Genetic Diversity and Population Structure of Local Chicken Ecotypes in Burkina Faso Using Microsatellite Markers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. DNA Amplification and Microsatellite Genotyping
2.3. Estimates of Genetic Diversity and Distance
2.4. Genetic Relationships and Structure
3. Results
3.1. Estimates of Genetic Diversity
3.2. Genetic Distances of Local Chicken Ecotypes
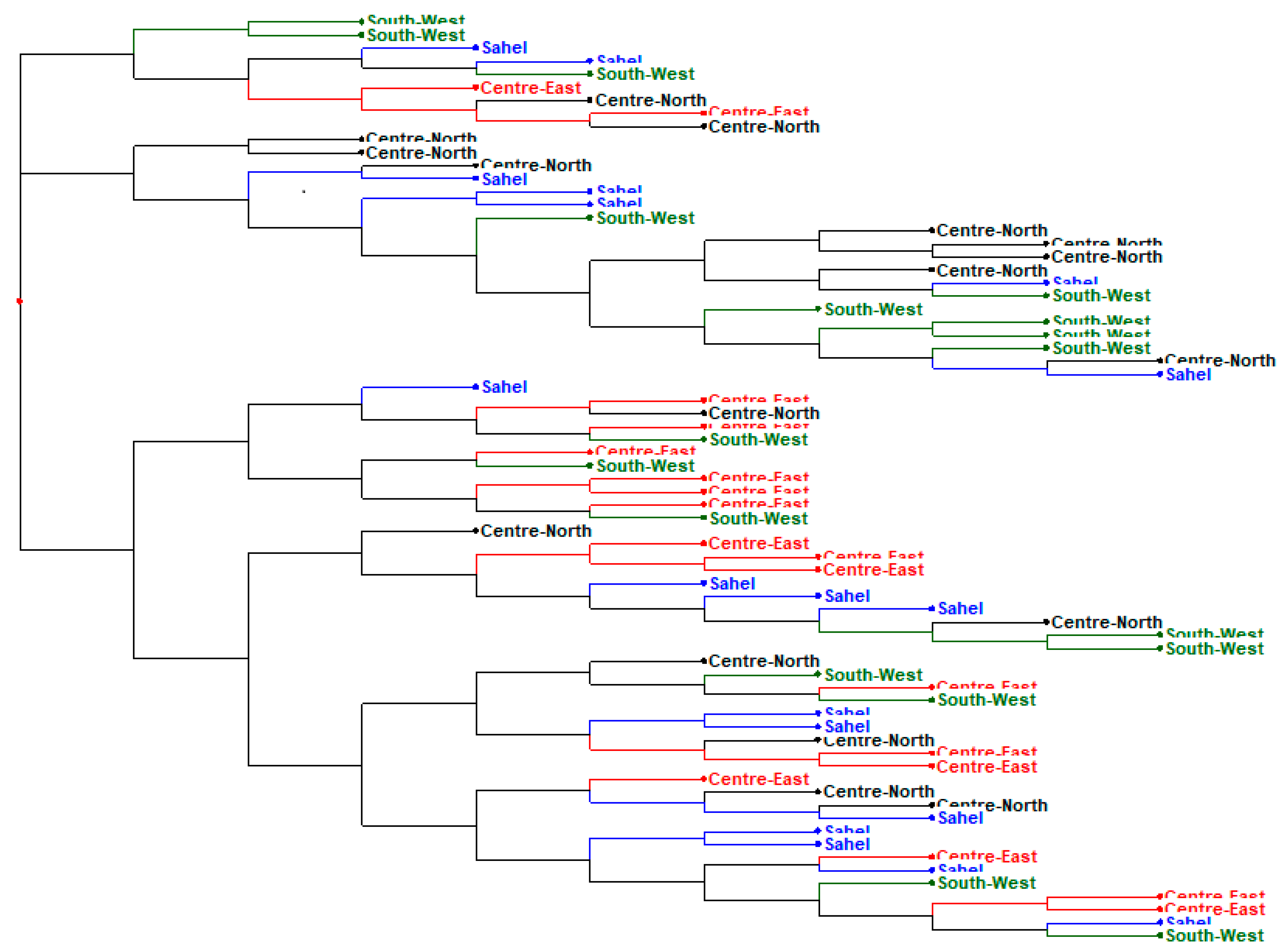
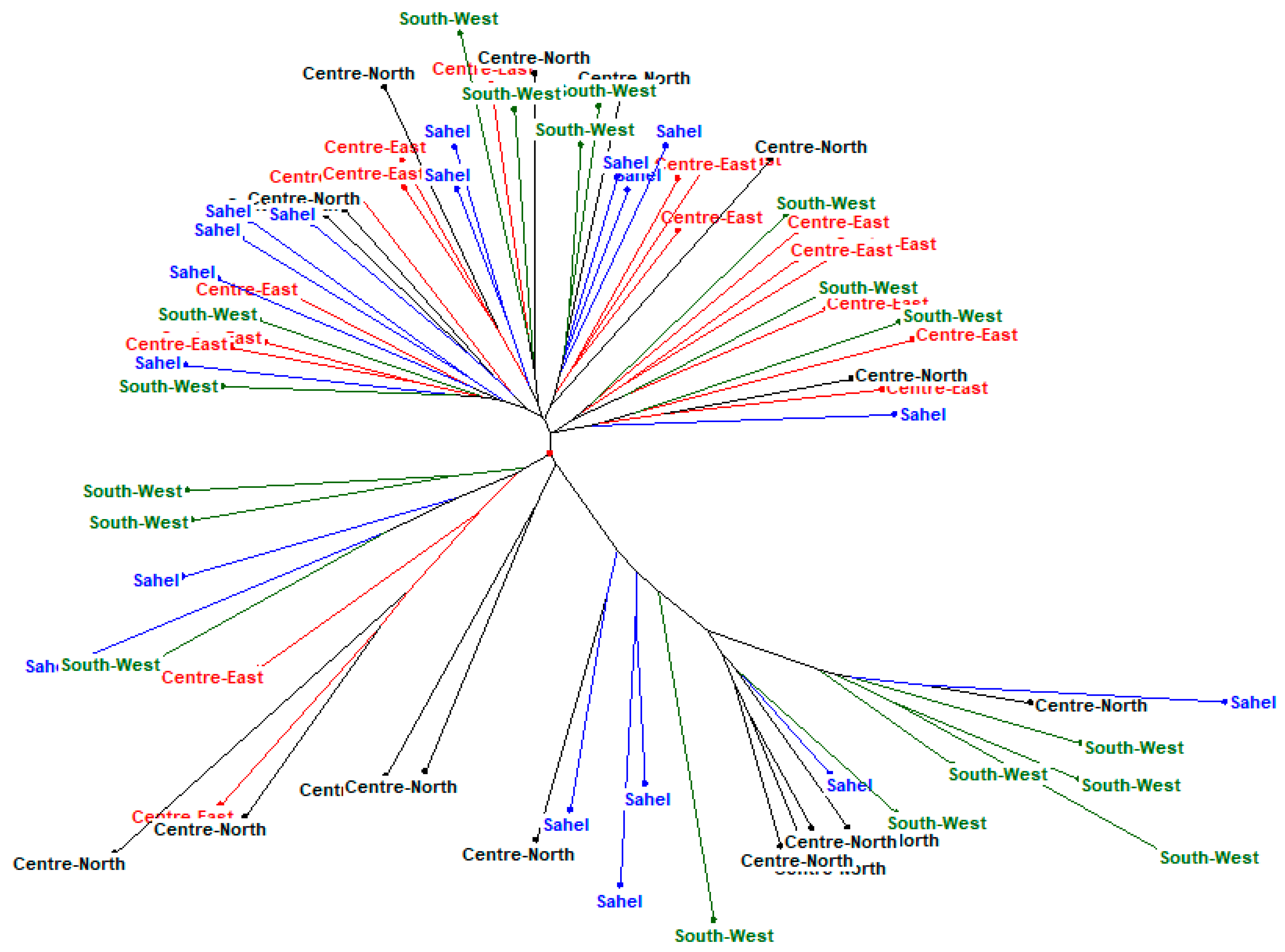
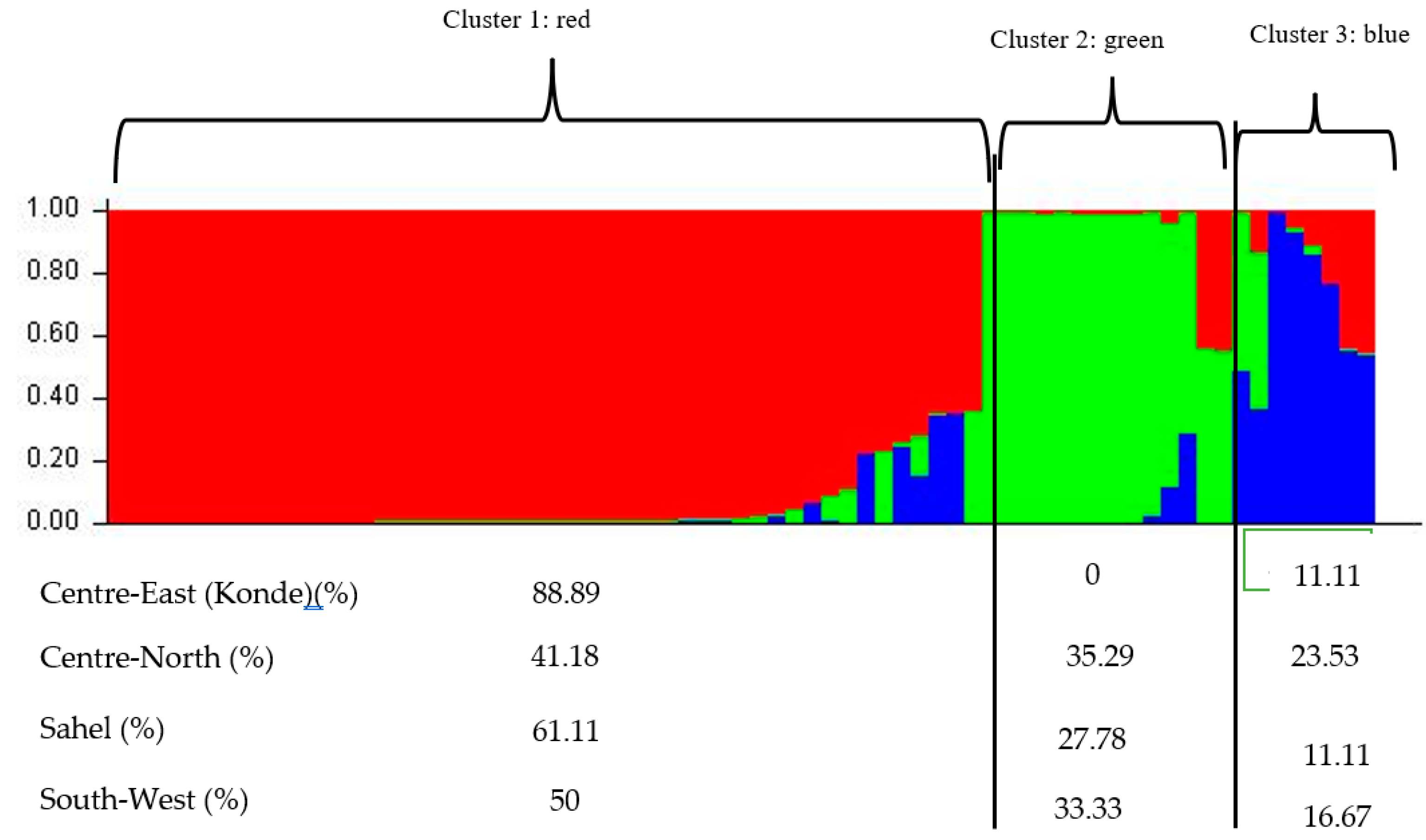
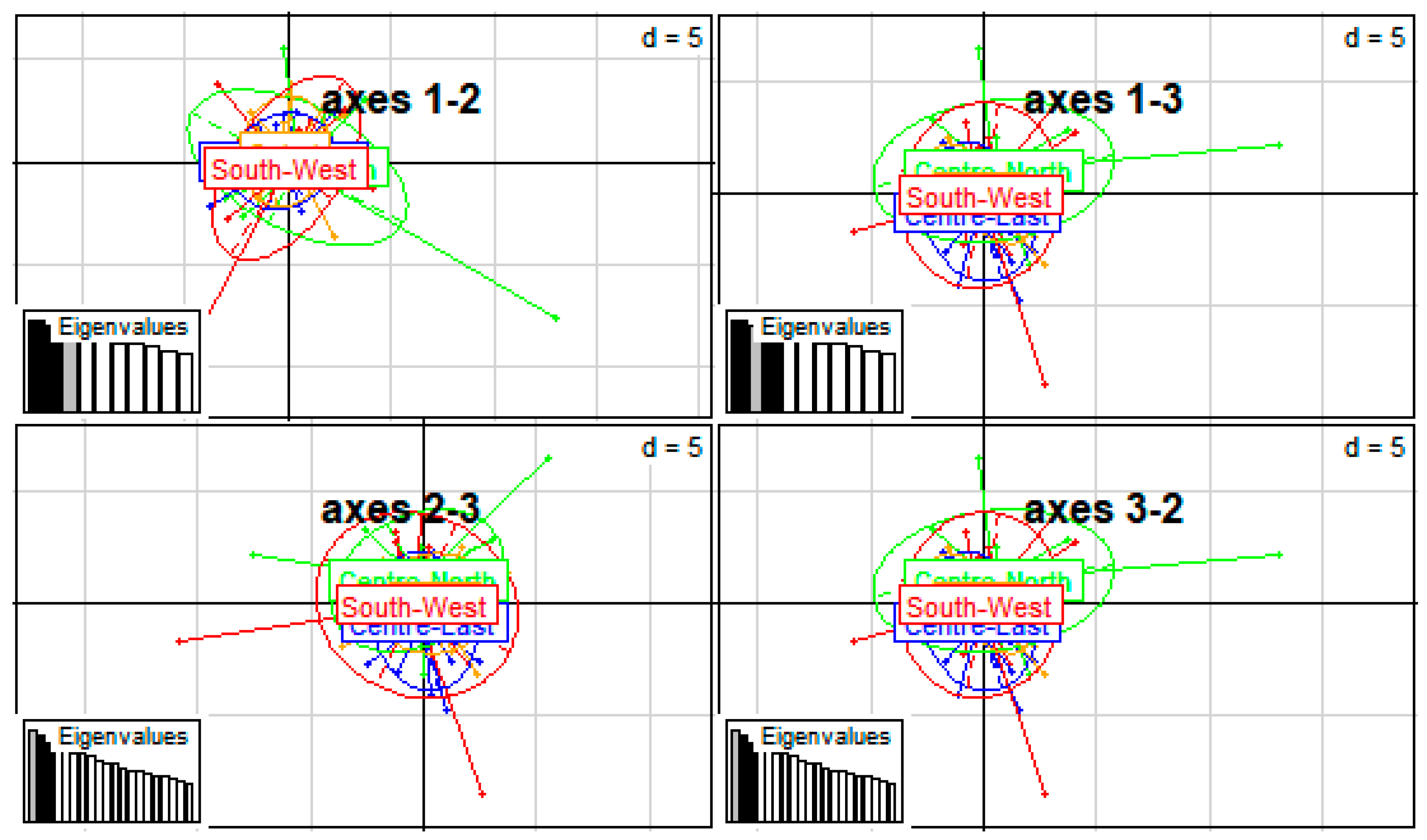
3.3. Phylogenetic Analysis and Cluster of Local Chicken Ecotypes
4. Discussion
4.1. Allelic Diversity
4.2. Heterozygosity Rate
4.3. Polymarphism Informative Content
4.4. Genetic Distance
4.5. Phylogenetic Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tadano, R.; Nagasaka, N.; Goto, N.; Rikimaru, K.; Tsudzuki, M. Genetic Characterization and Conservation Priorities of Chicken Lines. Poult. Sci. 2013, 92, 2860–2865. [Google Scholar] [CrossRef] [PubMed]
- Abebe, A.S.; Mikko, S.; Johansson, A.M. Genetic Diversity of Five Local Swedish Chicken Breeds Detected by Microsatellite Markers | Enhanced Reader. PLoS ONE 2015, 10, e0120580. [Google Scholar] [CrossRef] [PubMed]
