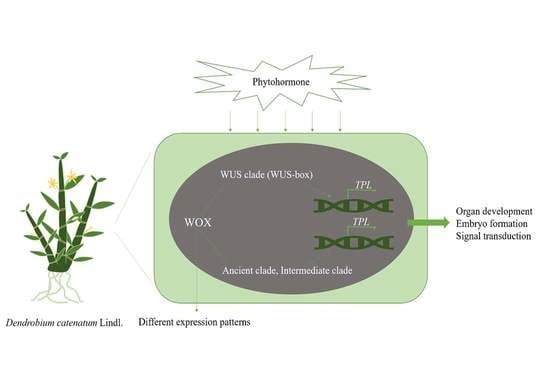
Genome-Wide Analysis of the WOX Transcription Factor Genes in Dendrobium catenatum Lindl.
Abstract
:1. Introduction
2. Materials and Methods
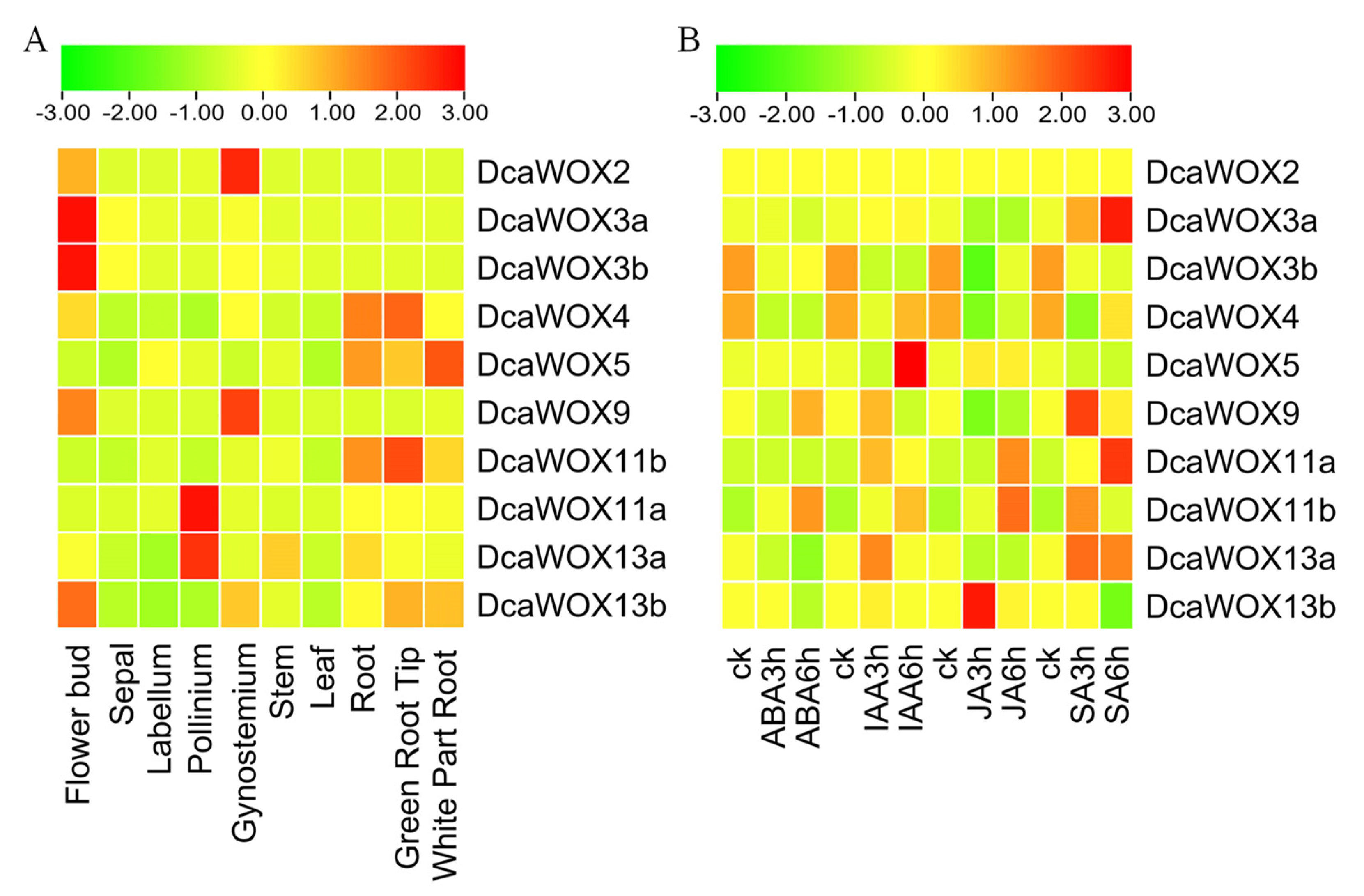
2.1. Plant Materials and Phytohormone Treatments
2.2. Identification of WOX Family Genes in D. catenatum
2.3. Sequence Characterization and Phylogenetic Analysis
2.4. Gene Structure and Conserved Motif Analyses
2.5. Subcellular Localization
2.6. Transient Expression in N. benthamiana
2.7. Transcriptomic Analysis
3. Results
3.1. Identification of WOX Family Genes in D. catenatum
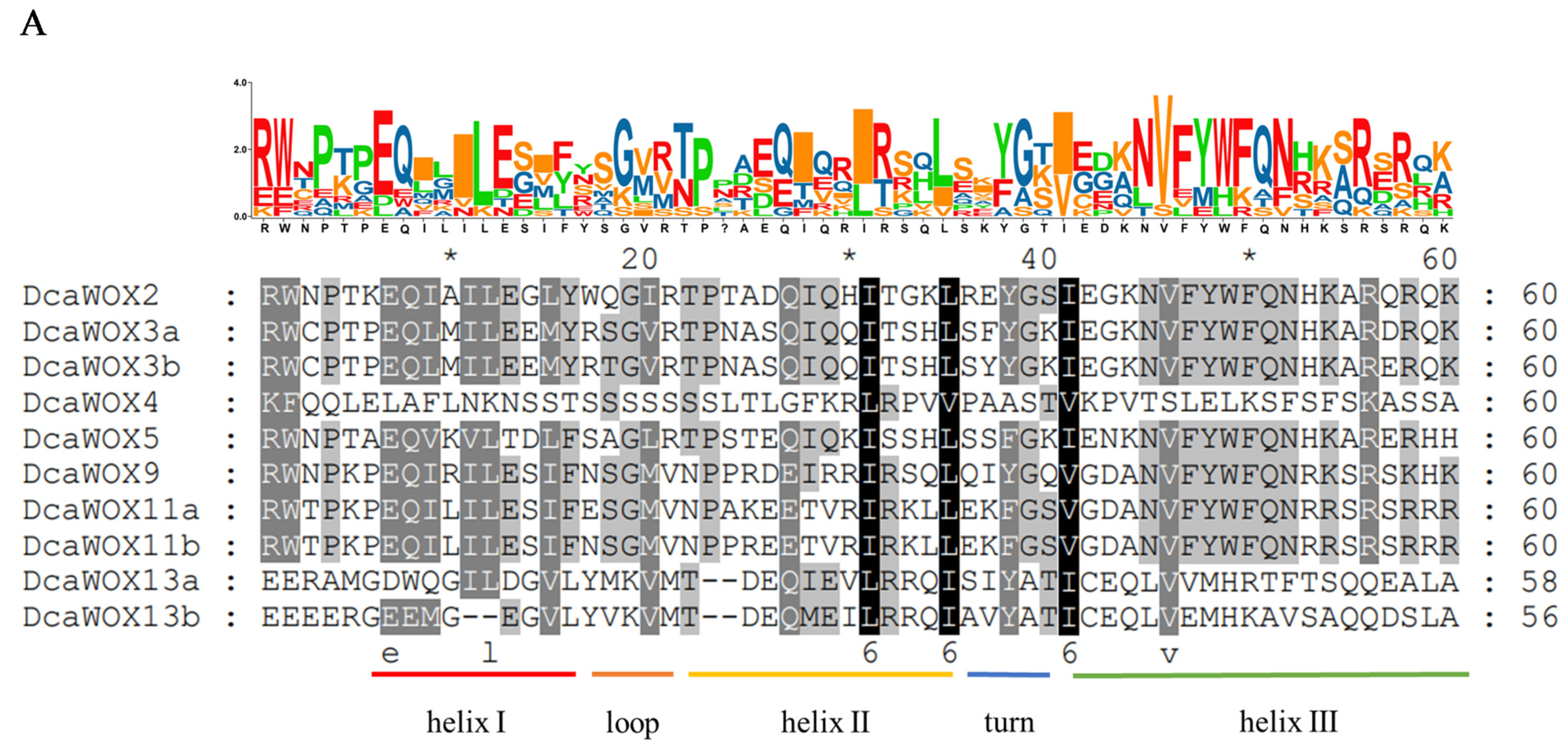
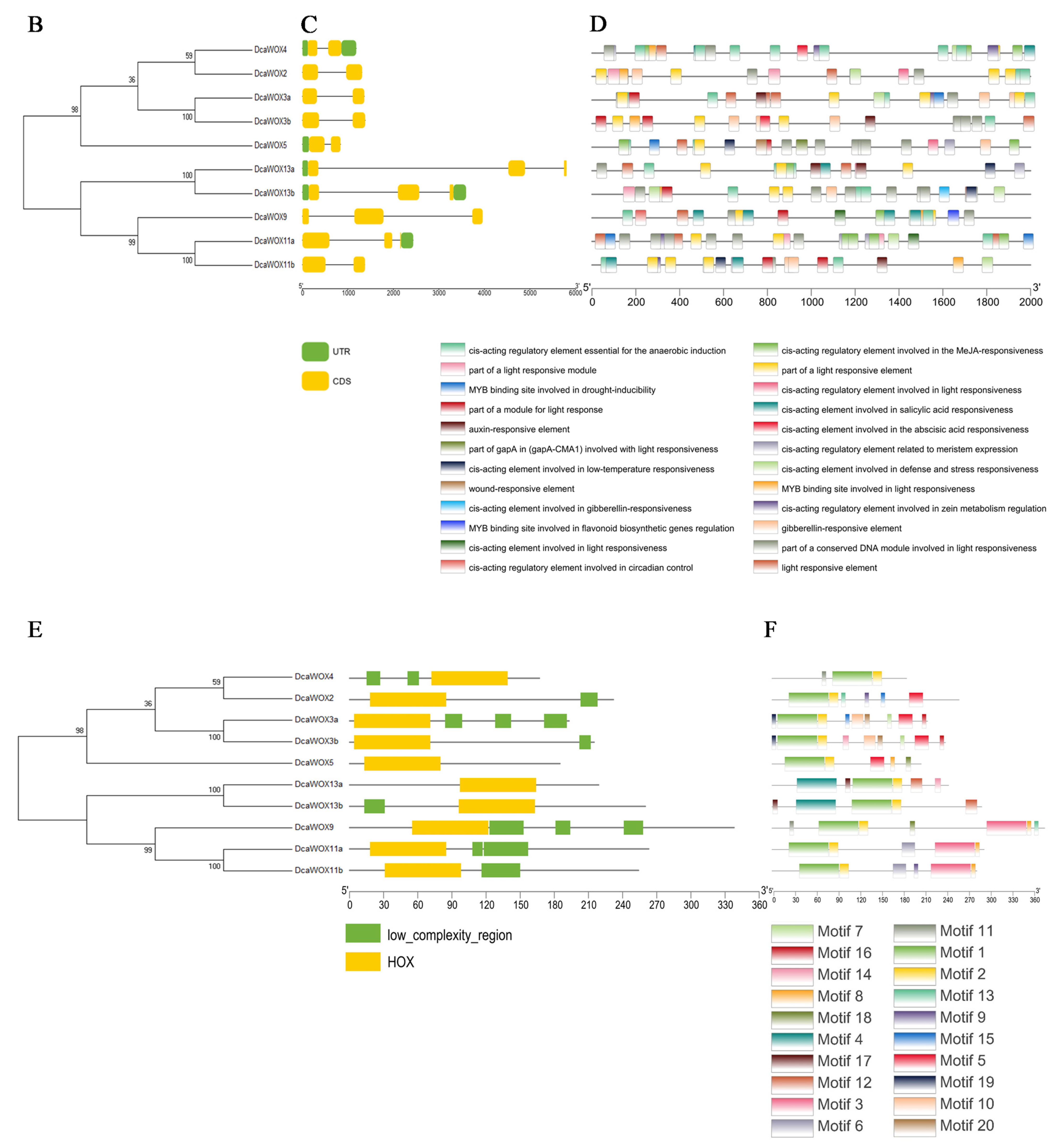
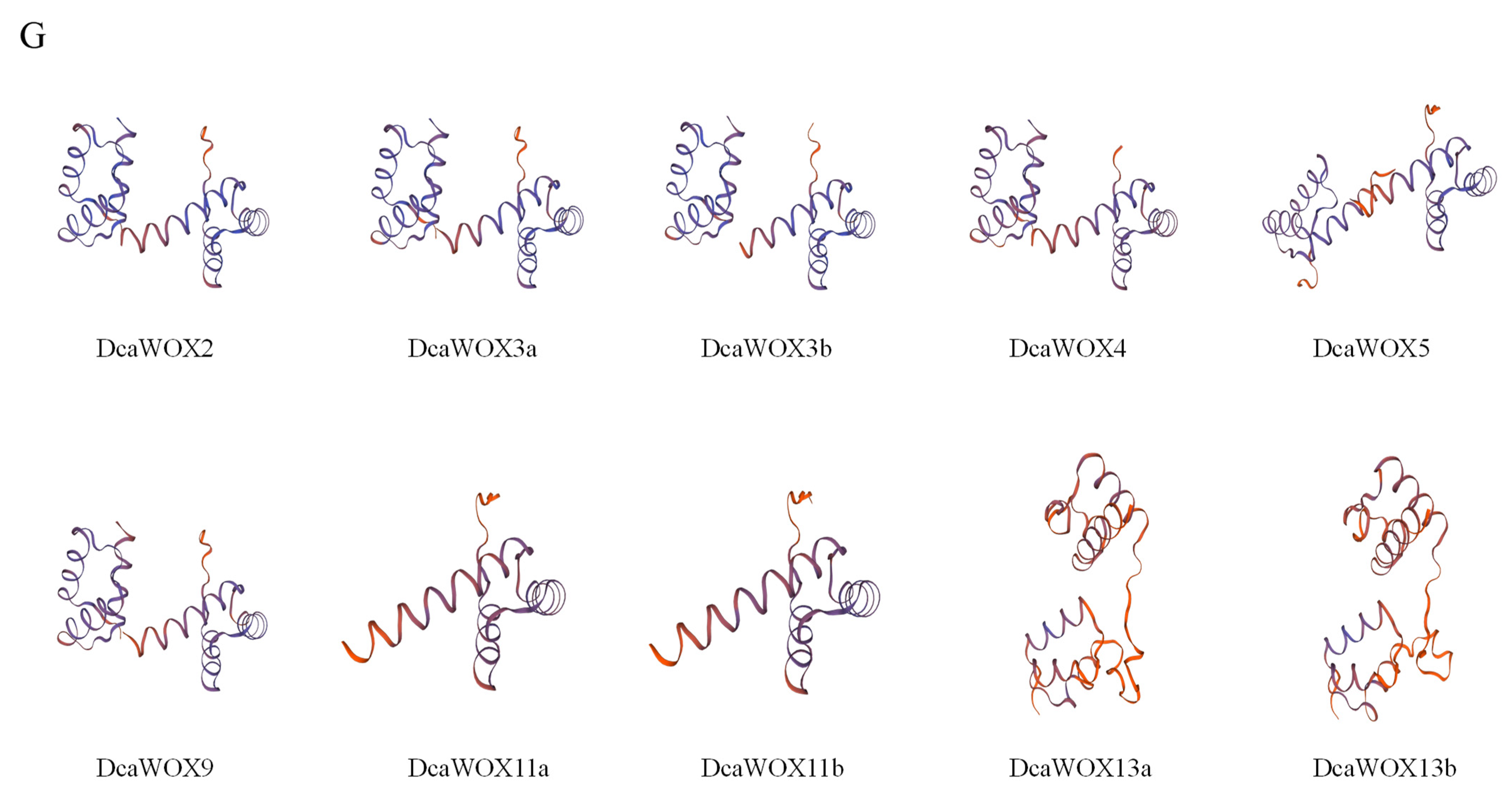
3.2. Analysis of D. catenatum WOX Phylogenetic Relationships, Genetic Structures, and Conserved Domains
3.3. Subcellular Localization
3.4. DcaWOX Regulates Plant Cell Division and Differentiation by Activating TPL Expression
3.5. WOX Gene Expression Profiles in D. catenatum
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Haecker, A.; Gross-Hardt, R.; Geiges, B.; Sarkar, A.; Breuninger, H.; Herrmann, M.; Laux, T. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 2004, 131, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Van, D.G.E.; Laux, T.; Rensing, S.A. The WUS homeobox-containing (WOX) protein family. Genome Biol. 2009, 10, 248. [Google Scholar] [CrossRef]
- Kamiya, N.; Nagasaki, H.; Morikami, A.; Sato, Y.; Matsuoka, M. Isolation and characterization of a rice WUSCHEL-type homeobox gene that is specifically expressed in the central cells of a quiescent center in the root apical meristem. Plant J. Cell Mol. Biol. 2003, 35, 429–441. [Google Scholar] [CrossRef]
