Prediction of the Effects of Missense Mutations on Human Myeloperoxidase Protein Stability Using In Silico Saturation Mutagenesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Sequence Selection and Structure Preparation
2.2. Saturated Computational Mutagenesis and Structure-Based Energy Calculation
2.3. Sequence-Based Prediction of PTM Sites
2.4. Sequence-Based Pathogenicity Prediction
2.5. Detection of PTM-SNPs
3. Results
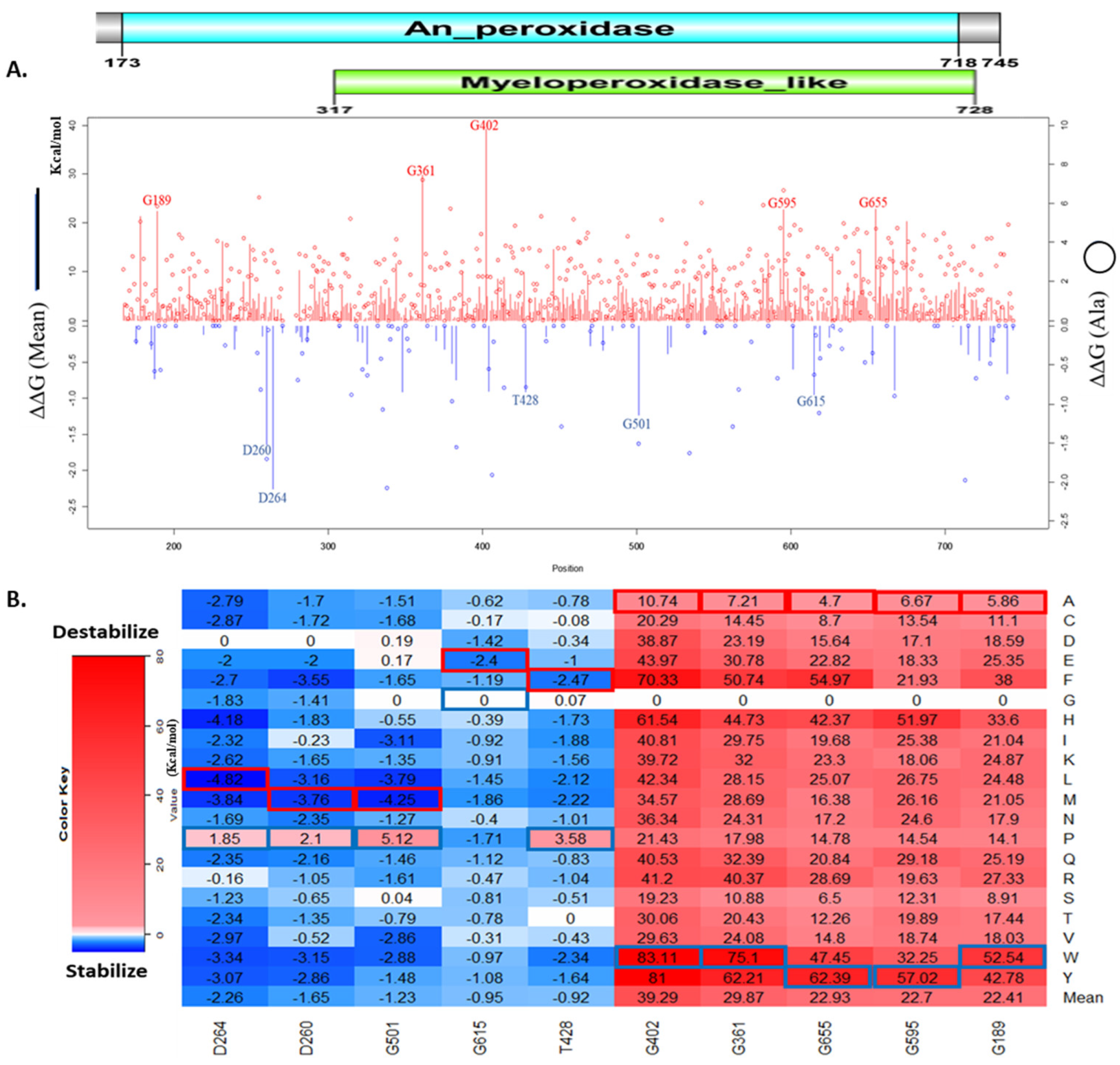
3.1. Analysis of Overall Computed Missense Mutations
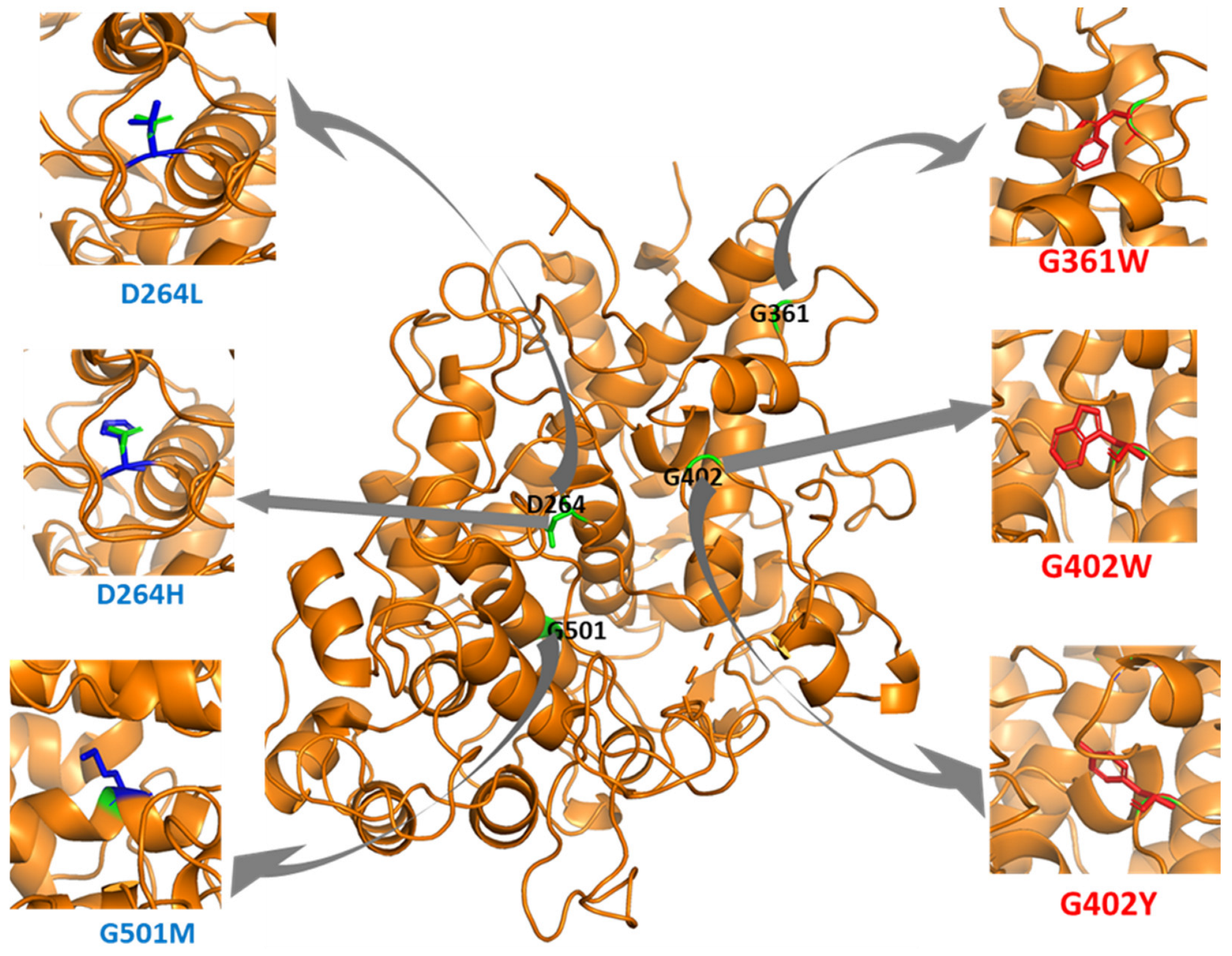
3.2. Top Missense Mutations Affecting MPO Stability
3.3. Sequence-Based Analysis of the Effects of Missense Mutations on MPO Stability
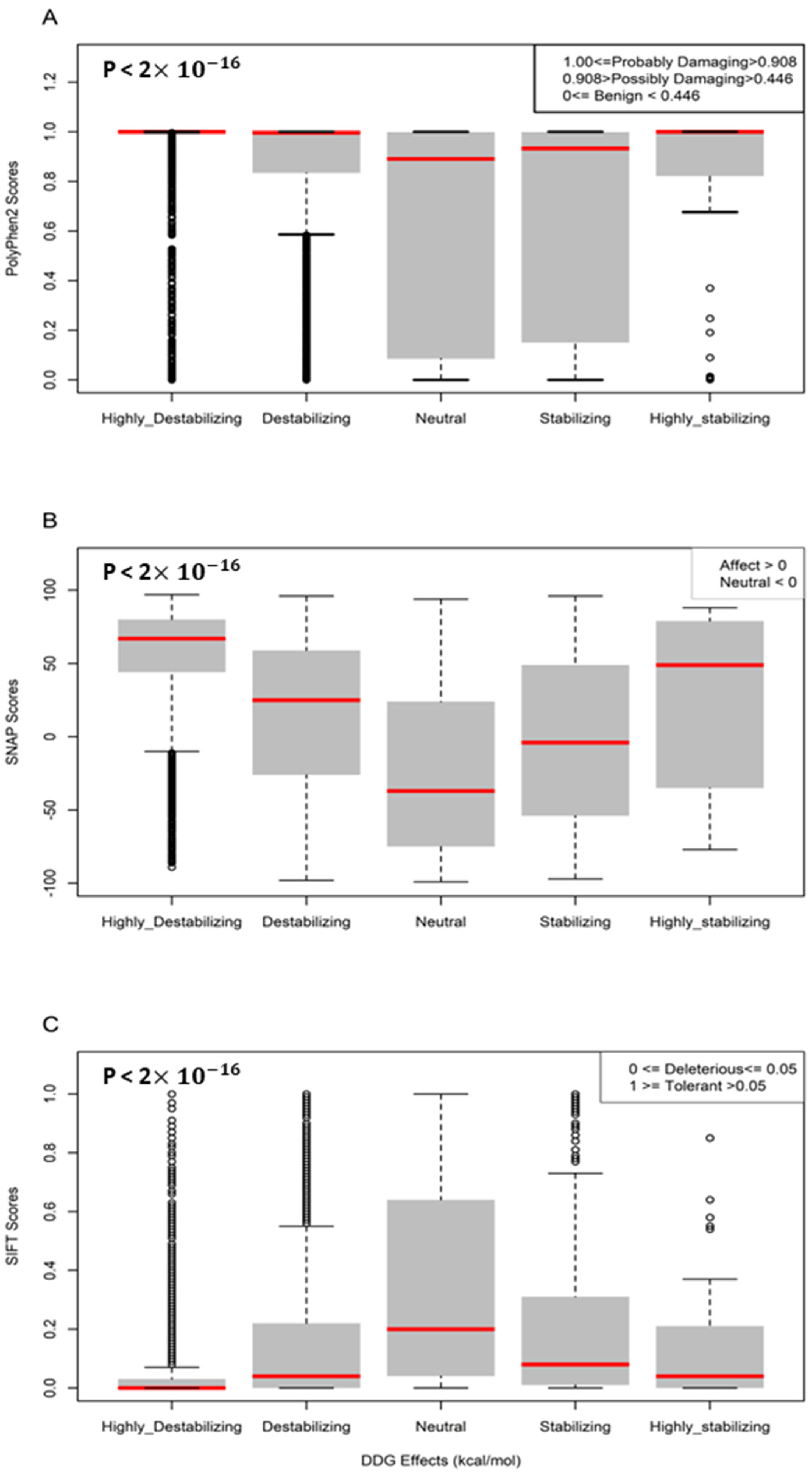
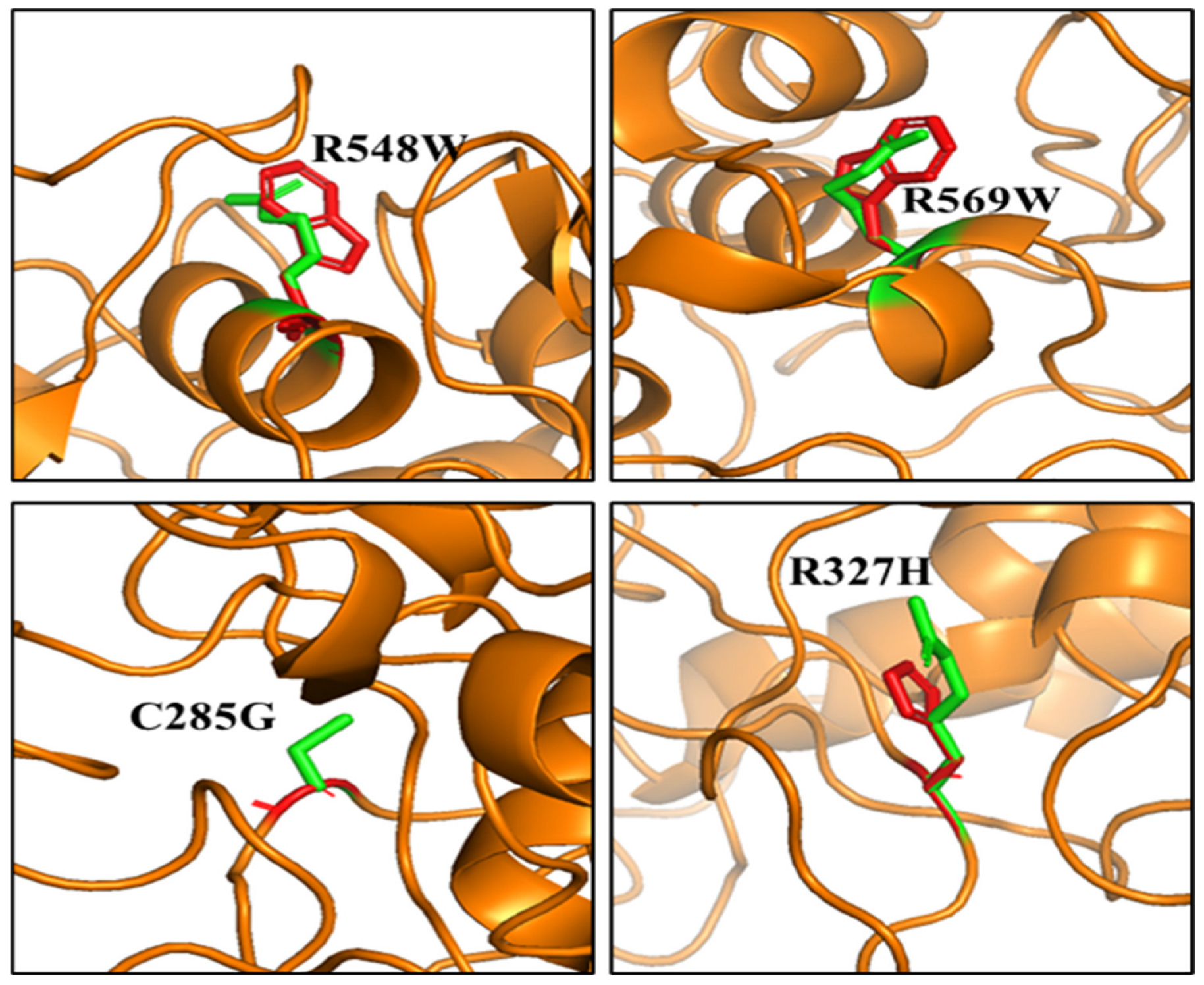
3.4. Computational Predictions of Disease Phenotypes
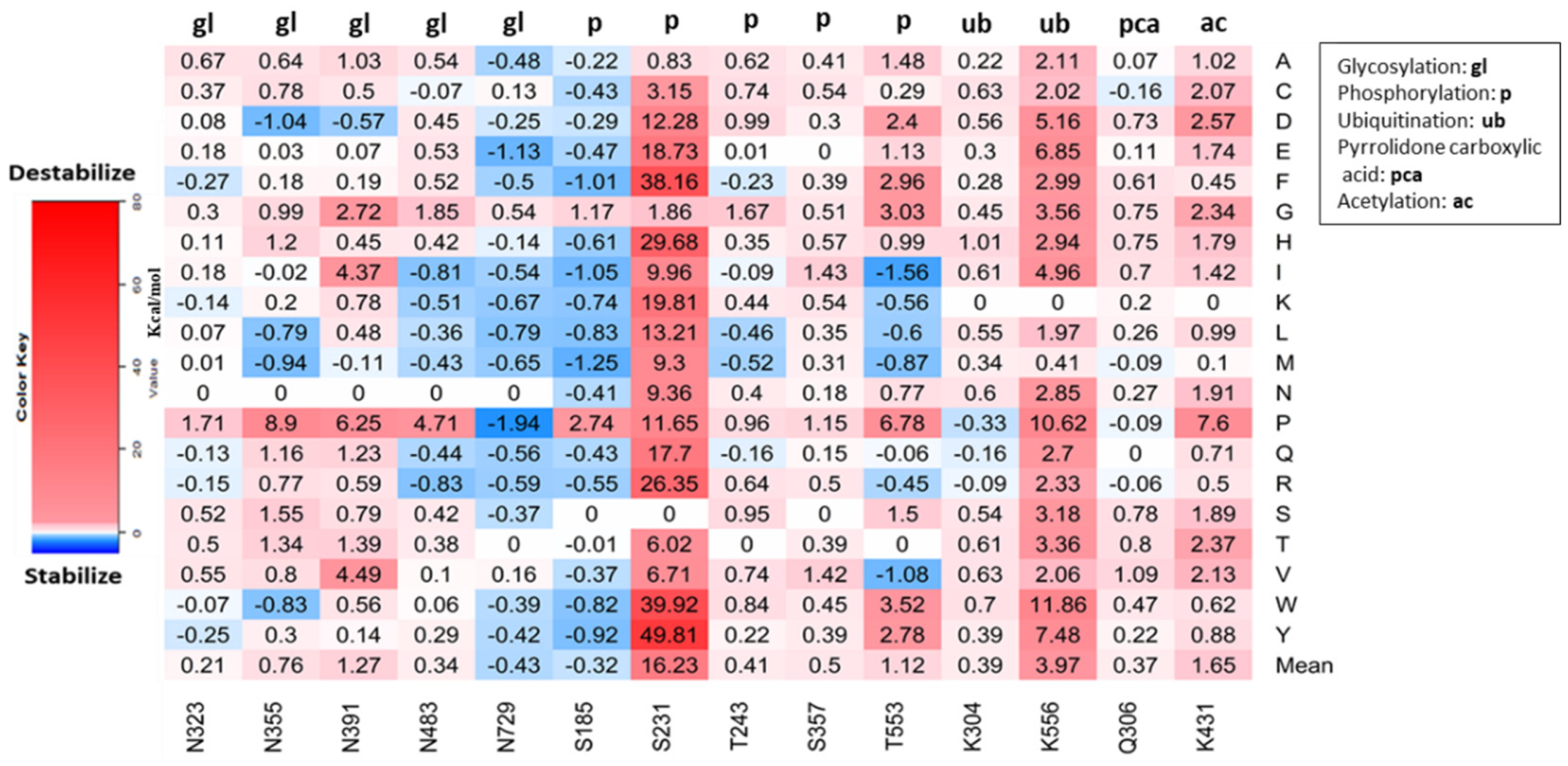
3.5. Effect of Missense Mutations on Post-Translational Modification Sites on MPO
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Aratani, Y. Myeloperoxidase: Its role for host defense, inflammation, and neutrophil function. In Archives of Biochemistry and Biophysics; Academic Press Inc.: Cambridge, MA, USA, 2018; Volume 640, pp. 47–52. [Google Scholar]
- Khan, A.; Alsahli, M.; Rahmani, A. Myeloperoxidase as an Active Disease Biomarker: Recent Biochemical and Pathological Perspectives. Med. Sci. 2018, 6, 33. [Google Scholar] [CrossRef] [PubMed]
- Gorudko, I.V.; Sokolov, A.V.; Shamova, E.V.; Grigorieva, D.V.; Mironova, E.V.; Kudryavtsev, I.V.; Gusev, S.A.; Gusev, A.A.; Chekanov, A.V.; Vasilyev, V.B.; et al. Binding of human myeloperoxidase to red blood cells: Molecular targets and biophysical consequences at the plasma membrane level. Arch. Biochem. Biophys. 2016, 591, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Nauseef, W.M.; McCormick, S.; Goedken, M. Redox Report Communications in Free Radical Research Impact of missense mutations on biosynthesis of myeloperoxidase. Redox Rep. 2013, 5, 187–206. [Google Scholar] [CrossRef]
