IbMYB308, a Sweet Potato R2R3-MYB Gene, Improves Salt Stress Tolerance in Transgenic Tobacco
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Cloning and Bioinformatics Analysis of IbMYB308 and Its Promoter
2.3. Bioinformatics Analysis
2.4. Expression Analysis of IbMYB308 in Sweet Potato and Transgenic Tobacco
2.5. Construction of Overexpression Vectors
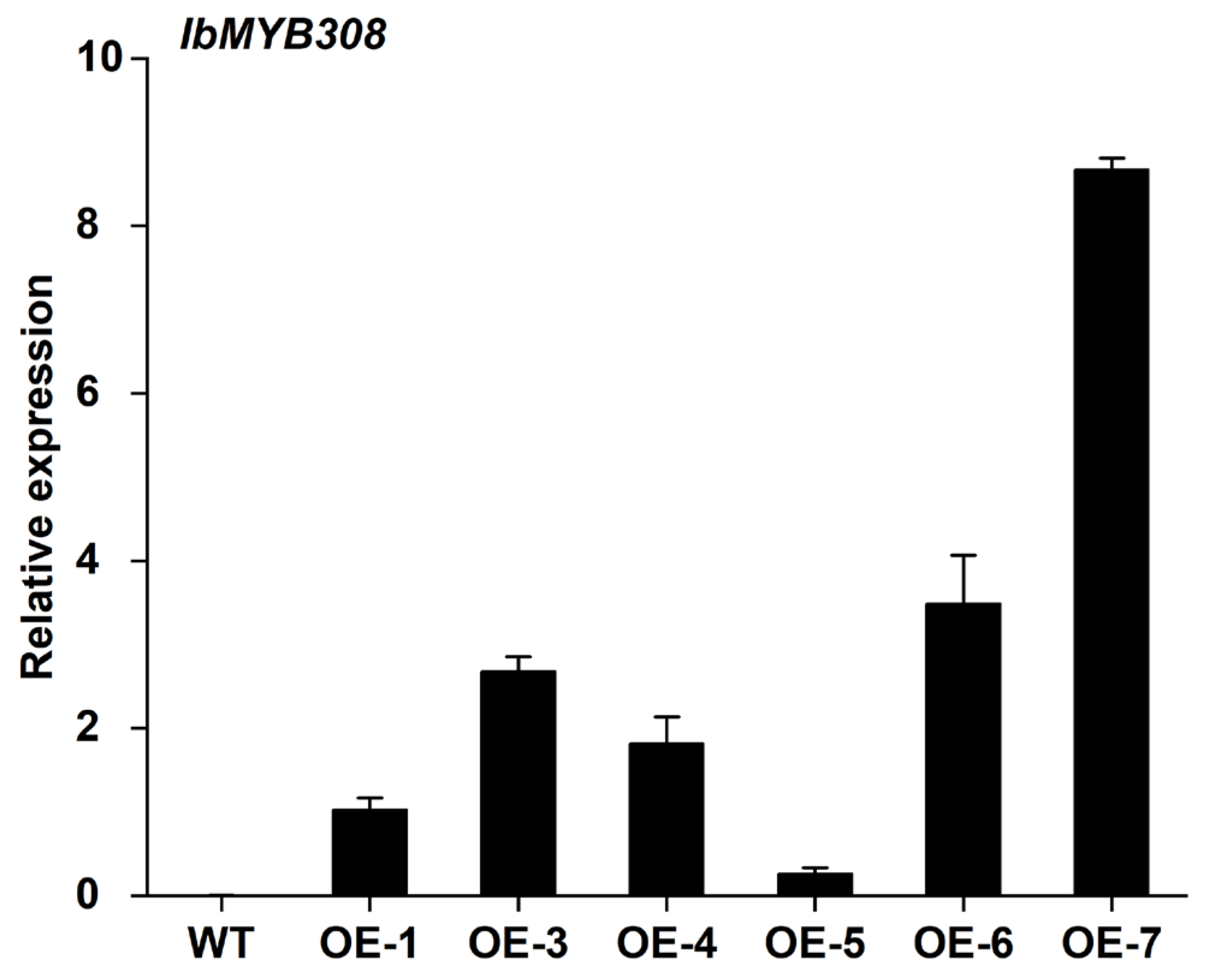
2.6. Generation of Transgenic Tobacco
2.7. Data Analysis
3. Results
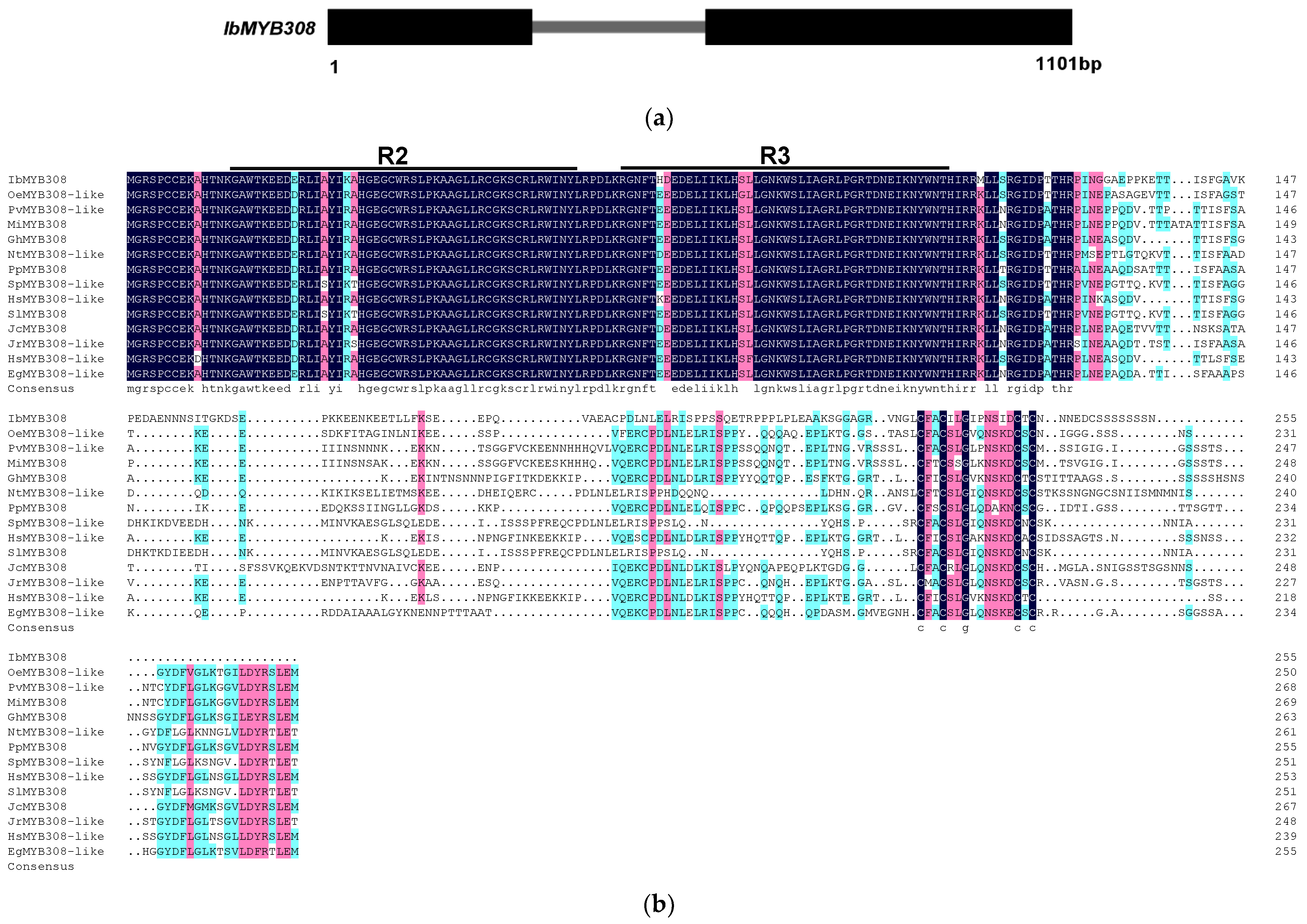
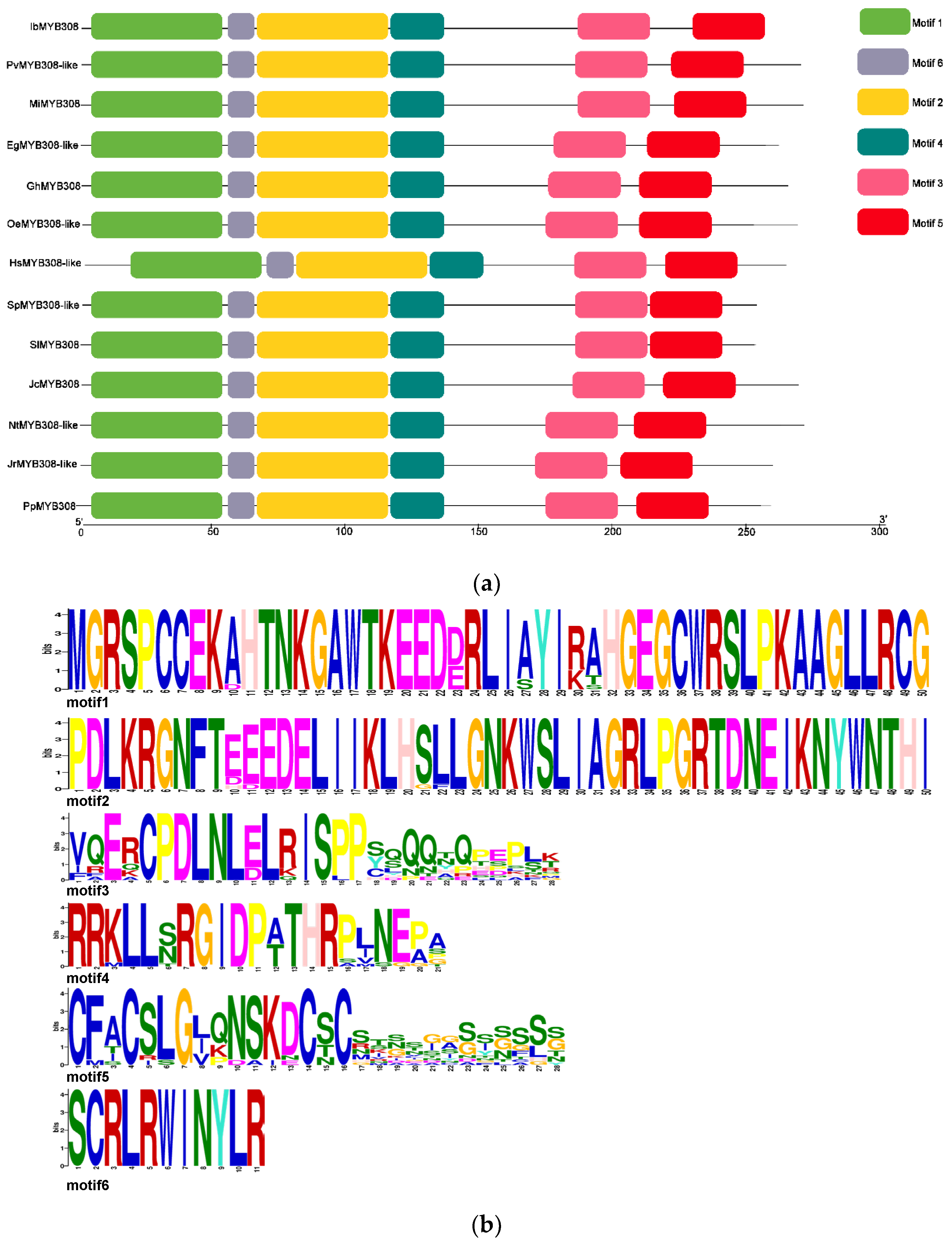
3.1. Isolation and Characterization of IbMYB308
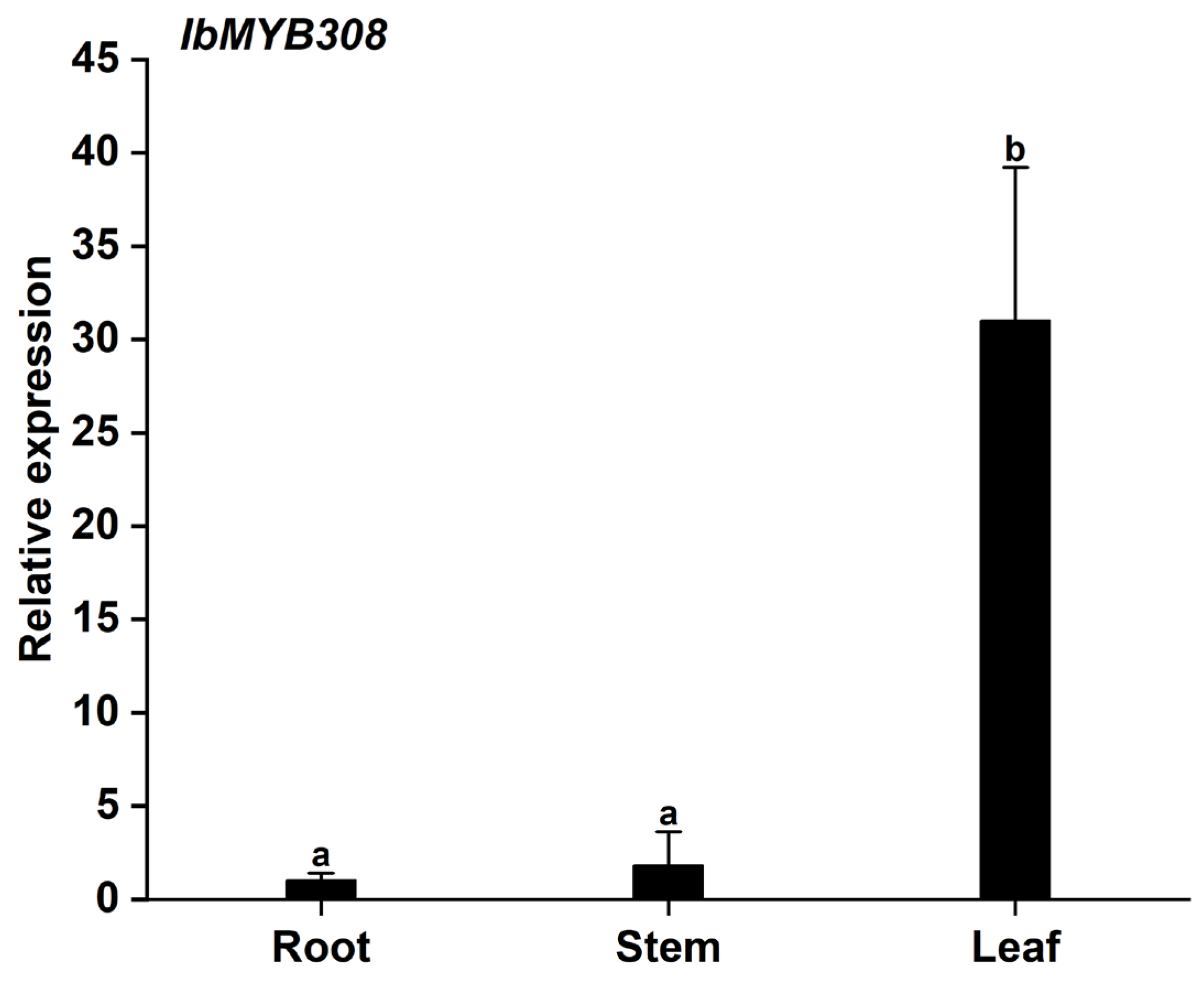
3.2. Expression Analysis of the IbMYB308 in Sweet Potato
3.3. Expression Profiles of IbMYB308 under Abiotic Stress
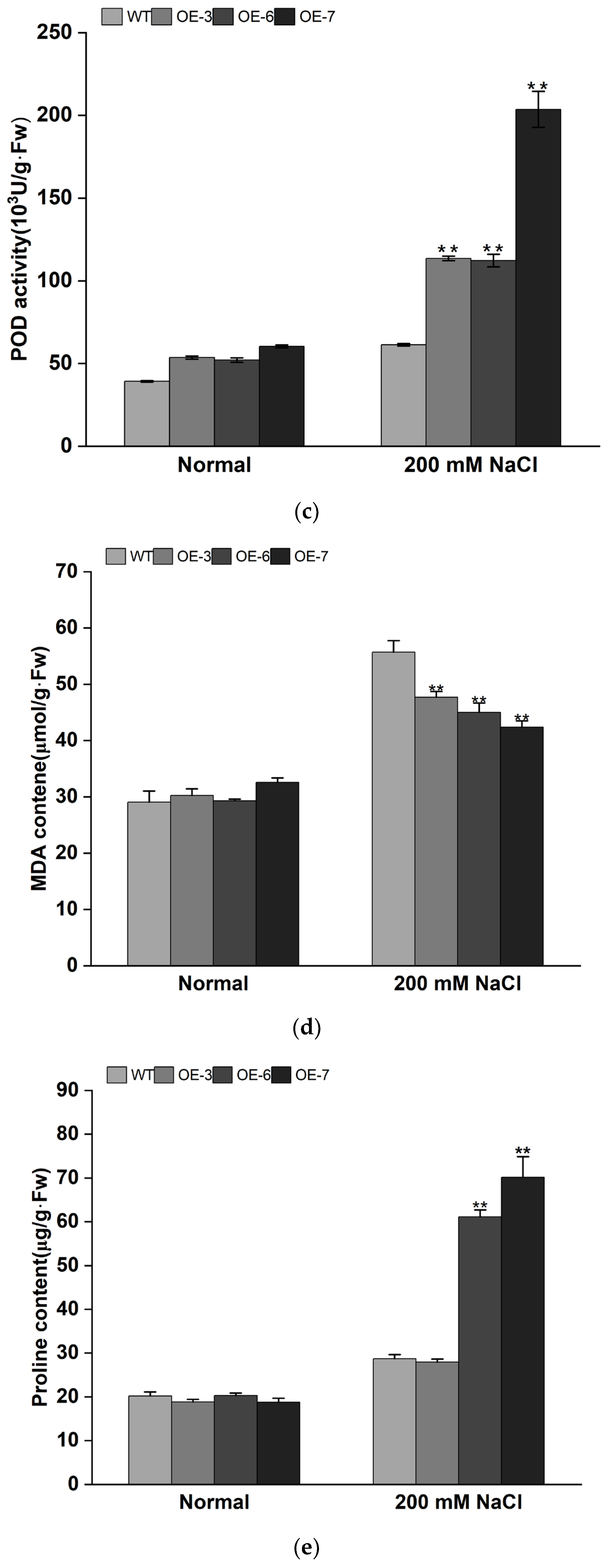
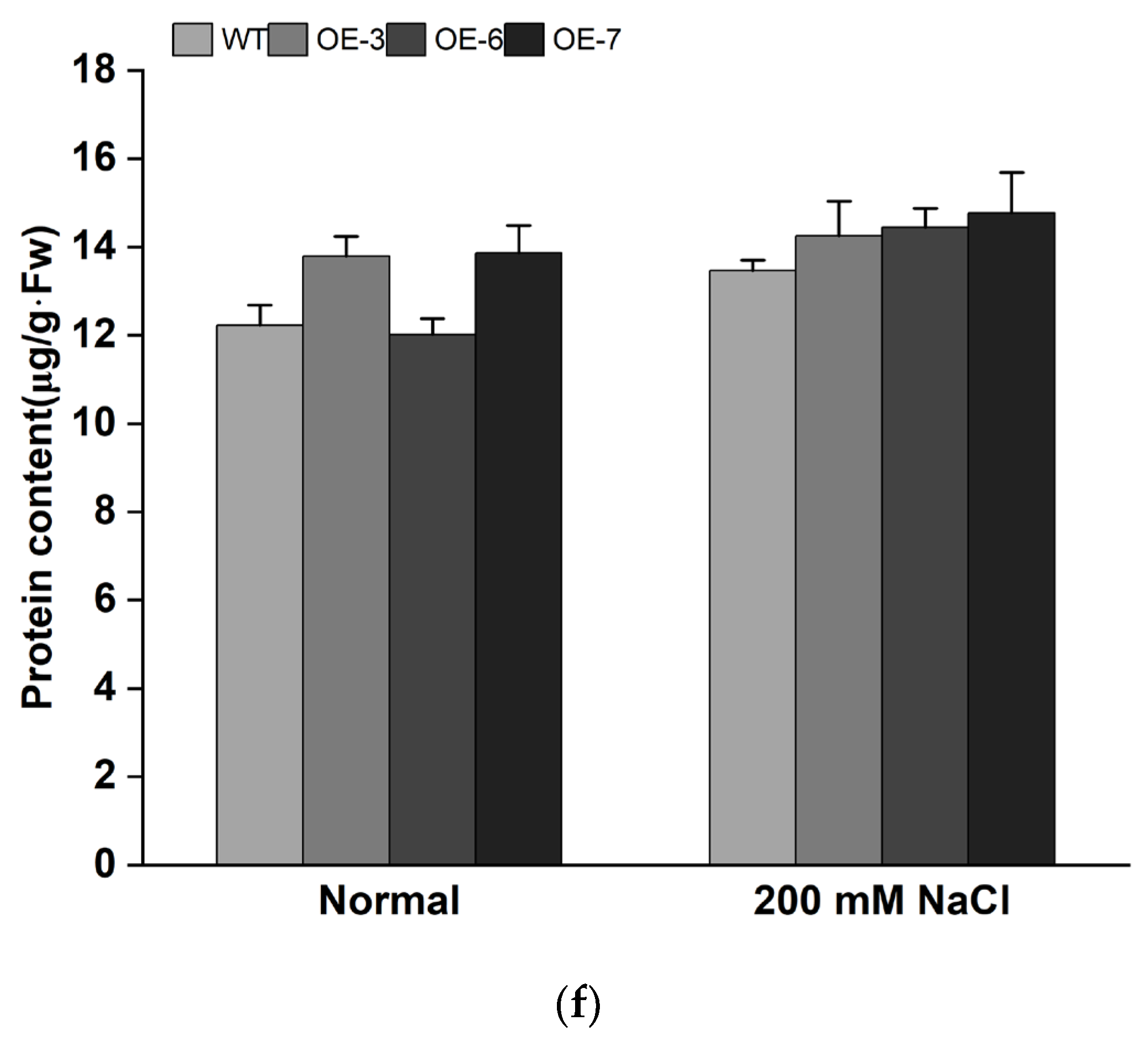
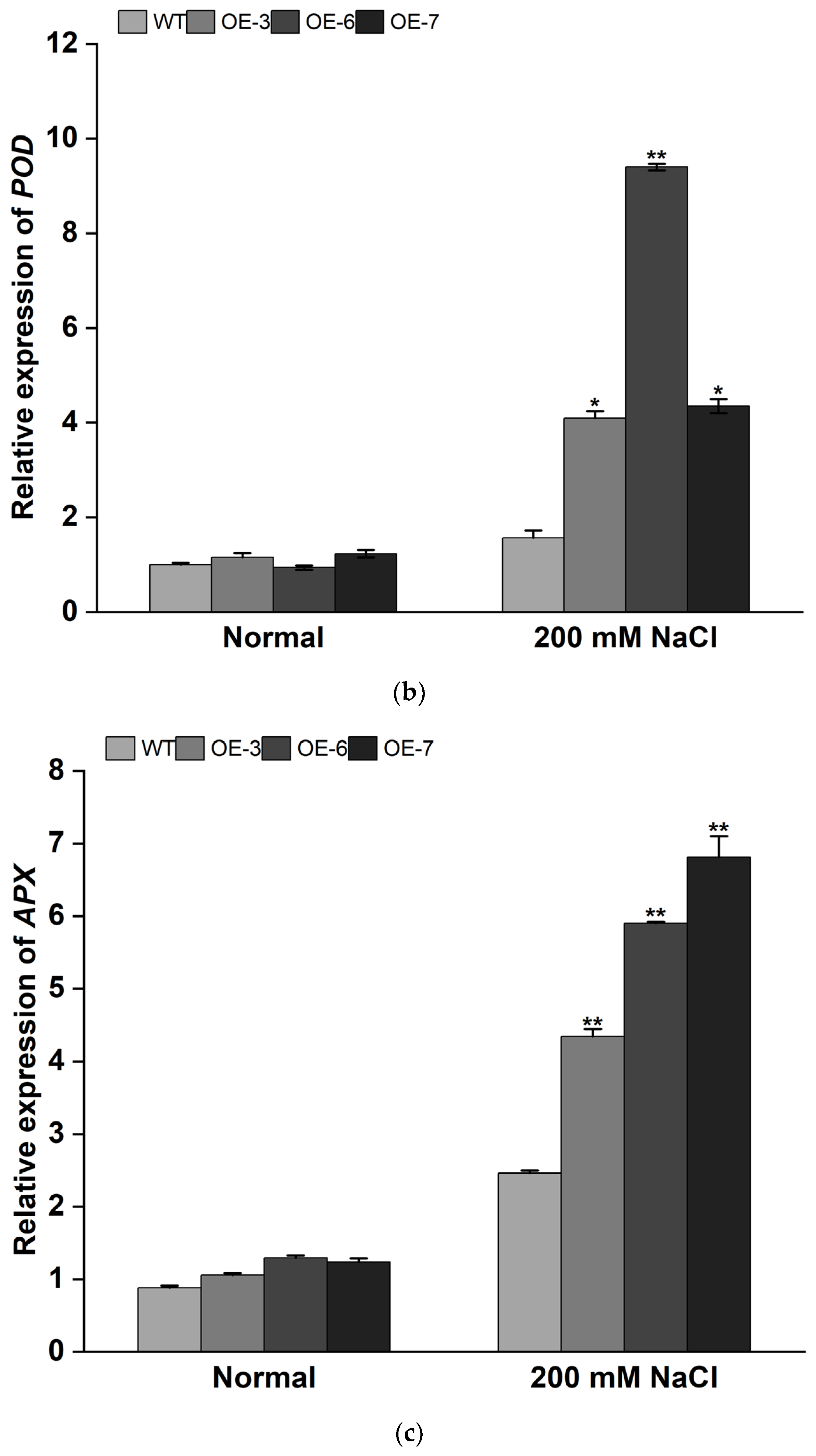
3.4. Overexpression of IbMYB308 Improves Tolerance to Salt Stress in Transgenic Plants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, J.; Moeinzadeh, M.H.; Kuhl, H.; Helmuth, J.; Xiao, P.; Haas, S.; Liu, G.L.; Zheng, J.L.; Sun, Z.; Fan, W.J.; et al. Haplotype-resolved sweet potato genome traces back its hexaploidization history. Nat. Plants 2017, 3, 696–703. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.J.; Kim, M.; Deng, X.P.; Kwak, S.O.; Chen, W. Enhanced salt stress tolerance in transgenic potato plants expressing IbMYB1, a sweet potato transcription factor. J. Mic. Bio. 2013, 23, 1737–1746. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.J.; Wang, H.X.; Wu, Y.L.; Yang, N.; Yang, J.; Zhang, P. H(+) -pyrophosphatase IbVP1 promotes efficient iron use in sweet potato [Ipomoea batatas (L.) Lam.]. Plant Biotech. J. 2017, 15, 698–712. [Google Scholar] [CrossRef]
- Zhai, H.; Wang, F.B.; Si, Z.Z.; Huo, J.X.; Xing, L.; An, Y.Y.; He, S.A.; Liu, Q.C. A myo-inositol-1-phosphate synthase gene, IbMIPS1, enhances salt and drought tolerance and stem nematode resistance in transgenic sweet potato. Plant Biotech. J. 2016, 14, 592–602. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Gao, X.R.; Zhi, Y.H.; Li, X.; Zhang, Q.; Niu, J.B.; Wang, J.; Zhai, H.; Zhao, N.; Li, J.G.; et al. A non-tandem CCCH-type zinc-finger protein, IbC3H18, functions as a nuclear transcriptional activator and enhances abiotic stress tolerance in sweet potato. New Phytol. 2019, 223, 1918–1936. [Google Scholar] [CrossRef]