- Leibfried, A.; To, J.P.C.; Busch, W.; Stehling, S.; Kehle, A.; Demar, M.; Kieber, J.J.; Lohmann, J.U. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 2005, 438, 1172–1175. [Google Scholar] [CrossRef] [PubMed]
- Lian, G.B.; Ding, Z.W.; Wang, Q.; Zhang, D.B.; Xu, J. Origins and evolution of WUSCHEL-related homeobox protein family in plant kingdom. Sci. World J. 2014, 534140. [Google Scholar] [CrossRef]
- Wu, C.C.; Li, F.W.; Kramer, E.M. Large-scale phylogenomic analysis suggests three ancient superclades of the WUSCHEL-RELATED HOMEOBOX transcription factor family in plants. PLoS ONE 2019, 14, e0223521. [Google Scholar] [CrossRef]
- Ikeda, M.; Mitsuda, N.; Ohme-Takagi, M. Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 2009, 21, 3493–3505. [Google Scholar] [CrossRef]
- Laux, T.; Mayer, K.F.; Berger, J.; Jürgens, G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Dev. 1996, 122, 87–96. [Google Scholar] [CrossRef]
- Zhang, X.; Zong, J.; Liu, J.H.; Yin, J.Y.; Zhang, D.B. Genome-wide analysis of WOX gene family in rice, sorghum, maize, Arabidopsis and poplar. J. Integr. Plant Biol. 2010, 52, 1016–1026. [Google Scholar] [CrossRef]
- Wang, X.L.; Bi, C.W.; Wang, C.Y.; Ye, Q.L.; Yin, T.M.; Ye, N. Genome-wide identification and characterization of WUSCHEL-related homeobox (WOX) genes in Salix suchowensis. J. For. Res. 2019, 30, 1811–1822. [Google Scholar] [CrossRef]
- Yang, Z.E.; Gong, Q.; Qin, W.Q.; Yang, Z.R.; Cheng, Y.; Lu, L.L.; Ge, X.Y.; Zhang, C.J.; Wu, Z.X.; Li, F.G. Genome-wide analysis of WOX genes in upland cotton and their expression pattern under different stresses. BMC Plant Biol. 2017, 17, 113. [Google Scholar] [CrossRef]