- Marchetti, C.; Patriarca, P.; Solero, G.P.; Baralle, F.E.; Romano, M. Genetic studies on myeloperoxidase deficiency in Italy. Jpn. J. Infect. Dis. 2004, 57, S10–S12. [Google Scholar]
- Parry, M.F.; Root, R.K.; Metcalf, J.A.; Delaney, K.K.; Kaplow, L.S.; Richar, W.J. Myeloperoxidase deficiency: Prevalence and clinical significance. Ann. Intern. Med. 1981, 95, 293–301. [Google Scholar] [CrossRef]
- Nauseef, W.M.; Cogley, M.; McCormick, S. Effect of the R569W missense mutation on the biosynthesis of myeloperoxidase. J. Biol. Chem. 1996, 271, 9546–9549. [Google Scholar] [CrossRef]
- Vergnano, M.; Grys, K.; Benzian-Olsson, N.; Mahil, S.K.; Chaloner, C.; Barbosa, I.A.; August, S.; Burden, A.D.; Choon, S.E.; Cooper, H.; et al. Loss-of-function myeloperoxidase mutations are associated with increased neutrophil counts and pustular skin disease. Am. J. Hum. Genet. 2020, 107, 539–543. [Google Scholar] [CrossRef]
- Haskamp, S.; Bruns, H.; Hahn, M.; Hoffmann, M.; Gregor, A.; Löhr, S.; Hahn, J.; Schauer, C.; Ringer, M.; Flamann, C.; et al. Myeloperoxidase Modulates Inflammation in Generalized Pustular Psoriasis and Additional Rare Pustular Skin Diseases. Am. J. Hum. Genet. 2020, 107, 527–538. [Google Scholar] [CrossRef]
- Volkman, R.; Ben-Zur, T.; Kahana, A.; Garty, B.Z.; Offen, D. Myeloperoxidase Deficiency Inhibits Cognitive Decline in the 5XFAD Mouse Model of Alzheimer’s Disease. Front. Neurosci. 2019, 13, 990. [Google Scholar] [CrossRef]
- Zappia, M.; Manna, I.; Serra, P.; Cittadella, R.; Andreoli, V.; Russa, A.L.; Annesi, F.; Spadafora, P.; Romeo, N.; Nicoletti, G.; et al. Increased Risk for Alzheimer Disease With the Interaction of MPO and A2M Polymorphisms. Arch. Neurol. 2004, 61, 341–344. [Google Scholar] [CrossRef]
- Jiang, Y.H.; Yuen, R.K.C.; Jin, X.; Wang, M.; Chen, N.; Wu, X.; Ju, J.; Mei, J.; Shi, Y.; He, M.; et al. Detection of clinically relevant genetic variants in autism spectrum disorder by whole-genome sequencing. Am. J. Hum. Genet. 2013, 93, 249–263. [Google Scholar] [CrossRef] [PubMed]
