Alteration of E2F2 Expression in Governing Endothelial Cell Senescence
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Mice
2.3. Quantitative Real-Time PCR (qPCR)
2.4. Transfection of siRNA and Adenovirus DNA
2.5. SA-β-gal Staining
2.6. Immunofluorescent Staining
2.7. Protein Extraction and Western Blotting
2.8. Cell Cycle Analysis
2.9. EdU Cell Proliferation Assay
2.10. ELISA
2.11. RNA-Sequencing Analysis
2.12. Statistical Analysis
3. Results
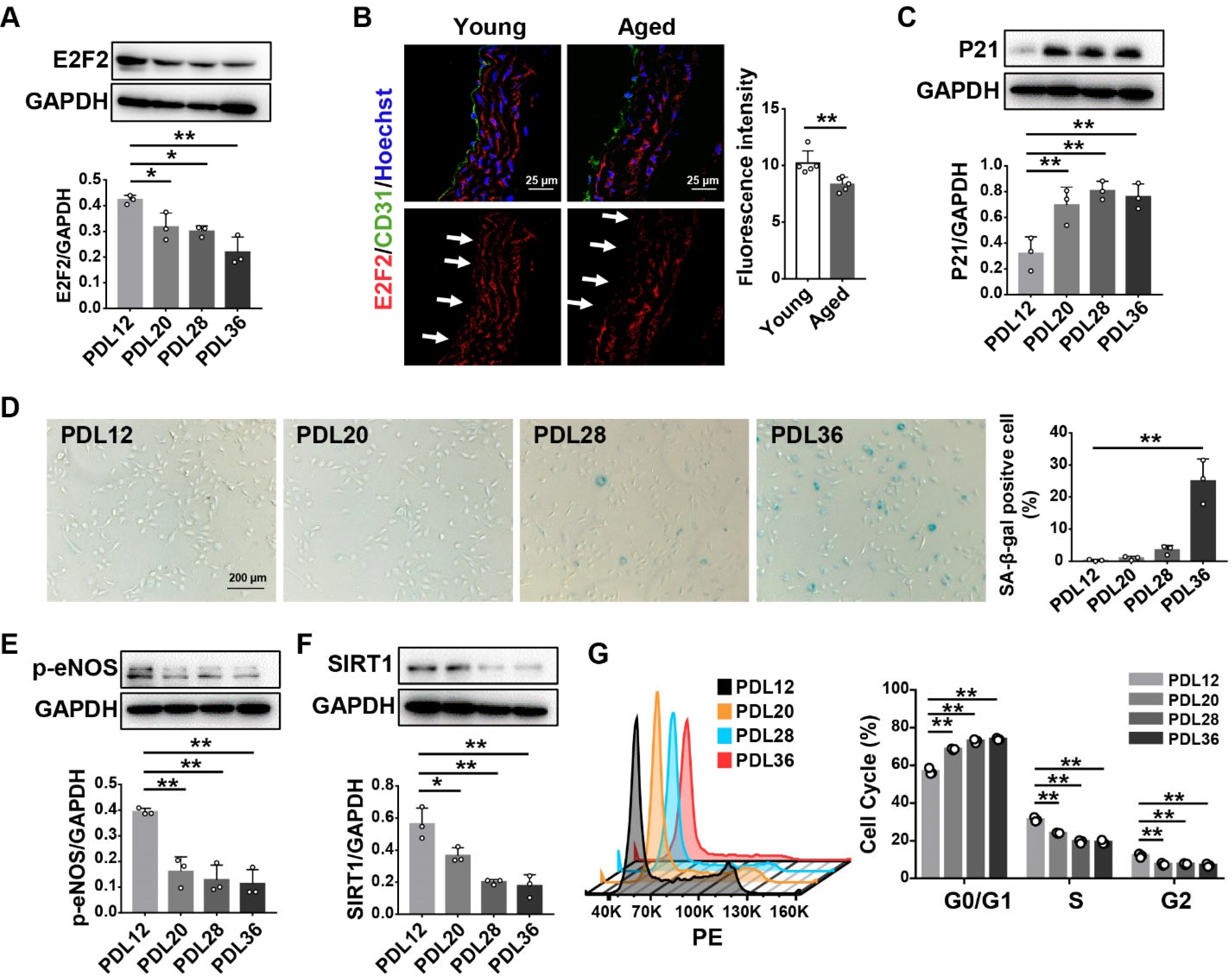
3.1. E2F2 Expression Is Decreased in the Senescent Endothelial Cells and the Aorta of Aged Mice
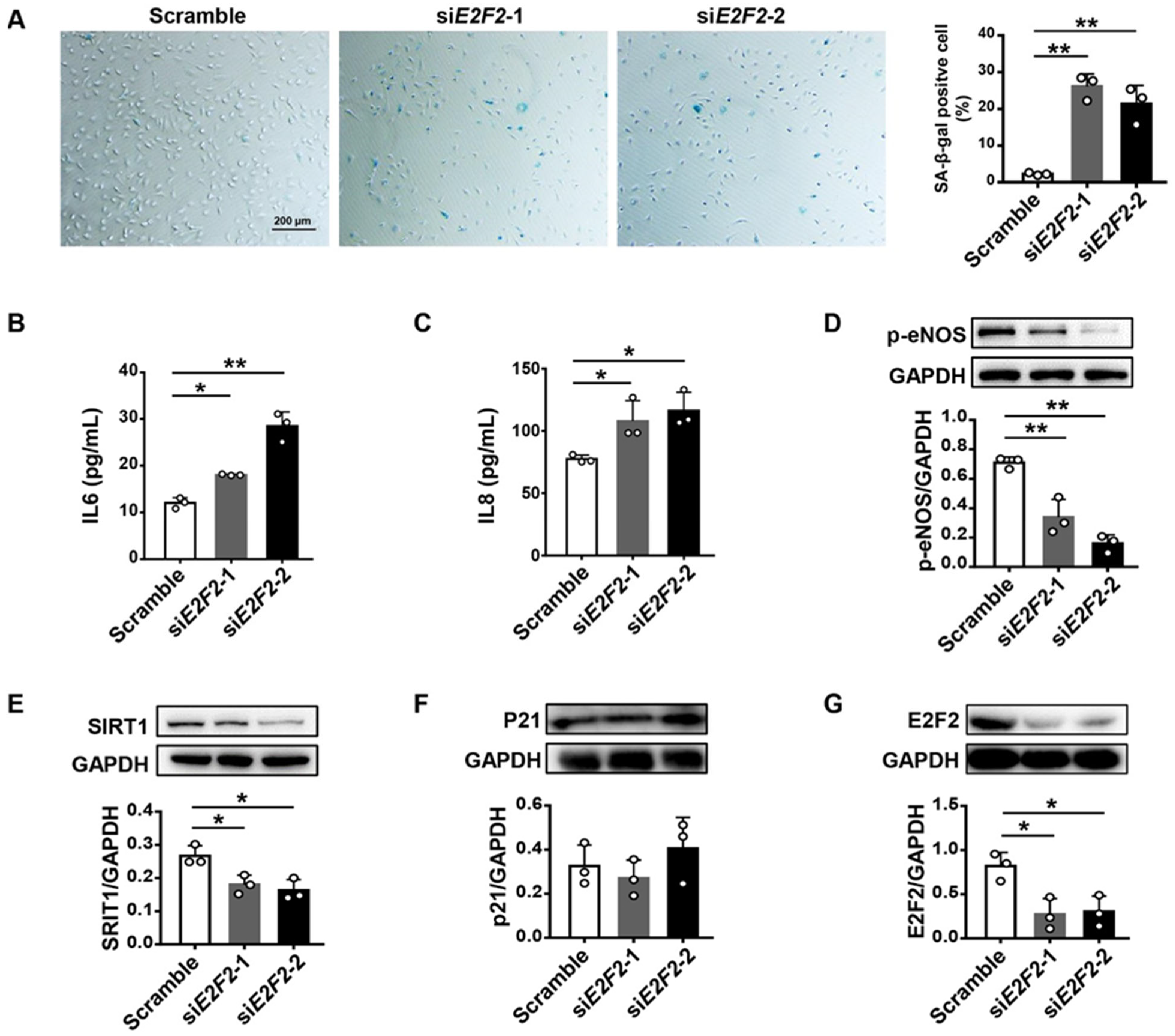
3.2. E2F2-Knockdown Induces Endothelial Cell Senescence
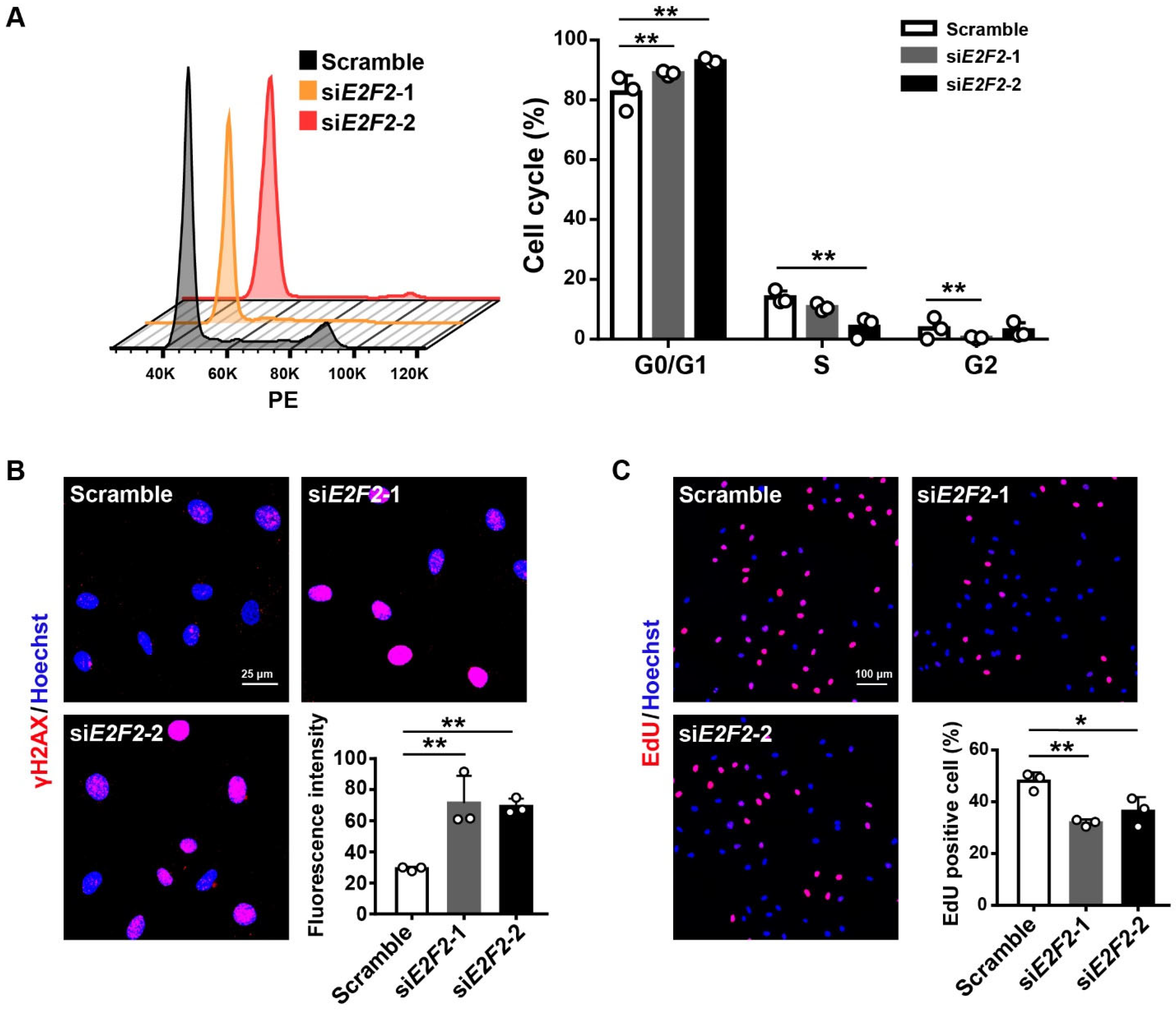
3.3. E2F2-Knockdown Impairs Endothelial Cellular Function
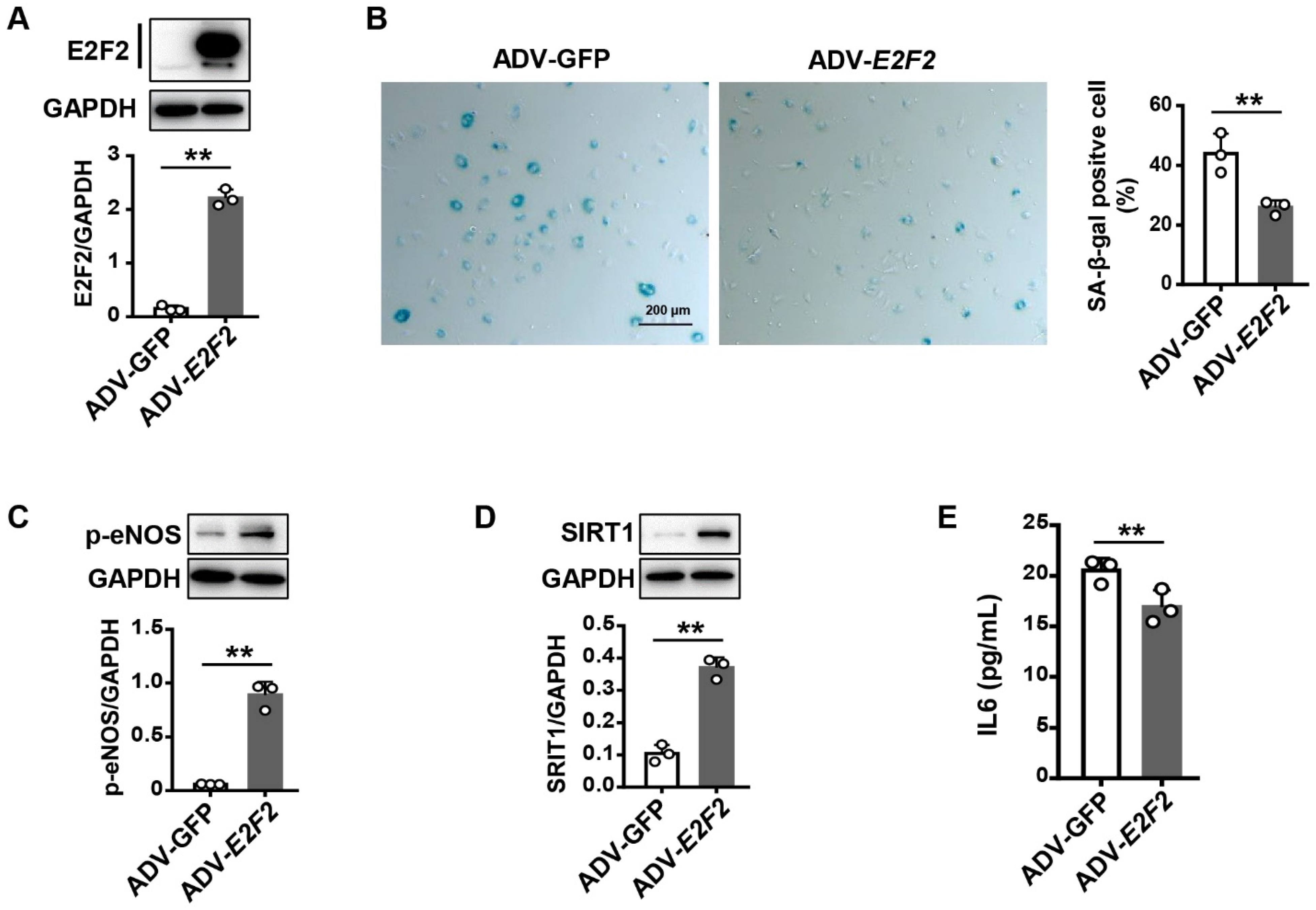
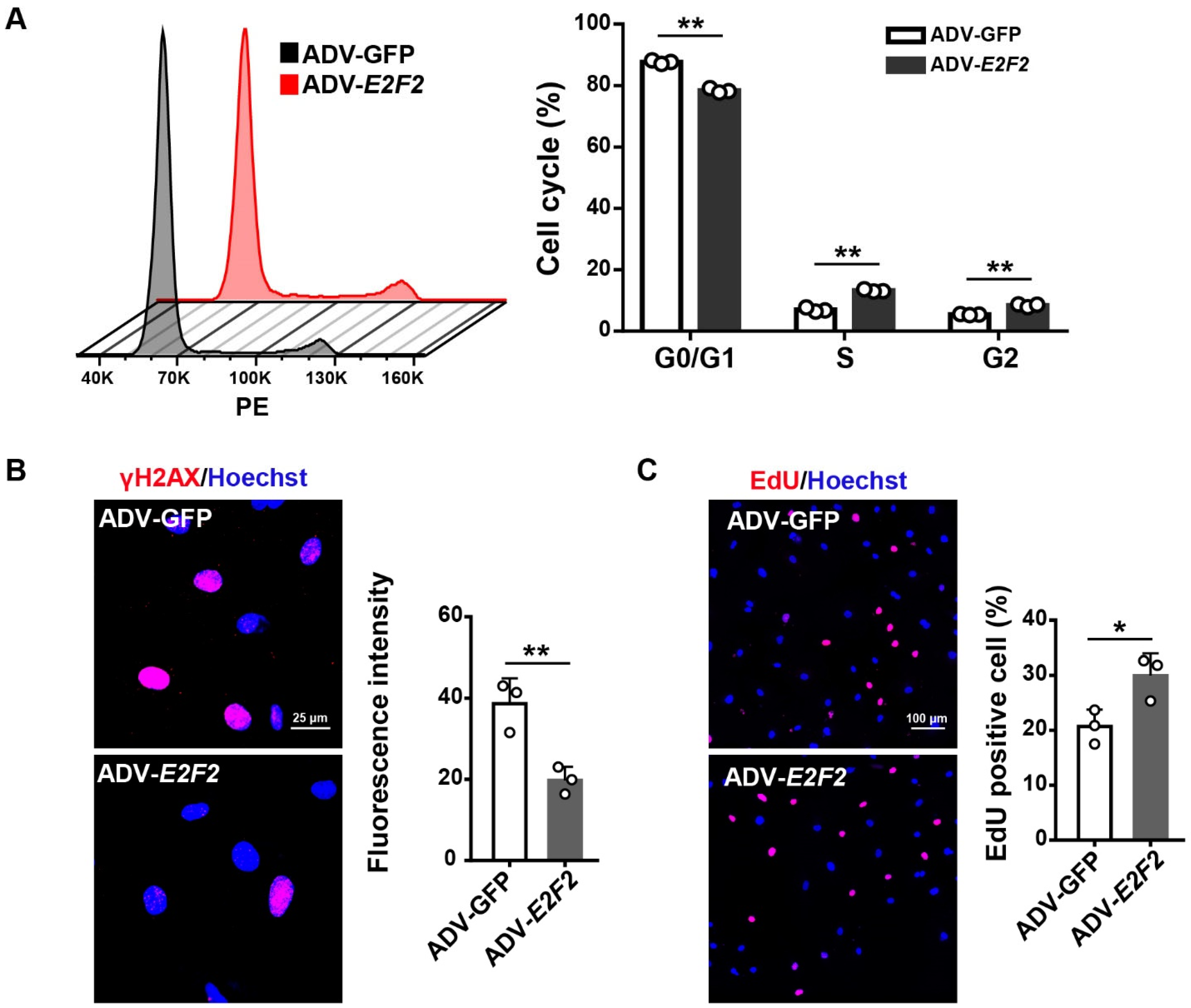
3.4. Overexpression of E2F2 Attenuates the Senescence Phenotype of Endothelial Cells
3.5. E2F2 Overexpression Reverses Endothelial Dysfunction in Senescent Endothelial Cells
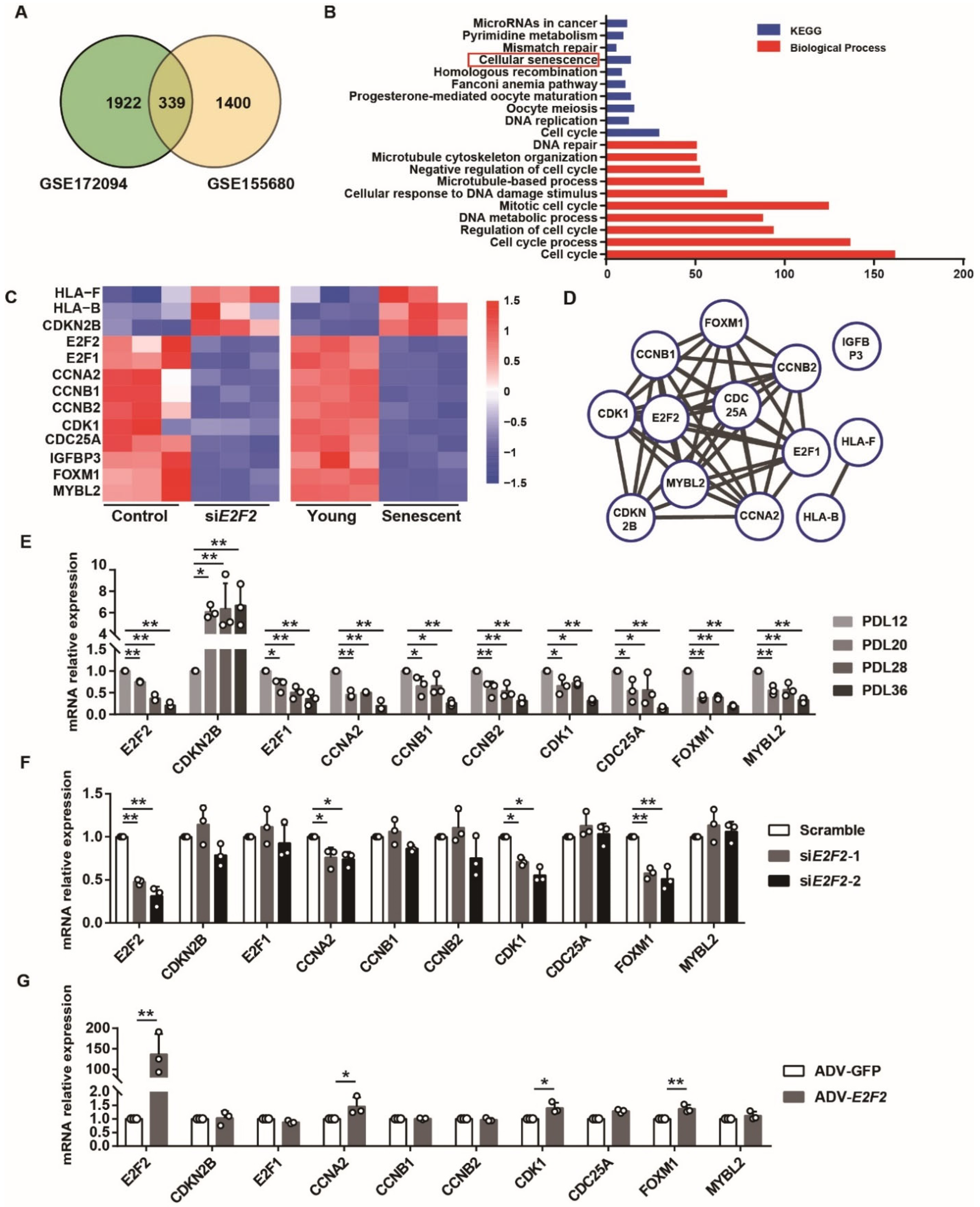
3.6. Differentially Expressed Genes (DEGs) Are Identified in E2F2-knockdown and Senescent Endothelial Cells
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kruger-Genge, A.; Blocki, A.; Franke, R.P.; Jung, F. Vascular Endothelial Cell Biology: An Update. Int. J. Mol. Sci. 2019, 20, 4411. [Google Scholar] [CrossRef] [PubMed]
- Carracedo, J.; Ramírez-Carracedo, R.; Alique, M.; Ramírez-Chamond, R. Endothelial Cell Senescence in the Pathogenesis of Endothelial Dysfunction. In Endothelial Dysfunction—Old Concepts and New Challenges; IntechOpen: London, UK, 2018; pp. 49–73. [Google Scholar] [CrossRef]