- Li, X.X.; Liu, C.; Li, W.; Zhang, Z.L.; Gao, X.M.; Zhou, H.; Guo, Y.F. Genome-wide identification, phylogenetic analysis and expression profiling of the WOX family genes in Solanum lycopersicum. Yi Chuan 2016, 38, 444–460. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.C.; Liu, J.; Zeng, M.H.; He, J.F.; Qin, P.; Huang, H.; Xu, L. Identification of WOX family genes in Selaginella kraussiana for studies on stem cells and regeneration in lycophytes. Front. Plant Sci. 2016, 7, 93. [Google Scholar] [CrossRef] [PubMed]
- Schoof, H.; Lenhard, M.; Haecker, A.; Mayer, K.F.; Jürgens, G.; Laux, T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 2000, 100, 635–644. [Google Scholar] [CrossRef]
- Tvorogova, V.E.; Krasnoperova, E.Y.; Potsenkovskaia, E.A.; Kudriashov, A.A.; Dodueva, I.E.; Lutova, L.A. What does the WOX say? review of regulators, targets, partners. Mol. Biol. 2021, 55, 311–337. [Google Scholar] [CrossRef]
- Yvonne, S.; René, H.W.; Gwyneth, C.I.; Rüdiger, S. A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr. Biol. 2009, 19, 909–914. [Google Scholar] [CrossRef]
- He, G.R.; Cao, Y.W.; Wang, J.; Song, M.; Bi, M.M.; Tang, Y.C.; Xu, L.F.; Ming, J.; Yang, P.P. WUSCHEL-Related homeobox genes cooperate with cytokinin to promote bulbil formation in Lilium lancifolium. Plant Physiol. 2022. [Google Scholar] [CrossRef]
- Wan, X.; Zou, L.H.; Zheng, B.Q.; Tian, Y.Q.; Wang, Y. Transcriptomic profiling for prolonged drought in Dendrobium catenatum. Sci. Data 2018, 5, 180233. [Google Scholar] [CrossRef]