- Pagel, K.A.; Pejaver, V.; Lin, G.N.; Nam, H.-J.; Mort, M.; Cooper, D.N.; Sebat, J.; Iakoucheva, L.M.; Mooney, S.D.; Radivojac, P. When loss-of-function is loss of function: Assessing mutational signatures and impact of loss-of-function genetic variants. Bioinformatics 2017, 33, i389–i398. [Google Scholar] [CrossRef]
- Antwerpen, P.V.; Slomianny, M.-C.; Boudjeltia, K.Z.; Delporte, C.; Faid, V.; Calay, D.; Rousseau, A.; Moguilevsky, N.; Raes, M.; Vanhamme, L.; et al. Glycosylation Pattern of Mature Dimeric Leukocyte and Recombinant Monomeric Myeloperoxidase: Glycosylation is required for optimal enzymatic activity. J. Biol. Chem. 2010, 285, 16351–16359. [Google Scholar] [CrossRef] [PubMed]
- Reiding, K.R.; Franc, V.; Huitema, M.G.; Brouwer, E.; Heeringa, P.; Heck, A.J.R. Neutrophil myeloperoxidase harbors distinct site-specific peculiarities in its glycosylation. J. Biol. Chem. 2019, 294, 20233–20245. [Google Scholar] [CrossRef] [PubMed]
- Cieutat, A.M.; Lobel, P.; August, J.T.; Kjeldsen, L.; Sengeløv, H.; Borregaard, N.; Bainton, D.F. Azurophilic Granules of Human Neutrophilic Leukocytes Are Deficient in Lysosome-Associated Membrane Proteins but Retain the Mannose 6-Phosphate Recognition Marker. Blood 1998, 91, 1044–1058. [Google Scholar] [CrossRef]
- UniProt, C. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Bateman, A. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Fiedler, T.J.; Davey, C.A.; Fenna, R.E. X-ray crystal structure and characterization of halide-binding sites of human myeloperoxidase at 1.8 A resolution. J. Biol. Chem. 2000, 275, 11964–11971. [Google Scholar] [CrossRef]
- Delgado, J.; Radusky, L.G.; Cianferoni, D.; Serrano, L. FoldX 5.0: Working with RNA, small molecules and a new graphical interface. Bioinformatics 2019, 35, 4168–4169. [Google Scholar] [CrossRef]