- Kondombo, S.R.; Nianogo, A.J.; Kwakkel, R.P.; Udo, H.M.Y.; Slingerland, M. Comparative Analysis of Village Chicken Production in Two Farming Systems in Burkina Faso. Trop. Anim. Health Prod. 2003, 35, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Pousga, S. Supplementation Strategies for Semi-Scavenging Chickens in Burkina Faso Evaluation of Some Local Feed Resources. Ph.D. Thesis, Swedish University of Agricultural Sciences, Uppsala, Sweden, 2007. [Google Scholar]
- Keambou, T.C.; Hako, B.A.; Ommeh, S.; Bembide, C.; Ngono, E.P.; Manjeli, Y.; Wamonje, F.; Nzuki; Wanjala, B.; Wamalwa, M.; et al. Genetic Diversity of the Cameroon Indigenous Chicken Ecotypes. Int. J. Poult. Sci. 2014, 13, 279–291. [Google Scholar] [CrossRef]
- Pinde, S.; Tapsoba, A.S.R.; Traore, F.G.; Ouedraogo, R.W.; Ba, S.; Sanou, M.; Traore, A.; Tamboura, H.H.; Simpore, J. Caractérisation et Typologie des Systèmes d’élevage de La Poule Locale Du Burkina Faso. Int. J. Biol. Chem. Sci. 2020, 46, 14. [Google Scholar] [CrossRef]