- Zhao, M.M.; Zhang, G.; Zhang, D.W.; Hsiao, Y.Y.; Guo, S.X. ESTs analysis reveals putative genes involved in symbiotic seed germination in Dendrobium officinale. PLoS ONE 2013, 8, e72705. [Google Scholar] [CrossRef]
- Zhang, T.T.; Li, Y.X.; Kang, Y.Q.; Wang, P.; Li, W.; Yu, W.G.; Wang, J.; Song, X.Q.; Jiang, X.; Zhou, Y. The Dendrobium catenatum DcCIPK24 increases drought and salt tolerance of transgenic Arabidopsis. Ind. Crops Prod. 2022, 187, 115375. [Google Scholar] [CrossRef]
- Zhang, T.T.; Xu, Y.; Ding, Y.D.; Yu, W.G.; Wang, J.; Lai, H.G.; Zhou, Y. Identification and expression analysis of WRKY gene family in response to abiotic stress in Dendrobium catenatum. Front. Genet. 2022, 13, 800019. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.T.; Cui, Z.; Li, Y.X.; Kang, Y.Q.; Song, X.Q.; Wang, J.; Zhou, Y. Genome-Wide identification and expression analysis of MYB transcription factor superfamily in Dendrobium catenatum. Front. Genet. 2021, 12, 714696. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.Q.; Teixeira, D.S.J.A.; Zhang, M.Z.; Yu, Z.M.; Si, C.; Zhao, C.H.; Dai, G.Y.; He, C.M.; Duan, J. Genome-Wide identification and analysis of the APETALA2 (AP2) transcription factor in Dendrobium officinale. Int. J. Mol. Sci. 2021, 22, 5221. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, A.Z. Genomic characterization and expression analysis of the SnRK family genes in Dendrobium officinale Kimura et Migo (Orchidaceae). Plants 2021, 10, 479. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y. Expression Analysis of DoWOX and DoSERK in Dendrobium Officinale during Protocorm Development; Southwest Jiaotong University: Chengdu, China, 2018; pp. 1–61. [Google Scholar]