- Higashi, Y.; Kihara, Y.; Noma, K. Endothelial dysfunction and hypertension in aging. Hypertens. Res. 2012, 35, 1039–1047. [Google Scholar] [CrossRef]
- Lopez-Otin, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed]
- Durik, M.; Kavousi, M.; van der Pluijm, I.; Isaacs, A.; Cheng, C.; Verdonk, K.; Loot, A.E.; Oeseburg, H.; Bhaggoe, U.M.; Leijten, F.; et al. Nucleotide excision DNA repair is associated with age-related vascular dysfunction. Circulation 2012, 126, 468–478. [Google Scholar] [CrossRef]
- Kennedy, B.K.; Berger, S.L.; Brunet, A.; Campisi, J.; Cuervo, A.M.; Epel, E.S.; Franceschi, C.; Lithgow, G.J.; Morimoto, R.I.; Pessin, J.E.; et al. Geroscience: Linking aging to chronic disease. Cell 2014, 159, 709–713. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.L.; Li, Y. Endothelial cell senescence and age-related vascular diseases. J. Genet. Genomics 2014, 41, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Harvey, A.; Montezano, A.C.; Touyz, R.M. Vascular biology of ageing-Implications in hypertension. J. Mol. Cell Cardiol. 2015, 83, 112–121. [Google Scholar] [CrossRef]
- Bochenek, M.L.; Schutz, E.; Schafer, K. Endothelial cell senescence and thrombosis: Ageing clots. Thromb. Res. 2016, 147, 36–45. [Google Scholar] [CrossRef]
- Buford, T.W. Hypertension and aging. Ageing Res. Rev. 2016, 26, 96–111. [Google Scholar] [CrossRef] [Green Version]
- Hammond, G.; Rich, M.W. Hypertensive Heart Failure in the Very Old. Heart Fail. Clin. 2019, 15, 477–485. [Google Scholar] [CrossRef]