- Chen, S.P.; Lin, I.W.; Chen, X.; Huang, Y.H.; Chang, S.C.; Lo, H.S.; Lu, H.H.; Yeh, K.W. Sweet potato NAC transcription factor, IbNAC1, upregulates sporamin gene expression by binding the SWRE motif against mechanical wounding and herbivore attack. Plant J. 2016, 86, 234–248. [Google Scholar] [CrossRef]
- Wang, X.P.; Niu, Y.L.; Zheng, Y. Multiple functions of MYB transcription factors in abiotic stress responses. Int. J. Mol. Sci. 2021, 22, 6125. [Google Scholar] [CrossRef]
- Baldoni, E.; Genga, A.; Cominelli, E. Plant MYB transcription factors: Their role in drought response mechanisms. Int. J. Mol. Sci. 2015, 16, 15811–15851. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, K.X.; Khurshid, M.; Li, J.B.; He, M.; Georgiev, M.I.; Zhang, X.Q.; Zhou, M.L. MYB transcription repressors regulate plant secondary metabolism. Crit. Rev. Plant Sci. 2019, 38, 159–170. [Google Scholar] [CrossRef]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Jung, C.; Seo, J.S.; Han, S.W.; Koo, Y.J.; Kim, C.H.; Song, S.I.; Nahm, B.H.; Choi, Y.D.; Cheong, J.J. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008, 146, 623–635. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.K.; Dubos, C.; Dodd, I.C.; Holroyd, G.H.; Hetherington, A.M.; Campbell, M.M. AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Curr. Biol. 2005, 15, 1201–1206. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Wang, Y.H.; Zheng, H.; Lu, W.; Wu, C.A.; Huang, J.H.; Yan, K.; Yang, G.D.; Zheng, C.C. Salt-induced transcription factor MYB74 is regulated by the RNA-directed DNA methylation pathway in Arabidopsis. J. Exp. Bot. 2015, 66, 5997–6008. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, D.Y.; Zhang, D.L.; Yin, D.D.; Zhao, Y.; Ji, C.J.; Zhao, X.F.; Li, X.B.; He, Q.; Chen, R.S.; et al. A novel antisense long noncoding RNA, TWISTED LEAF, maintains leaf blade flattening by regulating its associated sense R2R3-MYB gene in rice. New Phytol. 2018, 218, 774–788. [Google Scholar] [CrossRef]
- El-Kereamy, A.; Bi, Y.M.; Ranathunge, K.; Beatty, P.H.; Good, A.G.; Rothstein, S.J. The rice R2R3-MYB transcription factor OsMYB55 is involved in the tolerance to high temperature and modulates amino acid metabolism. PLoS ONE 2012, 7, e52030. [Google Scholar] [CrossRef]
- Wang, F.Z.; Chen, M.X.; Yu, L.J.; Xie, L.J.; Yuan, L.B.; Qi, H.; Xiao, M.; Guo, W.; Chen, Z.; Yi, K.; et al. OsARM1, an R2R3 MYB transcription factor, is involved in regulation of the response to arsenic stress in rice. Front. Plant Sci. 2017, 8, 1868. [Google Scholar] [CrossRef]