- Li, C.; Shen, Q.Y.; Cai, X.; Lai, D.N.; Wu, L.S.; Han, Z.G.; Zhao, T.L.; Chen, D.H.; Si, J.P. JA signal-mediated immunity of Dendrobium catenatum to necrotrophic Southern Blight pathogen. BMC Plant Biol. 2021, 21, 360. [Google Scholar] [CrossRef] [PubMed]
- Thakku, R.R.; Madhvi, K.; Santosh, K.U.; Jaspreet, K.S. Identification and characterization of WUSCHEL-related homeobox (WOX) gene family in economically important orchid species Phalaenopsis equestris and Dendrobium catenatum. Plant Gene 2018, 14, 37–45. [Google Scholar] [CrossRef]
- Gehring, W.J.; Affolter, M.; Burglin, T. Homeodomain proteins. Annu. Rev. Biochem. 1994, 63, 487–526. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, T.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Kieffer, M.; Stern, Y.; Cook, H.; Clerici, E.; Maulbetsch, C.; Laux, T.; Davies, B. Analysis of the transcription factor WUSCHEL and its functional homologue in Antirrhinum reveals a potential mechanism for their roles in meristem maintenance. Plant Cell 2006, 18, 560–573. [Google Scholar] [CrossRef]
- Busch, W.; Miotk, A.; Ariel, F.D.; Zhao, Z.; Forner, J.; Daum, G.; Suzaki, T.; Schuster, C.; Schultheiss, S.J.; Leibfried, A.; et al. Transcriptional control of a plant stem cell niche. Dev. Cell 2010, 18, 849–861. [Google Scholar] [CrossRef]
- Causier, B.; Ashworth, M.; Guo, W.J.; Davies, B. The TOPLESS interactome: A framework for gene repression in Arabidopsis. Plant Physiol. 2012, 158, 423–438. [Google Scholar] [CrossRef] [PubMed]