- Zare, Y.; Isidore, G.B.; Isidore, H.; Michel, K.; Boureima, T.; Moussa, Z.; Samuel, B.; Apollinair, T.P.; Mahamoudou, Z.; Romdhane, R.; et al. Morpho-Biometric Evaluation of the Genetic Diversity of Local Chicken Ecotypes in Four Regions (Centre-East, Sahel, Centre-North and South-West) of Burkina Faso. Int. J. Poult. Sci. 2021, 20, 231–242. [Google Scholar] [CrossRef]
- Pinde, S.; Tapsoba, A.S.R.; Sanou, M.; Traore, F.G.; Ouedraogo, R.W.; Ba, S.; Traore, A.; Tamboura, H.H.; Simpore, J. Profils Morpho-Biometriques de La Poule Locale Du Burkina Faso. Int. J. Biol. Chem. Sci. 2020, 14, 2240–2256. [Google Scholar] [CrossRef]
- Ouandaogo, Z.C. La Souche Kondé: Conservation-Amélioration-Vulgarisation, 1975.
- FAO—Food and Agriculture Organisation of the United Nations. Secondary Guidelines for Development of National Farm Animal Genetic Resources Management Plans. In Measurement of Domestic Animal Diversity (MoDAD): Recommended Microsatellite Markers; FAO: Rome, Italy, 1998. [Google Scholar]
- Noah, O.; Ngeranwa, J.J.N.; Binepal, Y.S.; Kahi, A.K.; Bramwel, W.W.; Ateya, L.O.; Wekesa, F.C. Genetic Diversity of Indigenous Chickens from Selected Areas in Kenya Using Microsatellite Markers. J. Genet. Eng. Biotechnol. 2017, 15, 489–495. [Google Scholar] [CrossRef]