- Li, M.; Lin, L.; Zhang, Y.; Sui, N. ZmMYB31, a R2R3-MYB transcription factor in maize, positively regulates the expression of CBF genes and enhances resistance to chilling and oxidative stress. Mol. Biol. Rep. 2019, 46, 3937–3944. [Google Scholar] [CrossRef]
- Du, H.; Feng, B.R.; Yang, S.S.; Huang, Y.B.; Tang, Y.X. The R2R3-MYB transcription factor gene family in maize. PLoS ONE 2012, 7, e37463. [Google Scholar] [CrossRef]
- Wu, J.D.; Jiang, Y.L.; Liang, Y.N.; Chen, L.; Chen, W.J.; Cheng, B.J. Expression of the maize MYB transcription factor ZmMYB3R enhances drought and salt stress tolerance in transgenic plants. Plant Physiol. Biochem. 2019, 137, 179–188. [Google Scholar] [CrossRef]
- Yu, Y.H.; Ni, Z.Y.; Chen, Q.J.; Qu, Y.Y. The wheat salinity-induced R2R3-MYB transcription factor TaSIM confers salt stress tolerance in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2017, 491, 642–648. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.H.; Chen, R.; Wei, X.; Liu, Y.H.; Zhao, S.J.; Yin, X.P.; Xie, T. Genome-wide identification of R2R3-MYB family in wheat and functional characteristics of the abiotic stress responsive gene TaMYB344. BMC Genom. 2020, 21, 792. [Google Scholar] [CrossRef] [PubMed]
- He, Y.N.; Li, W.; Lv, J.; Jia, Y.B.; Wang, M.C.; Xia, G.M. Ectopic expression of a wheat MYB transcription factor gene, TaMYB73, improves salinity stress tolerance in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 1511–1522. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Tang, J.; Hu, R.; Wu, P.; Hou, X.L.; Song, X.M.; Xiong, A.S. Genome-wide analysis of the R2R3-MYB transcription factor genes in Chinese cabbage (Brassica rapa ssp. pekinensis) reveals their stress and hormone responsive patterns. BMC Genom. 2015, 16, 17. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Niu, F.F.; Liu, W.Z.; Yang, B.; Zhang, J.X.; Ma, J.Y.; Cheng, H.; Han, F.; Jiang, Y.Q. Identification, cloning and characterization of R2R3-MYB gene family in canola (Brassica napus L.) identify a novel member modulating ROS accumulation and hypersensitive-like cell death. DNA Res. 2016, 23, 101–114. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Li, R.; Qu, F.J.; You, C.X.; Wang, X.F.; Hao, Y.J. R2R3-MYB transcription factor MdMYB23 is involved in the cold tolerance and proanthocyanidin accumulation in apple. Plant J. 2018, 96, 562–577. [Google Scholar] [CrossRef]
- Wang, R.K.; Cao, Z.H.; Hao, Y.J. Overexpression of a R2R3 MYB gene MdSIMYB1 increases tolerance to multiple stresses in transgenic tobacco and apples. Physiol. Plant. 2014, 150, 76–87. [Google Scholar] [CrossRef]