- Dolzblasz, A.; Nardmann, J.; Clerici, E.; Causier, B.; Graaff, E.V.D.; Chen, J.H.; Davies, B.; Werr, W.; Laux, T. Stem cell regulation by Arabidopsis WOX genes. Mol. Plant 2016, 9, 1028–1039. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.Y.; Liu, H.; Wang, H.; Liu, Y.; Zhu, B.T.; Wang, Z.Y.; Hou, Y.L.; Zhang, P.C.; Wen, J.Q.; Yang, H.S.; et al. HEADLESS, a WUSCHEL homolog, uncovers novel aspects of shoot meristem regulation and leaf blade development in Medicago truncatula. J. Exp. Bot. 2019, 70, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.D.; Sun, L.D.; Zhou, Y.Z.; Yang, W.R.; Cheng, T.R.; Wang, J.; Zhang, Q.X. Identification and expression analysis of the SQUAMOSA promoter-binding protein (SBP)-box gene family in Prunus mume. Mol. Genet. Genom. 2015, 290, 1701–1715. [Google Scholar] [CrossRef]
- Liu, H.F.; Liang, J.X.; Zhong, Y.M.; Xiao, G.S.; Efferth, T.; Georgiev, M.I.; VargasDeLaCruz, C.; Bajpai, V.K.; Caprioli, G.; Liu, J.L.; et al. Dendrobium officinale polysaccharide alleviates intestinal inflammation by promoting small extracellular vesicle packaging of miR-433-3p. J. Agric. Food Chem. 2021, 69, 13510–13523. [Google Scholar] [CrossRef]
- Yuan, Y.D.; Zuo, J.J.; Zhang, H.Y.; Zu, M.T.; Yu, M.Y.; Liu, S.A. Transcriptome and metabolome profiling unveil the accumulation of flavonoids in Dendrobium officinale. Genomics 2022, 114, 110324. [Google Scholar] [CrossRef]
- Breuninger, H.; Rikirsch, E.; Hermann, M.; Ueda, M.; Laux, T. Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev. Cell 2008, 14, 867–876. [Google Scholar] [CrossRef]
- Cannon, S.; Mitra, A.; Baumgarten, A.; Young, N.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Yoo, S.C.; Cho, S.H.; Paek, N.C. Rice WUSCHEL-related homeobox 3A (OsWOX3A) modulates auxin-transport gene expression in lateral root and root hair development. Plant Signal. Behav. 2013, 8, e25929. [Google Scholar] [CrossRef]