- Ota, H.; Eto, M.; Ogawa, S.; Iijima, K.; Akishita, M.; Ouchi, Y. SIRT1/eNOS axis as a potential target against vascular senescence, dysfunction and atherosclerosis. J. Atheroscler. Thromb. 2010, 17, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Matjusaitis, M.; Chin, G.; Sarnoski, E.A.; Stolzing, A. Biomarkers to identify and isolate senescent cells. Ageing Res. Rev. 2016, 29, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ivey-Hoyle, M.; Conroy, R.; Huber, H.E.; Goodhart, P.J.; Oliff, A.; Heimbrook, D.C. Cloning and characterization of E2F-2, a novel protein with the biochemical properties of transcription factor E2F. Mol. Cell Biol. 1993, 13, 7802–7812. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Timmers, C.; Maiti, B.; Saavedra, H.I.; Sang, L.; Chong, G.T.; Nuckolls, F.; Giangrande, P.; Wright, F.A.; Field, S.J.; et al. The E2F1-3 transcription factors are essential for cellular proliferation. Nature 2001, 414, 457–462. [Google Scholar] [CrossRef]
- Gołąbek, K.; Biernacki, K.; Gaździcka, J.; Strzelczyk, J.K.; Miśkiewicz-Orczyk, K.; Krakowczyk, Ł.; Zięba, N.; Kiczmer, P.; Ostrowska, Z.; Misiołek, M. Selected E2F2 Polymorphisms in Oral and Oropharyngeal Squamous Cell Carcinoma. Biomed. Res. Int. 2021, 2021, 8098130. [Google Scholar] [CrossRef]
- Azkargorta, M.; Fullaondo, A.; Laresgoiti, U.; Aloria, K.; Infante, A.; Arizmendi, J.M.; Zubiaga, A.M. Differential proteomics analysis reveals a role for E2F2 in the regulation of the Ahr pathway in T lymphocytes. Mol. Cell. Proteom. MCP 2010, 9, 2184–2194. [Google Scholar] [CrossRef]
- Delgado, I.; Fresnedo, O.; Iglesias, A.; Rueda, Y.; Syn, W.K.; Zubiaga, A.M.; Ochoa, B. A role for transcription factor E2F2 in hepatocyte proliferation and timely liver regeneration. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 301, G20–G31. [Google Scholar] [CrossRef]