- Wang, S.S.; Shi, M.Y.; Zhang, Y.; Xie, X.B.; Sun, P.P.; Fang, C.B.; Zhao, J. FvMYB24, a strawberry R2R3-MYB transcription factor, improved salt stress tolerance in transgenic Arabidopsis. Biochem. Biophys. Res. Commun. 2021, 569, 93–99. [Google Scholar] [CrossRef]
- Cui, M.H.; Yoo, K.S.; Hyoung, S.; Nguyen, H.T.; Kim, Y.Y.; Kim, H.J.; Ok, S.H.; Yoo, S.D.; Shin, J.S. An Arabidopsis R2R3-MYB transcription factor, AtMYB20, negatively regulates type 2C serine/threonine protein phosphatases to enhance salt tolerance. FEBS Lett. 2013, 587, 1773–1778. [Google Scholar] [CrossRef]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef]
- Wang, N.; Zhang, W.X.; Qin, M.Y.; Li, S.; Qiao, M.; Liu, Z.H.; Xiang, F.N. Drought tolerance conferred in soybean (Glycine max. L) by GmMYB84, a Novel R2R3-MYB transcription factor. Plant Cell Physiol. 2017, 58, 1764–1776. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.Q.; Guo, Y. Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 2018, 60, 796–804. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Cheng, S.; Liu, X.; Du, H.; Dai, M.; Zhou, D.X.; Yang, W.; Zhao, Y. The R2R3-type MYB gene OsMYB91 has a function in coordinating plant growth and salt stress tolerance in rice. Plant Sci. 2015, 236, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Huo, J.; Bing, D.U.; Sun, S.; He, S.Z.; Zhao, N.; Liu, Q.C.; Zhai, H. A novel aldo-keto reductase gene, IbAKR, from sweet potato confers higher tolerance to cadmium stress in tobacco. Front. Agric. Sci. Eng. 2018, 5, 206–213. [Google Scholar]
- He, Y.X.; Dong, Y.S.; Yang, X.D.; Guo, D.Q.; Qian, X.Y.; Yan, F.; Wang, Y.; Li, J.W.; Wang, Q.Y. Functional activation of a novel R2R3-MYB protein gene, GmMYB68, confers salt-alkali resistance in soybean (Glycine max L.). Genom 2020, 63, 13–26. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Waszczak, C.; Carmody, M.; Kangasjarvi, J. Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol. 2018, 69, 209–236. [Google Scholar] [CrossRef]
- He, J.; Liu, Y.Q.; Yuan, D.Y.; Duan, M.J.; Liu, Y.L.; Shen, Z.J.; Yang, C.Y.; Qiu, Z.Y.; Liu, D.M.; Wen, P.Z.; et al. An R2R3 MYB transcription factor confers brown planthopper resistance by regulating the phenylalanine ammonia-lyase pathway in rice. Proc. Natl. Acad. Sci. USA 2020, 117, 271–277. [Google Scholar] [CrossRef]
- Liu, C.; Xie, T.; Chen, C.; Luan, A.; Long, J.; Li, C.; Ding, Y.; He, Y. Genome-wide organization and expression profiling of the R2R3-MYB transcription factor family in pineapple (Ananas comosus). BMC Genom. 2017, 18, 503. [Google Scholar] [CrossRef]