- Buß, O.; Rudat, J.; Ochsenreither, K. FoldX as Protein Engineering Tool: Better Than Random Based Approaches? Comput. Struct. Biotechnol. J. 2018, 16, 25–33. [Google Scholar] [CrossRef]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX web server: An online force field. Nucleic Acids Res. 2005, 33, W382–W388. [Google Scholar] [CrossRef] [PubMed]
- Bromberg, Y.; Rost, B. Correlating protein function and stability through the analysis of single amino acid substitutions. BMC Bioinform. 2009, 10, S8. [Google Scholar] [CrossRef]
- Steentoft, C.; Vakhrushev, S.Y.; Joshi, H.J.; Kong, Y.; Vester-Christensen, M.B.; Schjoldager, K.T.; Lavrsen, K.; Dabelsteen, S.; Pedersen, N.B.; Marcos-Silva, L.; et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013, 32, 1478–1488. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Brunak, S. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac. Symp. Biocomput. 2002, 7, 310–322. [Google Scholar]
- Wang, D.; Liu, D.; Yuchi, J.; He, F.; Jiang, Y.; Cai, S.; Li, J.; Xu, D. MusiteDeep: A deep-learning based webserver for protein post-translational modification site prediction and visualization. Nucleic Acids Res. 2020, 48, W140–W146. [Google Scholar] [CrossRef]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. PolyPhen-2: Prediction of functional effects of human nsSNPs. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef]
- Sim, N.-L.; Kumar, P.; Hu, J.; Henikoff, S.; Schneider, G.; Ng, P.C. SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012, 40, W452–W457. [Google Scholar] [CrossRef]
- Bromberg, Y.; Rost, B. SNAP: Predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007, 35, 3823–3835. [Google Scholar] [CrossRef]
- Liu, X.; Li, C.; Mou, C.; Dong, Y.; Tu, Y. dbNSFP v4: A comprehensive database of transcript-specific functional predictions and annotations for human nonsynonymous and splice-site SNVs. Genome Med. 2020, 12, 103. [Google Scholar] [CrossRef]
- Fenna, R.; Zeng, J.; Davey, C. Structure of the Green Heme in Myeloperoxidase. Arch. Biochem. Biophys. 1995, 316, 653–656. [Google Scholar] [CrossRef]