- Ebelt, H.; Zhang, Y.; Kampke, A.; Xu, J.; Schlitt, A.; Buerke, M.; Müller-Werdan, U.; Werdan, K.; Braun, T. E2F2 expression induces proliferation of terminally differentiated cardiomyocytes in vivo. Cardiovasc. Res. 2008, 80, 219–226. [Google Scholar] [CrossRef]
- Infante, A.; Laresgoiti, U.; Fernández-Rueda, J.; Fullaondo, A.; Galán, J.; Díaz-Uriarte, R.; Malumbres, M.; Field, S.J.; Zubiaga, A.M. E2F2 represses cell cycle regulators to maintain quiescence. Cell Cycle 2008, 7, 3915–3927. [Google Scholar] [CrossRef] [Green Version]
- Bao, X.; Hu, X. Up-regulated expression of E2F2 is necessary for p16INK4a-induced cartilage injury. BMC Musculoskelet. Disord. 2018, 19, 334. [Google Scholar] [CrossRef]
- Pusapati, R.V.; Weaks, R.L.; Rounbehler, R.J.; McArthur, M.J.; Johnson, D.G. E2F2 suppresses Myc-induced proliferation and tumorigenesis. Mol. Carcinog. 2010, 49, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Yuasa, K.; Suzue, K.; Nagahama, M.; Matsuda, Y.; Tsuji, A. Transcriptional regulation of subtilisin-like proprotein convertase PACE4 by E2F: Possible role of E2F-mediated upregulation of PACE4 in tumor progression. Gene 2007, 402, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Zhan, W.; Chen, L.; Liu, H.; Long, C.; Liu, J.; Ding, S.; Wu, Q.; Chen, S. Pcsk6 Deficiency Promotes Cardiomyocyte Senescence by Modulating Ddit3-Mediated ER Stress. Genes 2022, 13, 711. [Google Scholar] [CrossRef]
- Zhou, J.; Zhu, Y.; Cheng, M.; Dinesh, D.; Thorne, T.; Poh, K.K.; Liu, D.; Botros, C.; Tang, Y.L.; Reisdorph, N.; et al. Regulation of vascular contractility and blood pressure by the E2F2 transcription factor. Circulation 2009, 120, 1213–1221. [Google Scholar] [CrossRef]
- Li, R.; Mi, X.; Yang, S.; Yang, Y.; Zhang, S.; Hui, R.; Chen, Y.; Zhang, W. Long-term stimulation of angiotensin II induced endothelial senescence and dysfunction. Exp. Gerontol. 2019, 119, 212–220. [Google Scholar] [CrossRef]