- Yang, J.H.; Zhang, B.H.; Gu, G.; Yuan, J.Z.; Shen, S.J.; Jin, L.; Lin, Z.Q.; Lin, J.F.; Xie, X.F. Genome-wide identification and expression analysis of the R2R3-MYB gene family in tobacco (Nicotiana tabacum L.). BMC Genom. 2022, 23, 432. [Google Scholar] [CrossRef]
- Chen, Q.C.; Zhang, X.D.; Fang, Y.X.; Wang, B.Y.; Xu, S.S.; Zhao, K.; Zhang, J.S.; Fang, J.P. Genome-wide identification and expression analysis of the R2R3-MYB transcription factor family revealed their potential roles in the flowering process in longan (Dimocarpus longan). Front. Plant Sci. 2022, 13, 820439. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.J.; Wang, P.J.; Gu, M.Y.; Lin, X.Y.; Hou, B.H.; Zheng, Y.C.; Sun, Y.; Jin, S.; Ye, N.X. R2R3-MYB transcription factor family in tea plant (Camellia sinensis): Genome-wide characterization, phylogeny, chromosome location, structure and expression patterns. Genomics 2021, 113, 1565–1578. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Ahmad, N.; Zhang, J.; Lv, Y.; Zhang, X.; Ma, X.; Xiuming, L.; Na, Y. Genome-wide analysis and transcriptional reprogrammings of MYB superfamily revealed positive insights into abiotic stress responses and anthocyanin accumulation in Carthamus tinctorius L. Mol. Genet. Genom. 2022, 297, 125–145. [Google Scholar] [CrossRef] [PubMed]
- Bao, X.M.; Zong, Y.; Hu, N.; Li, S.M.; Liu, B.L.; Wang, H.L. Functional R2R3-MYB transcription factor NsMYB1, regulating anthocyanin biosynthesis, was relative to the fruit color differentiation in Nitraria sibirica Pall. BMC Plant Biol. 2022, 22, 186. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.Y.; Shi, Y.Y.; Wang, R.; Feng, Y.; Wang, L.S.; Zhang, H.S.; Shi, X.Y.; Jing, G.Q.; Deng, P.; Song, T.Z.; et al. The transcription factor OsMYBc and an E3 ligase regulate expression of a K+ transporter during salt stress. Plant Physiol. 2022, kiac283. [Google Scholar] [CrossRef]
- Zhong, C.M.; Tang, Y.; Pang, B.; Li, X.K.; Yang, Y.P.; Deng, J.; Feng, C.Y.; Li, L.F.; Ren, G.P.; Wang, Y.Q.; et al. The R2R3-MYB transcription factor GhMYB1a regulates flavonol and anthocyanin accumulation in Gerbera hybrida. Hortic. Res. 2020, 7, 78. [Google Scholar] [CrossRef]
- Dhakarey, R.; Yaritz, U.; Tian, L.; Amir, R. A Myb transcription factor, PgMyb308-like, enhances the level of shikimate, aromatic amino acids, and lignins, but represses the synthesis of flavonoids and hydrolyzable tannins, in pomegranate (Punica granatum L.). Hortic. Res. 2022, 9, 1–13. [Google Scholar]