- Grayson, P.C.; Kaplan, M.J. At the Bench: Neutrophil extracellular traps (NETs) highlight novel aspects of innate immune system involvement in autoimmune diseases. J. Leukoc. Biol. 2016, 99, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Odobasic, D.; Yang, Y.; Muljadi, R.C.M.; O’Sullivan, K.M.; Kao, W.; Smith, M.; Morand, E.F.; Holdsworth, S.R. Endogenous myeloperoxidase is a mediator of joint inflammation and damage in experimental arthritis. Arthritis Rheumatol. 2014, 66, 907–917. [Google Scholar] [CrossRef] [PubMed]
- Ndrepepa, G. Myeloperoxidase—A bridge linking inflammation and oxidative stress with cardiovascular disease. Clin. Chim. Acta Int. J. Clin. Chem. 2019, 493, 36–51. [Google Scholar] [CrossRef] [PubMed]
- Matreyek, K.A.; Starita, L.M.; Stephany, J.J.; Martin, B.; Chiasson, M.A.; Gray, V.E.; Kircher, M.; Khechaduri, A.; Dines, J.N.; Hause, R.J.; et al. Multiplex Assessment of Protein Variant Abundance by Massively Parallel Sequencing. Nat. Genet. 2018, 50, 874–882. [Google Scholar] [CrossRef]
- Bloom, J.D.; Labthavikul, S.T.; Otey, C.R.; Arnold, F.H. Protein stability promotes evolvability. Proc. Natl. Acad. Sci. USA 2006, 103, 5869–5874. [Google Scholar] [CrossRef]
- Post, K.L.; Belmadani, M.; Ganguly, P.; Meili, F.; Dingwall, R.; McDiarmid, T.A.; Meyers, W.M.; Herrington, C.; Young, B.P.; Callaghan, D.B.; et al. Multi-model functionalization of disease-associated PTEN missense mutations identifies multiple molecular mechanisms underlying protein dysfunction. Nat. Commun. 2020, 11, 2073. [Google Scholar] [CrossRef]
- Hasnain, M.J.U.; Amin, F.; Ghani, A.; Ahmad, S.; Rehman, Z.; Aslam, T.; Pervez, M.T. Structural and Functional Impact of Damaging Nonsynonymous Single Nucleotide Polymorphisms (nsSNPs) on Human VPS35 Protein Using Computational Approaches. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021, 1. [Google Scholar] [CrossRef]