- Boisen, L.; Drasbek, K.R.; Pedersen, A.S.; Kristensen, P. Evaluation of endothelial cell culture as a model system of vascular ageing. Exp. Gerontol. 2010, 45, 779–787. [Google Scholar] [CrossRef]
- Dimri, G.P.; Lee, X.; Basile, G.; Acosta, M.; Scott, G.; Roskelley, C.; Medrano, E.E.; Linskens, M.; Rubelj, I.; Pereira-Smith, O.; et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 1995, 92, 9363–9367. [Google Scholar] [CrossRef]
- Mah, L.J.; El-Osta, A.; Karagiannis, T.C. gammaH2AX: A sensitive molecular marker of DNA damage and repair. Leukemia 2010, 24, 679–686. [Google Scholar] [CrossRef]
- Foreman, K.E.; Tang, J. Molecular mechanisms of replicative senescence in endothelial cells. Exp. Gerontol. 2003, 38, 1251–1257. [Google Scholar] [CrossRef] [PubMed]
- Ogrodnik, M. Cellular aging beyond cellular senescence: Markers of senescence prior to cell cycle arrest in vitro and in vivo. Aging Cell 2021, 20, e13338. [Google Scholar] [CrossRef]
- Zampino, M.; Ferrucci, L.; Semba, R.D. Biomarkers in the path from cellular senescence to frailty. Exp. Gerontol. 2020, 129, 110750. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R.; Jat, P. Mechanisms of Cellular Senescence: Cell Cycle Arrest and Senescence Associated Secretory Phenotype. Front. Cell Dev. Biol. 2021, 9, 645593. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, S.; Zhang, Y.; Pan, J. The E2F transcription factor 2: What do we know? Biosci. Trends 2021, 15, 83–92. [Google Scholar] [CrossRef]
- Wagner, M.; Hampel, B.; Bernhard, D.; Hala, M.; Zwerschke, W.; Jansen-Dürr, P. Replicative senescence of human endothelial cells in vitro involves G1 arrest, polyploidization and senescence-associated apoptosis. Exp. Gerontol. 2001, 36, 1327–1347. [Google Scholar] [CrossRef]