- Jiang, W.; Zhou, S.L.; Zhang, Q.; Song, H.Z.; Zhou, D.X.; Zhao, Y. Transcriptional regulatory network of WOX11 is involved in the control of crown root development, cytokinin signals, and redox in rice. J. Exp. Bot. 2017, 68, 2787–2798. [Google Scholar] [CrossRef]
- Jin, J.P.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.C.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Ling, L.; Zhang, W.R.; An, Y.M.; Du, B.H.; Wang, D.; Guo, C.H. Genome-wide analysis of the TCP transcription factor genes in five legume genomes and their response to salt and drought stresses. Funct. Integr. Genom. 2020, 20, 537–550. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef]
- Du, J.C.; Hu, S.M.; Yu, Q.; Wang, C.D.; Yang, Y.P.; Sun, H.; Sun, X.D. Genome-Wide identification and characterization of BrrTCP transcription factors in Brassica rapa ssp. rapa. Front. Plant Sci. 2017, 8, 1588. [Google Scholar] [CrossRef]
- Chen, H.; Zou, Y.; Shang, Y.L.; Lin, H.Q.; Wang, Y.J.; Cai, R.; Tang, X.Y.; Zhou, J.M. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008, 146, 368–376. [Google Scholar] [CrossRef]
- Zhang, L.; Li, C.; Yang, D.N.; Wang, Y.H.; Yang, Y.P.; Sun, X.D. Genome-Wide analysis of the TCP transcription factor genes in Dendrobium catenatum Lindl. Int. J. Mol. Sci. 2021, 22, 10269. [Google Scholar] [CrossRef]

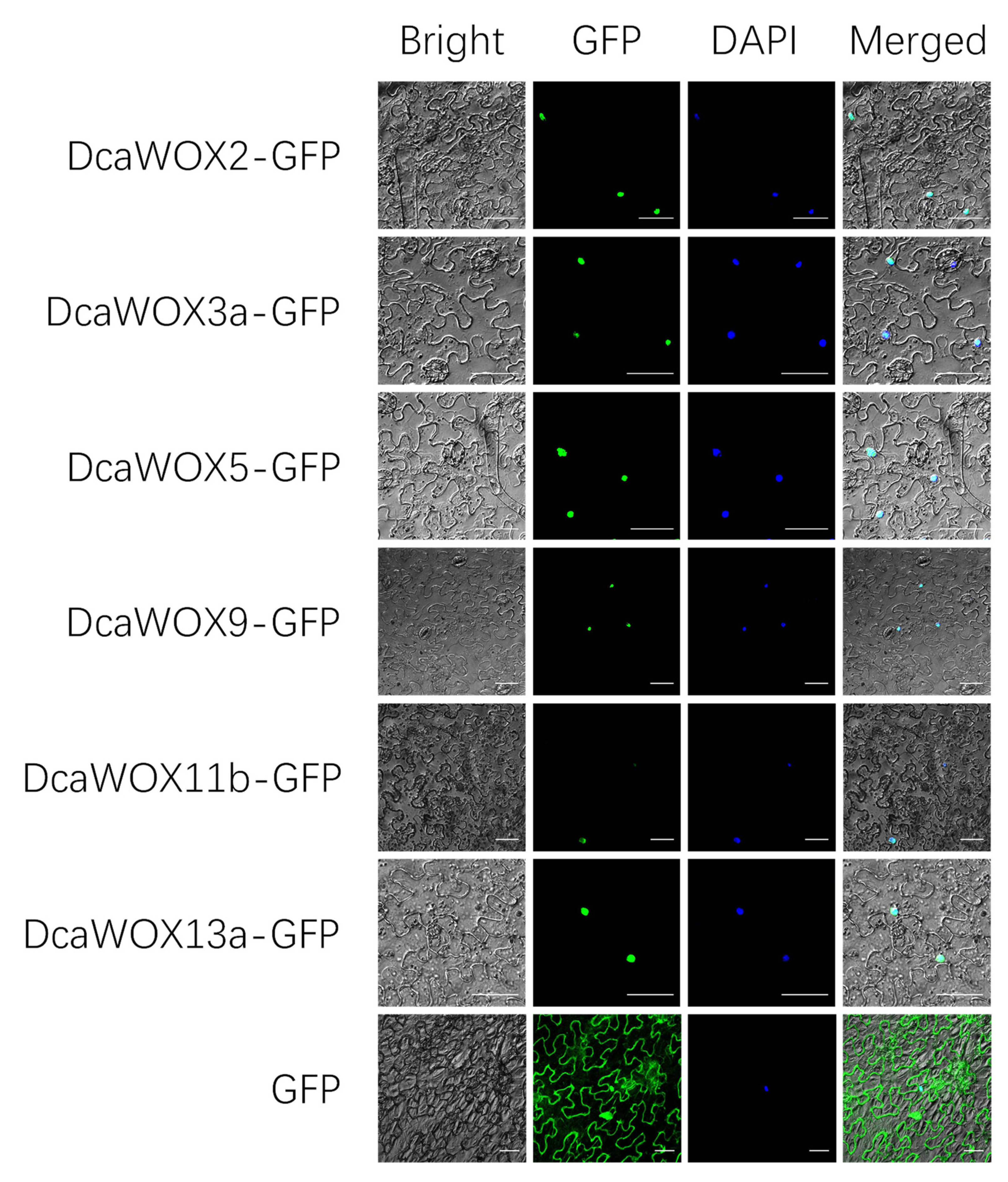

| Gene Name | Accession Number | CDS Length (bp) | Protein Size (aa) | MW (kD) | PI | GRAVY | Protein Localization |
|---|---|---|---|---|---|---|---|