- Pandey, S.; Dhusia, K.; Katara, P.; Singh, S.; Gautam, B. An in silico analysis of deleterious single nucleotide polymorphisms and molecular dynamics simulation of disease linked mutations in genes responsible for neurodegenerative disorder. J. Biomol. Struct. Dyn. 2020, 38, 4259–4272. [Google Scholar] [CrossRef]
- Sobitan, A.; Mahase, V.; Rhoades, R.; Williams, D.; Liu, D.; Xie, Y.; Li, L.; Tang, Q.; Teng, S. Computational Saturation Mutagenesis of SARS-CoV-1 Spike Glycoprotein: Stability, Binding Affinity, and Comparison With SARS-CoV-2. Front. Mol. Biosci. 2021, 8, 784303. [Google Scholar] [CrossRef]
- Iossifov, I.; O’Roak, B.J.; Sanders, S.J.; Ronemus, M.; Krumm, N.; Levy, D.; Stessman, H.A.; Witherspoon, K.T.; Vives, L.; Patterson, K.E.; et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 2014, 515, 216–221. [Google Scholar] [CrossRef]
- Lim, E.T.; Uddin, M.; De Rubeis, S.; Chan, Y.; Kamumbu, A.S.; Zhang, X.; D’Gama, A.M.; Kim, S.N.; Hill, R.S.; Goldberg, A.P.; et al. Rates, distribution and implications of postzygotic mosaic mutations in autism spectrum disorder. Nat. Neurosci. 2017, 20, 1217–1224. [Google Scholar] [CrossRef] [PubMed]
- DeLeo, F.R.; Goedken, M.; McCormick, S.J.; Nauseef, W.M. A novel form of hereditary myeloperoxidase deficiency linked to endoplasmic reticulum/proteasome degradation. J. Clin. Investig. 1998, 101, 2900–2909. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Romano, M.; Dri, P.; Dadalt, L.; Patriarca, P.; Baralle, F.E. Biochemical and Molecular Characterization of Hereditary Myeloperoxidase Deficiency. Blood 1997, 90, 4126–4134. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Liu, X.; Shen, J.; Tian, W.; Fang, R.; Li, B.; Ma, J.; Cao, L.; Chen, S.; Li, G.; et al. The Whole Exome Sequencing Clarifies the Genotype- Phenotype Correlations in Patients with Early-Onset Dementia. Aging Dis. 2018, 9, 696–705. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, C.; Patriarca, P.; Solero, G.P.; Baralle, F.E.; Romano, M. Genetic Characterization of Myeloperoxidase Deficiency in Italy. Hum. Mutat. 2004, 23, 496–505. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, Y.Y.; Kameoka, Y.; Persad, A.S.; Koi, F.; Yamagoe, S.; Hashimoto, K.; Suzuki, K. Novel missense mutation found in a Japanese patient with myeloperoxidase deficiency. Gene 2004, 327, 195–200. [Google Scholar] [CrossRef]