- Ji, X.T.; Wang, M.L.; Xu, Z.Z.; Wang, K.; Sun, D.Y.; Niu, L.X. PlMYB308 regulates flower senescence by modulating ethylene biosynthesis in Herbaceous Peony. Front. Plant Sci. 2022, 13, 872442. [Google Scholar] [CrossRef]
- Zhang, D.W.; Tan, Y.J.; Dong, F.; Zhang, Y.; Huang, Y.L.; Zhou, Y.Z.; Zhao, Z.J.; Yin, Q.; Xie, X.H.; Gao, X.W.; et al. The expression of IbMYB1 is essential to maintain the purple color of leaf and storage root in sweet potato [Ipomoea batatas (L.) Lam]. Front. Plant Sci. 2021, 12, 688707. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhu, H.; He, S.; Zhai, H.; Liu, Q. A novel sweetpotato transcription factor gene IbMYB116 enhances drought tolerance in transgenic Arabidopsis. Front. Plant Sci. 2019, 10, 1025. [Google Scholar] [CrossRef]
- Zhu, H.; Yang, X.; Wang, X.; Li, Q.; Guo, J.; Ma, T.; Zhao, C.; Tang, Y.; Qiao, L.; Wang, J.; et al. The sweetpotato β-amylase gene IbBAM1.1 enhances drought and salt stress resistance by regulating ROS homeostasis and osmotic balance. Plant Physiol. Biochem. 2021, 168, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Hussey, S.G.; Grima-Pettenati, J.; Myburg, A.A.; Mizrachi, E.; Brady, S.M.; Yoshikuni, Y.; Deutsch, S. A standardized synthetic eucalyptus transcription factor and promoter panel for re-engineering secondary cell wall regulation in biomass and bioenergy crops. ACS. Synth. Biol. 2019, 8, 463–465. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Ma, B.Q.; Yang, N.X.; Jin, L.; Wang, L.; Ma, S.Y.; Ruan, Y.L.; Ma, F.W.; Li, M.J. Variation in the promoter of the sorbitol dehydrogenase gene MdSDH2 affects binding of the transcription factor MdABI3 and alters fructose content in apple fruit. Plant J. 2022, 109, 1183–1198. [Google Scholar] [CrossRef]
- Zou, C.; Sun, K.L.; Mackaluso, J.D.; Seddon, A.E.; Jin, R.; Thomashow, M.F.; Shiu, S.H. Cis-regulatory code of stress-responsive transcription in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2011, 108, 14992–14997. [Google Scholar] [CrossRef]
- Wang, X.P.; Xu, L.L.; Liu, X.; Xin, L.; Wu, S.J.; Chen, X.S. Development of potent promoters that drive the efficient expression of genes in apple protoplasts. Hortic. Res. 2021, 8, 211. [Google Scholar] [CrossRef] [PubMed]
- Halter, T.; Wang, J.; Amesefe, D.; Lastrucci, E.; Charvin, M.; Singla Rastogi, M.; Navarro, L. The Arabidopsis active demethylase ROS1 cis-regulates defence genes by erasing DNA methylation at promoter-regulatory regions. eLife 2021, 10, e62994. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, S.; Li, H.Q.; Wang, L.T.; Liu, Y.; Niu, L.L.; Yang, Q.; Meng, D.; Fu, Y.J. Genome-wide analysis and characterization of R2R3-MYB family in pigeon pea (Cajanus cajan) and their functional identification in phenylpropanoids biosynthesis. Planta 2021, 254, 64. [Google Scholar] [CrossRef]