- Medcalf, A.S.; Klein-Szanto, A.J.; Cristofalo, V.J. Expression of p21 is not required for senescence of human fibroblasts. Cancer Res. 1996, 56, 4582–4585. [Google Scholar]
- Dulić, V.; Beney, G.E.; Frebourg, G.; Drullinger, L.F.; Stein, G.H. Uncoupling between phenotypic senescence and cell cycle arrest in aging p21-deficient fibroblasts. Mol. Cell Biol. 2000, 20, 6741–6754. [Google Scholar] [CrossRef]
- Zhu, L.; Sun, C.; Ren, J.; Wang, G.; Ma, R.; Sun, L.; Yang, D.; Gao, S.; Ning, K.; Wang, Z.; et al. Stress-induced precocious aging in PD-patient iPSC-derived NSCs may underlie the pathophysiology of Parkinson’s disease. Cell Death Dis. 2019, 10, 105. [Google Scholar] [CrossRef]
- Pham, T.H.; Park, H.M.; Kim, J.; Hong, J.T.; Yoon, D.Y. Interleukin-32θ Triggers Cellular Senescence and Reduces Sensitivity to Doxorubicin-Mediated Cytotoxicity in MDA-MB-231 Cells. Int. J. Mol. Sci. 2021, 22, 4974. [Google Scholar] [CrossRef]
- Baroni, M.; Yi, C.; Choudhary, S.; Lei, X.; Kosti, A.; Grieshober, D.; Velasco, M.; Qiao, M.; Burns, S.S.; Araujo, P.R.; et al. Musashi1 Contribution to Glioblastoma Development via Regulation of a Network of DNA Replication, Cell Cycle and Division Genes. Cancers 2021, 13, 1494. [Google Scholar] [CrossRef]
- Zhang, R.; Chen, H.Z.; Liu, D.P. The Four Layers of Aging. Cell Syst. 2015, 1, 180–186. [Google Scholar] [CrossRef] [Green Version]
- Birch, J.; Gil, J. Senescence and the SASP: Many therapeutic avenues. Genes Dev. 2020, 34, 1565–1576. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Bloom, S.I.; Donato, A.J. The role of senescence, telomere dysfunction and shelterin in vascular aging. Microcirculation 2019, 26, e12487. [Google Scholar] [CrossRef] [PubMed]