- Fotsa, J.C.; Kamdem, D.P.; Bordas, A.; Tixier-Boichard, M.; Rognon, X. Assessment of the Genetic Diversity of Cameroon Indigenous Chickens by the Use of Microsatellites. Livest. Res. Rural Dev. 2011, 23, 5. [Google Scholar]
- FAO. Molecular Genetic Characterization of Animal Genetic Resources; FAO: Rome, Italy, 2011. [Google Scholar]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the Analysis of Population Structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef]
- Nei, M. Genetic Distance Between Populations. Mol. Evol. Genet. 1972, 106, 208–253. [Google Scholar] [CrossRef]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring Weak Population Structure with the Assistance of Sample Group Information. Mol. Ecol. Resour. 2009, 9, 1322–1332. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A Website and Program for Visualizing STRUCTURE Output and Implementing the Evanno Method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software STRUCTURE: A Simulation Study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T. Exploratory Analysis of Genetic and Genomic Data: Package ‘Adegenet’; CRAN: Vienna, Austria, 2021; p. 188. [Google Scholar]
- Babar, M.E.; Nadeem, A.; Hussain, T.; Wajid, A.; Shah, S.A.; Iqbal, A.; Sarfraz, Z.; Akram, M. Microsatellite Marker Based Genetic Diversity among Four Varieties of Pakistani Aseel Chicken. Pak. Vet. J. 2012, 32, 237–241. [Google Scholar]
- Pirany, N.; Romanov, M.N.; Ganpule, S.P.; Devegowda, G.; Prasad, D.T. Microsatellite Analysis of Genetic Diversity in Indian Chicken Populations. Poult. Sci. 2007, 44, 19–28. [Google Scholar] [CrossRef]
- Dorji, N.; Duangjinda, M.; Phasuk, Y. Genetic Characterization of Bhutanese Native Chickens Based on an Analysis of Red Junglefowl (Gallus Gallus Gallus and Gallus Gallus Spadecieus), Domestic Southeast Asian and Commercial Chicken Lines (Gallus Gallus Domesticus). Genet. Mol. Biol. 2012, 35, 603–609. [Google Scholar] [CrossRef]
- Azimu, W.; Manatbay, B.; Li, Y.; Kaimaerdan, D.; Wang, H.E.; Reheman, A.; Muhatai, G. Genetic Diversity and Population Structure Analysis of Eight Local Chicken Breeds of Southern Xinjiang. Br. Poult. Sci. 2018, 59, 629–635. [Google Scholar] [CrossRef]
- Pandey, A.K.; Tantia, M.S.; Chaudhary, P.; Vijh, K.; Box, O. Three Poultry, 1999; 1536–1542.