- Chen, G.Q.; He, W.Z.; Guo, X.X.; Pan, J.S. Genome-wide identification, classification and expression analysis of the MYB transcription factor family in Petunia. Int. J. Mol. Sci. 2021, 22, 4838. [Google Scholar] [CrossRef]
- Zhou, W.; Zhang, Q.; Sun, Y.; Yang, L.; Wang, Z. Genome-wide identification and characterization of R2R3-MYB family in Hypericum perforatum under diverse abiotic stresses. Int. J. Biol. Macromol. 2020, 145, 341–354. [Google Scholar] [CrossRef]
- Ali, F.; Li, Y.H.; Li, F.G.; Wang, Z. Genome-wide characterization and expression analysis of cystathionine β-synthase genes in plant development and abiotic stresses of cotton (Gossypium spp.). Int. J. Biol. Macromol. 2021, 193, 823–837. [Google Scholar] [CrossRef]
- Yang, X.Y.; Guo, T.; Li, J.; Chen, Z.; Guo, B.; An, X.M. Genome-wide analysis of the MYB-related transcription factor family and associated responses to abiotic stressors in Populus. Int. J. Biol. Macromol. 2021, 191, 359–376. [Google Scholar] [CrossRef] [PubMed]
- Hao, S.X.; Lu, Y.F.; Peng, Z.; Wang, E.Y.; Chao, L.K.; Zhong, S.L.; Yao, Y.C. McMYB4 improves temperature adaptation by regulating phenylpropanoid metabolism and hormone signaling in apple. Hortic. Res. 2021, 8, 182. [Google Scholar] [CrossRef] [PubMed]
- Aslam, M.; Greaves, J.G.; Jakada, B.H.; Fakher, B.; Wang, X.M.; Qin, Y. AcCIPK5, a pineapple CBL-interacting protein kinase, confers salt, osmotic and cold stress tolerance in transgenic Arabidopsis. Plant Sci. 2022, 320, 111284. [Google Scholar] [CrossRef]
- Gupta, P.K.; Balyan, H.S.; Sharma, S.; Kumar, R. Genetics of yield, abiotic stress tolerance and biofortification in wheat (Triticum aestivum L.). Theor. Appl. Genet. 2020, 133, 1569–1602. [Google Scholar] [CrossRef] [PubMed]
- Castro, B.; Citterico, M.; Kimura, S.; Stevens, D.M.; Wrzaczek, M.; Coaker, G. Stress-induced reactive oxygen species compartmentalization, perception and signalling. Nat. Plants 2021, 7, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Markham, K.K.; Greenham, K. Abiotic stress through time. New Phytol. 2021, 231, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.M.; Yang, H.L.; Fang, Y.S.; Guo, W.; Chen, H.F.; Zhang, X.J.; Dai, W.J.; Chen, S.L.; Hao, Q.N.; Yuan, S.L.; et al. Overexpression of GmMYB14 improves high-density yield and drought tolerance of soybean through regulating plant architecture mediated by the brassinosteroid pathway. Plant Biotechnol. J. 2021, 19, 702–716. [Google Scholar] [CrossRef] [PubMed]