- Kameoka, Y.; Persad, A.S.; Suzuki, K. Genomic variations in myeloperoxidase gene in the Japanese population. Jpn. J. Infect. Dis. 2004, 57, S12–S13. [Google Scholar]
- Mishra, O.P.; Popov, A.V.; Pietrofesa, R.A.; Hwang, W.T.; Andrake, M.; Nakamaru-Ogiso, E.; Christofidou-Solomidou, M. Radiation Activates Myeloperoxidase (MPO) to Generate Active Chlorine Species (ACS) via a Dephosphorylation Mechanism—Inhibitory Effect of LGM2605. Biochim. Biophys. Acta. Gen. Subj. 2020, 1864, 129548. [Google Scholar] [CrossRef]
- Kim, Y.; Kang, C.; Min, B.; Yi, G.S. Detection and analysis of disease-associated single nucleotide polymorphism influencing post-translational modification. BMC Med. Genom. 2015, 8, S7. [Google Scholar] [CrossRef]

| Structure-Based Prediction (Protein Stability) | Sequence-Based Prediction (Mutation Pathogenicity) | |||||||
|---|---|---|---|---|---|---|---|---|
| FoldX | PolyPhen2 | SIFT | SNAP | |||||
| Missense Mutations | ΔΔG (Kcal/mol) |
Mutation Effect | Score | Effect | Score | Effect | Score | Effect |
| G402W | 83.11 | H_Des | 1 | Pro_d | 0 | Del | 80 | Aff |
| G402Y | 81 | H_Des | 1 | Pro_d | 0 | Del | 75 | Aff |
| G361W | 75.1 | H_Des | 1 | Pro_d | 0 | Del | 93 | Aff |
| G402F | 70.33 | H_Des | 1 | Pro_d | 0 | Del | 77 | Aff |
| G655Y | 62.39 | H_Des | 1 | Pro_d | 0.01 | Del | 84 | Aff |
| D264L | −4.82 | H_Sta | 0.972 | Pro_d | 0.11 | Tol | 20 | Aff |
| G501M | −4.25 | H_Sta | 1 | Pro_d | 0 | Del | 85 | Aff |
| D264H | −4.18 | H_Sta | 0.191 | B | 0.09 | Tol | 11 | Aff |
| D264M | −3.84 | H_Sta | 0.995 | Pro_d | 0.05 | Del | 44 | Aff |
| G501L | −3.79 | H_Sta | 1 | Pro_d | 0 | Del | 84 | Aff |