| DcaWOX2 | XP_020689224.1 | 696 | 231 | 26.02 | 6.66 | −0.68 | Nucleus |
| DcaWOX3a | XP_020698032.1 | 579 | 192 | 21.65 | 9.11 | −0.70 | Nucleus |
| DcaWOX3b | XP_020698705.1 | 645 | 214 | 24.34 | 6.79 | −0.83 | Nucleus |
| DcaWOX4 | XP_028548805.1 | 501 | 166 | 18.64 | 10.25 | −0.64 | Nucleus |
| DcaWOX5 | XP_020673683.1 | 555 | 184 | 21.26 | 8.85 | −0.72 | Nucleus |
| DcaWOX9 | XP_028553243.1 | 1014 | 337 | 36.91 | 7.79 | −0.26 | Nucleus |
| DcaWOX11a | XP_020695082.1 | 789 | 262 | 27.81 | 5.78 | −0.23 | Nucleus |
| DcaWOX11b | XP_028555924.1 | 762 | 253 | 27.33 | 6.73 | −0.24 | Nucleus |
| DcaWOX13a | XP_020676225.2 | 657 | 218 | 25.16 | 6.98 | −0.68 | Nucleus |
| DcaWOX13b | XP_020685087.1 | 780 | 259 | 29.92 | 5.26 | −0.88 | Nucleus |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Li, C.; Wang, Y.; Qin, X.; Meng, L.; Sun, X. Genome-Wide Analysis of the WOX Transcription Factor Genes in Dendrobium catenatum Lindl. Genes 2022, 13, 1481. https://doi.org/10.3390/genes13081481
Li H, Li C, Wang Y, Qin X, Meng L, Sun X. Genome-Wide Analysis of the WOX Transcription Factor Genes in Dendrobium catenatum Lindl. Genes. 2022; 13(8):1481. https://doi.org/10.3390/genes13081481
Chicago/Turabian StyleLi, Hefan, Cheng Li, Yuhua Wang, Xiangshi Qin, Lihua Meng, and Xudong Sun. 2022. "Genome-Wide Analysis of the WOX Transcription Factor Genes in Dendrobium catenatum Lindl." Genes 13, no. 8: 1481. https://doi.org/10.3390/genes13081481
APA StyleLi, H., Li, C., Wang, Y., Qin, X., Meng, L., & Sun, X. (2022). Genome-Wide Analysis of the WOX Transcription Factor Genes in Dendrobium catenatum Lindl. Genes, 13(8), 1481. https://doi.org/10.3390/genes13081481