- Emenecker, R.J.; Holehouse, A.S.; Strader, L.C. Biological phase separation and biomolecular condensates in plants. Annu. Rev. Plant Biol. 2021, 72, 17–46. [Google Scholar] [CrossRef]
- Xu, Y.J.; Xu, H.X.; Wall, M.M.; Yang, J.Z. Roles of transcription factor SQUAMOSA promoter binding protein-like gene family in papaya (Carica papaya) development and ripening. Genomics 2020, 112, 2734–2747. [Google Scholar] [CrossRef]
- Zhao, Y.; Yang, Z.; Ding, Y.; Liu, L.; Han, X.; Zhan, J.; Wei, X.; Diao, Y.; Qin, W.; Wang, P.; et al. Over-expression of an R2R3 MYB gene, GhMYB73, increases tolerance to salt stress in transgenic Arabidopsis. Plant Sci. 2019, 286, 28–36. [Google Scholar] [CrossRef]
- Liu, Z.Y.; Li, X.P.; Zhang, T.Q.; Wang, Y.Y.; Wang, C.; Gao, C.Q. Overexpression of ThMYB8 mediates salt stress tolerance by directly activating stress-responsive gene expression. Plant Sci. 2021, 302, 110668. [Google Scholar] [CrossRef]
- Wen, X.F.; Geng, F.; Cheng, Y.J.; Wang, J.Q. Ectopic expression of CsMYB30 from Citrus sinensis enhances salt and drought tolerance by regulating wax synthesis in Arabidopsis thaliana. Plant Physiol. Biochem. 2021, 166, 777–788. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Wang, L.; Lei, J.; Chai, S.; Jin, X.; Zou, Y.; Sun, X.; Mei, Y.; Cheng, X.; Yang, X.; et al. IbMYB308, a Sweet Potato R2R3-MYB Gene, Improves Salt Stress Tolerance in Transgenic Tobacco. Genes 2022, 13, 1476. https://doi.org/10.3390/genes13081476
Wang C, Wang L, Lei J, Chai S, Jin X, Zou Y, Sun X, Mei Y, Cheng X, Yang X, et al. IbMYB308, a Sweet Potato R2R3-MYB Gene, Improves Salt Stress Tolerance in Transgenic Tobacco. Genes. 2022; 13(8):1476. https://doi.org/10.3390/genes13081476
Chicago/Turabian StyleWang, Chong, Lianjun Wang, Jian Lei, Shasha Chai, Xiaojie Jin, Yuyan Zou, Xiaoqiong Sun, Yuqin Mei, Xianliang Cheng, Xinsun Yang, and et al. 2022. "IbMYB308, a Sweet Potato R2R3-MYB Gene, Improves Salt Stress Tolerance in Transgenic Tobacco" Genes 13, no. 8: 1476. https://doi.org/10.3390/genes13081476
APA StyleWang, C., Wang, L., Lei, J., Chai, S., Jin, X., Zou, Y., Sun, X., Mei, Y., Cheng, X., Yang, X., Jiao, C., & Tian, X. (2022). IbMYB308, a Sweet Potato R2R3-MYB Gene, Improves Salt Stress Tolerance in Transgenic Tobacco. Genes, 13(8), 1476. https://doi.org/10.3390/genes13081476