- Lyimo, C.M.; Weigend, A.; Msoffe, P.L.; Simianer, H.; Weigend, S. Évaluation de La Diversité Génétique de Cinq Écotypes de Poulet Tanzaniens à l’ Aide d’ Outils Moléculaires Résumé. S. Afr. J. Anim. Sci. 2013, 43, 499–510. [Google Scholar] [CrossRef]
- Hassen, H.; Neser, F.W.C.; De Kock, A.; Van Marle-Köster, E. Study on the Genetic Diversity of Native Chickens in Northwest Ethiopia Using Microsatellite Markers. Afr. J. Biotechnol. 2009, 8, 1347–1353. [Google Scholar] [CrossRef]
- Hillel, J.; Groenen, M.A.M.; Tixier-Boichard, M.; Korol, A.B.; David, L.; Kirzhner, V.M.; Burke, T.; Barre-Dirie, A.; Crooijmans, R.P.M.A.; Elo, K.; et al. Biodiversity of 52 Chicken Populations Assessed by Microsatellite Typing of DNA Pools. Genet. Sel. Evol. 2003, 35, 533–557. [Google Scholar] [CrossRef] [PubMed]
- Ceccobelli, S.; Di Lorenzo, P.; Lancioni, H.; Monteagudo Ibáñez, L.V.; Tejedor, M.T.; Castellini, C.; Landi, V.; Martínez Martínez, A.; Delgado Bermejo, J.V.; Vega Pla, J.L.; et al. Genetic Diversity and Phylogeographic Structure of Sixteen Mediterranean Chicken Breeds Assessed with Microsatellites and Mitochondrial DNA. Livest. Sci. 2015, 175, 27–36. [Google Scholar] [CrossRef]
- Rashid, M.A.; Manjula, P.; Faruque, S.; Fazlul Haque Bhuiyan, A.K.; Seo, D.; Alam, J.; Lee, J.H.; Bhuiyan, M.S.A. Genetic Diversity and Population Structure of Indigenous Chicken of Bangladesh Using Microsatellite Markers. Asian-Australas. J. Anim. Sci. 2020, 33, 1732–1740. [Google Scholar] [CrossRef] [PubMed]
- Seo, D.W.; Hoque, M.R.; Choi, N.R.; Sultana, H.; Park, H.B.; Heo, K.N.; Kang, B.S.; Lim, H.T.; Lee, S.H.; Jo, C.; et al. Discrimination of Korean Native Chicken Lines Using Fifteen Selected Microsatellite Markers. Asian-Australas. J. Anim. Sci. 2013, 26, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.H.; Wu, X.S.; Wang, D.Q.; Qin, J.; Wu, S.L.; Zhou, Q.L.; Xie, F.; Cheng, R.; Xu, Q.; Liu, B.; et al. Cluster Analysis of 12 Chinese Native Chicken Populations Using Microsatellite Markers. Asian-Australas. J. Anim. Sci. 2004, 17, 1047–1052. [Google Scholar] [CrossRef]
- Kaya, M.; Yıldız, M.A. Genetic Diversity among Turkish Native Chickens, Denizli and Gerze, Estimated by Microsatellite Markers. Biochem. Genet. 2008, 46, 480–491. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of Genetic Linkage Map in Man Using Restriction Fragment Length Polymorphism. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- Vanhala, T.; Tuiskula-Haavisto, M.; Elo, K.; Vilkki, J.; Mäki-Tanila, A. Evaluation of Genetic Variability and Genetic Distances between Eight Chicken Lines Using Microsatellite Markers. Poult. Sci. 1998, 77, 783–790. [Google Scholar] [CrossRef]
- Ding, F.X.; Zhang, G.X.; Wang, J.Y.; Li, Y.; Zhang, L.J.; Wei, Y.; Wang, H.H.; Zhang, L.; Hou, Q.R. Genetic Diversity of a Chinese Native Chicken Breed, Bian Chicken, Based on Twenty-Nine Microsatellite Markers. Asian-Australas. J. Anim. Sci. 2010, 23, 154–161. [Google Scholar] [CrossRef]