- Rusanova, I.; Diaz-Casado, M.E.; Fernandez-Ortiz, M.; Aranda-Martinez, P.; Guerra-Librero, A.; Garcia-Garcia, F.J.; Escames, G.; Manas, L.; Acuna-Castroviejo, D. Analysis of Plasma MicroRNAs as Predictors and Biomarkers of Aging and Frailty in Humans. Oxid. Med. Cell Longev. 2018, 2018, 7671850. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Ishida, M.; Tashiro, S.; Takeishi, Y. DNA Damage and Senescence-Associated Inflammation in Cardiovascular Disease. Biol. Pharm. Bull. 2019, 42, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Bautista-Niño, P.K.; Portilla-Fernandez, E.; Vaughan, D.E.; Danser, A.H.; Roks, A.J. DNA Damage: A Main Determinant of Vascular Aging. Int. J. Mol. Sci. 2016, 17, 748. [Google Scholar] [CrossRef]
- Hernandez-Segura, A.; Nehme, J.; Demaria, M. Hallmarks of Cellular Senescence. Trends Cell Biol. 2018, 28, 436–453. [Google Scholar] [CrossRef]
- Castillo, D.S.; Campalans, A.; Belluscio, L.M.; Carcagno, A.L.; Radicella, J.P.; Cánepa, E.T.; Pregi, N. E2F1 and E2F2 induction in response to DNA damage preserves genomic stability in neuronal cells. Cell Cycle 2015, 14, 1300–1314. [Google Scholar] [CrossRef]
- Rennhack, J.P.; Andrechek, E.R. Low E2F2 activity is associated with high genomic instability and PARPi resistance. Sci. Rep. 2020, 10, 17948. [Google Scholar] [CrossRef]
- Shelton, D.N.; Chang, E.; Whittier, P.S.; Choi, D.; Funk, W.D. Microarray analysis of replicative senescence. Curr. Biol. 1999, 9, 939–945. [Google Scholar] [CrossRef]
- Quadri, R.A.; Arbogast, A.; Phelouzat, M.A.; Boutet, S.; Plastre, O.; Proust, J.J. Age-associated decline in cdk1 activity delays cell cycle progression of human T lymphocytes. J. Immunol. 1998, 161, 5203–5209. [Google Scholar]
