 2022. "Variation in Ovine DGAT1 and Its Association with Carcass Muscle Traits in Southdown Sheep" Genes 13, no. 9: 1670. https://doi.org/10.3390/genes13091670
APA StyleDai, R., Zhou, H., Fang, Q., Zhou, P., Yang, Y., Jiang, S., & Hickford, J. G. H. (2022). Variation in Ovine DGAT1 and Its Association with Carcass Muscle Traits in Southdown Sheep. Genes, 13(9), 1670. https://doi.org/10.3390/genes13091670