- Wu, S.X.; Chen, G.H.; Wang, D.Q.; Zhang, X.Y.; Wang, K.H.; Cheng, R.; Liu, B.; Xu, Q.; Zhou, Q.L. Analysis of Genetic Relationship among Chinese Native Chicken Breeds Using Microsatellite Markers. Acta Genet. Sin. 2004, 31, 43–50. [Google Scholar] [PubMed]
- Muchadeyi, F.C.; Eding, H.; Wollny, C.B.A.; Groeneveld, E.; Makuza, S.M.; Shamseldin, R.; Simianer, H.; Weigend, S. Absence of Population Substructuring in Zimbabwe Chicken Ecotypes Inferred Using Microsatellite Analysis. Anim. Genet. 2007, 38, 332–339. [Google Scholar] [CrossRef] [PubMed]
- Cuc, N.T.K.; Simianer, H.; Eding, H.; Tieu, H.V.; Cuong, V.C.; Wollny, C.B.A.; Groeneveld, L.F.; Weigend, S. Assessing Genetic Diversity of Vietnamese Local Chicken Breeds Using Microsatellites. Anim. Genet. 2010, 41, 545–547. [Google Scholar] [CrossRef] [PubMed]


| Na | PIC | Ho | He | uHe | F | FIS | FIT | Fst | Prob | Signif (HW) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ADL0268 | 8 | 0.633 | 0.593 | 0.619 | 0.64 | 0.038 | 0.042 | 0.064 | 0.023 | 0 | *** |
| MCW0206 | 6 | 0.618 | 0.635 | 0.603 | 0.621 | −0.053 | −0.052 | −0.027 | 0.024 | 0 | *** |
| ADL0278 | 7 | 0.7 | 0.516 | 0.676 | 0.699 | 0.225 | 0.236 | 0.262 | 0.035 | 0 | *** |
| MCW0103 | 2 | 0.297 | 0.333 | 0.296 | 0.304 | −0.133 | −0.127 | −0.123 | 0.004 | 0.305 | ns |
| MCW0037 | 3 | 0.549 | 0.336 | 0.511 | 0.53 | 0.35 | 0.341 | 0.387 | 0.069 | 0.009 | ** |
| MCW0183 | 11 | 0.602 | 0.516 | 0.594 | 0.611 | 0.125 | 0.132 | 0.144 | 0.013 | 0 | *** |
| MCW0069 | 6 | 0.562 | 0.449 | 0.554 | 0.571 | 0.178 | 0.189 | 0.201 | 0.014 | 0 | *** |
| MCW0081 | 8 | 0.578 | 0.595 | 0.569 | 0.588 | −0.052 | −0.046 | −0.031 | 0.015 | 0 | *** |
| MCW0222 | 5 | 0.553 | 0.172 | 0.531 | 0.552 | 0.679 | 0.676 | 0.689 | 0.04 | 0 | *** |
| MCW0216 | 4 | 0.605 | 0.276 | 0.533 | 0.552 | 0.467 | 0.481 | 0.544 | 0.12 | 0 | *** |
| MCW0098 | 2 | 0.391 | 0.083 | 0.368 | 0.383 | 0.831 | 0.774 | 0.787 | 0.057 | 0 | *** |
| MCW0111 | 6 | 0.734 | 0.37 | 0.691 | 0.717 | 0.469 | 0.465 | 0.496 | 0.059 | 0 | *** |
| MCW0330 | 7 | 0.541 | 0.22 | 0.535 | 0.558 | 0.595 | 0.59 | 0.594 | 0.01 | 0 | *** |
| MCW0067 | 4 | 0.31 | 0.343 | 0.302 | 0.314 | −0.115 | −0.136 | −0.106 | 0.026 | 0.664 | ns |
| MCW0295 | 7 | 0.548 | 0.589 | 0.529 | 0.546 | −0.116 | −0.113 | −0.075 | 0.034 | 0.996 | ns |
| MCW0248 | 3 | 0.103 | 0.078 | 0.101 | 0.104 | 0.125 | 0.222 | 0.237 | 0.019 | 0.116 | ns |
| ADL0112 | 6 | 0.637 | 0.467 | 0.615 | 0.635 | 0.25 | 0.241 | 0.267 | 0.035 | 0 | *** |
| MCW0034 | 10 | 0.512 | 0.319 | 0.496 | 0.512 | 0.34 | 0.356 | 0.376 | 0.03 | 0 | *** |
| MCW0078 | 4 | 0.588 | 0.348 | 0.572 | 0.59 | 0.396 | 0.391 | 0.408 | 0.028 | 0 | *** |
| LEI0192 | 18 | 0.773 | 0.583 | 0.727 | 0.75 | 0.189 | 0.198 | 0.245 | 0.059 | 0 | *** |