- Kümper, S.; Mardakheh, F.K.; McCarthy, A.; Yeo, M.; Stamp, G.W.; Paul, A.; Worboys, J.; Sadok, A.; Jørgensen, C.; Guichard, S.; et al. Rho-associated kinase (ROCK) function is essential for cell cycle progression, senescence and tumorigenesis. eLife 2016, 5, e12994. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, R.; Macedo, J.C.; Costa, M.; Ustiyan, V.; Shindyapina, A.V.; Tyshkovskiy, A.; Gomes, R.N.; Castro, J.P.; Kalin, T.V.; Vasques-Nóvoa, F.; et al. In vivo cyclic induction of the FOXM1 transcription factor delays natural and progeroid aging phenotypes and extends healthspan. Nat. Aging 2022, 2, 397–411. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Chen, L.; Xiao, W.; Liu, J.; Long, C.; Zhan, W.; Cui, C.; Yang, L.; Chen, S. Alteration of E2F2 Expression in Governing Endothelial Cell Senescence. Genes 2022, 13, 1522. https://doi.org/10.3390/genes13091522
Liu H, Chen L, Xiao W, Liu J, Long C, Zhan W, Cui C, Yang L, Chen S. Alteration of E2F2 Expression in Governing Endothelial Cell Senescence. Genes. 2022; 13(9):1522. https://doi.org/10.3390/genes13091522
Chicago/Turabian StyleLiu, Hongfei, Liping Chen, Wanli Xiao, Jiankun Liu, Changkun Long, Wenxing Zhan, Cui Cui, Lin Yang, and Shenghan Chen. 2022. "Alteration of E2F2 Expression in Governing Endothelial Cell Senescence" Genes 13, no. 9: 1522. https://doi.org/10.3390/genes13091522
APA StyleLiu, H., Chen, L., Xiao, W., Liu, J., Long, C., Zhan, W., Cui, C., Yang, L., & Chen, S. (2022). Alteration of E2F2 Expression in Governing Endothelial Cell Senescence. Genes, 13(9), 1522. https://doi.org/10.3390/genes13091522