| Mean | 6.35 | 0.541 | 0.391 | 0.521 | 0.539 | 0.239 | 0.243 | 0.267 | 0.036 |
| Population | Ne | I | Ho | He | uHe | |
|---|---|---|---|---|---|---|
| Centre-East(Konde) | Mean | 2.246 | 0.938 | 0.45 | 0.518 | 0.533 |
| SE | 0.13 | 0.069 | 0.041 | 0.036 | 0.037 | |
| Centre-North | Mean | 2.356 | 1.008 | 0.367 | 0.53 | 0.55 |
| SE | 0.165 | 0.086 | 0.042 | 0.037 | 0.038 | |
| Sahel | Mean | 2.161 | 0.894 | 0.364 | 0.489 | 0.506 |
| SE | 0.183 | 0.083 | 0.047 | 0.035 | 0.036 | |
| South-West | Mean | 2.454 | 1.054 | 0.384 | 0.547 | 0.567 |
| SE | 0.173 | 0.086 | 0.047 | 0.036 | 0.037 | |
| Total | Mean | 2.304 | 0.973 | 0.391 | 0.521 | 0.539 |
| SE | 0.081 | 0.04 | 0.022 | 0.018 | 0.018 |
| Centre-East | Centre-North | Sahel | South-West | |
|---|---|---|---|---|
| Centre-East | 0.000 | 0.032 | 0.020 | 0.026 |
| Centre-North | 0.026 ** | 0.000 | 0.002 | 0.020 |
| Sahel | 0.018 * | 0.003 ns | 0.000 | 0.021 |
| South-West | 0.021 ** | 0.016 ** | 0.020 ** | 0.000 |
| Source | Degree of Freedom | Sum Square | Mean Square | Estimated Variances | % of Estimated Variances | F-Statistics Value | p (Rand ≥ Data) |
|---|---|---|---|---|---|---|---|
| Among ecotype | 3 | 26.39 | 8.80 | 0.06 | 1 | FST = 0.011 | 0.005 |
| Among populations within ecotype | 67 | 445.82 | 6.65 | 1.23 | 22 | FIT = 0.227 | 0.000 |
| Within Individual | 71 | 297.67 | 4.19 | 4.19 | 77 | FIS = 0.235 | 0.000 |
| Total | 141 | 769.88 | 5.48 | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yacouba, Z.; Isidore, H.; Michel, K.; Isidore, G.B.; Boureima, T.; Vinsoun, M.; Maurice, K.; Ousseni, B.; Moussa, Z.; Valerie, B.-Y.M.C.; et al. Genetic Diversity and Population Structure of Local Chicken Ecotypes in Burkina Faso Using Microsatellite Markers. Genes 2022, 13, 1523. https://doi.org/10.3390/genes13091523
Yacouba Z, Isidore H, Michel K, Isidore GB, Boureima T, Vinsoun M, Maurice K, Ousseni B, Moussa Z, Valerie B-YMC, et al. Genetic Diversity and Population Structure of Local Chicken Ecotypes in Burkina Faso Using Microsatellite Markers. Genes. 2022; 13(9):1523. https://doi.org/10.3390/genes13091523
Chicago/Turabian StyleYacouba, Zare, Houaga Isidore, Kere Michel, Gnanda B. Isidore, Traore Boureima, Millogo Vinsoun, Konkobo Maurice, Bandaogo Ousseni, Zongo Moussa, Bougouma-Yameogo M. C. Valerie, and et al. 2022. "Genetic Diversity and Population Structure of Local Chicken Ecotypes in Burkina Faso Using Microsatellite Markers" Genes 13, no. 9: 1523. https://doi.org/10.3390/genes13091523
APA StyleYacouba, Z., Isidore, H., Michel, K., Isidore, G. B., Boureima, T., Vinsoun, M., Maurice, K., Ousseni, B., Moussa, Z., Valerie, B.-Y. M. C., Romdhane, R., & Joseph, N. A. (2022). Genetic Diversity and Population Structure of Local Chicken Ecotypes in Burkina Faso Using Microsatellite Markers. Genes, 13(9), 1523. https://doi.org/10.3390/genes13091523