| HGMD | Structure-Based Prediction (Protein Stability) | Sequence-Based Prediction (Mutation Pathogenicity) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| FoldX | PolyPhen2 | SIFT | SNAP | ||||||
| Missense Mutations | Phenotype | ΔΔG (Kcal/mol) | Mutation Effect | Score | Damaging Effect | Score | Effect | Score | Effect |
| R548W | GPP | 11.22 | H_Des | 1 | Pro_d | 0 | Del | 84 | Aff |
| R569W | MPOD | 7.84 | H_Des | 1 | Pro_d | 0.04 | Del | 71 | Aff |
| W643R | MPOD | 5.92 | H_Des | 1 | Pro_d | 0 | Del | 93 | Aff |
| M251T | MPOD | 4.81 | H_Des | 0.032 | B | 0.01 | Del | 35 | Aff |
| R327H | FTD | 4.72 | H_Des | 0.986 | Pro_d | 0 | Del | 57 | Aff |
| C285G | ASD | 3.31 | H_Des | 0.239 | B | 0 | Del | 74 | Aff |
| Y173C | MPOD | 3.08 | H_Des | 0.995 | Pro_d | 0 | Del | 71 | Aff |
| D371G | MPOD | 2.25 | Des | 0.999 | Pro_d | 0.12 | Tol | 64 | Aff |
| R499C | MPOD | 1.62 | Des | 0.999 | Pro_d | 0 | Del | 62 | Aff |
| R590C | GPP | 1.32 | Des | 1 | Pro_d | 0 | Del | 39 | Aff |
| D403E | ASD | 1.27 | Des | 1 | Pro_d | 0 | Del | 13 | Aff |
| A332V | MPOD | 1.07 | Des | 0.059 | B | 0.1 | Tol | −26 | N |
| L572W | MPOD | 0.51 | Des | 1 | Pro_d | 0 | Del | 73 | Aff |
| G501S | MPOD | 0.04 | N | 0.938 | Pro_d | 0 | Del | 23 | Aff |
| rs_dbSNP151 | Missense SNP | PTM SITE | PEPTIDE | DISTANCE APART (PTM ↔ SNP SITE) | PTM TYPE | Phenotype |
|---|---|---|---|---|---|---|
| rs760619802 | R327H | N323 | GSNITIRNQI | +4 | N-LINKED GLYCOSYLATION | FTD |
| rs148802625 | R548W | T553 | ILRGLMATPA | −5 | PHOSPHORYLATION | GPP |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sobitan, A.; Edwards, W.; Jalal, M.S.; Kolawole, A.; Ullah, H.; Duttaroy, A.; Li, J.; Teng, S. Prediction of the Effects of Missense Mutations on Human Myeloperoxidase Protein Stability Using In Silico Saturation Mutagenesis. Genes 2022, 13, 1412. https://doi.org/10.3390/genes13081412
Sobitan A, Edwards W, Jalal MS, Kolawole A, Ullah H, Duttaroy A, Li J, Teng S. Prediction of the Effects of Missense Mutations on Human Myeloperoxidase Protein Stability Using In Silico Saturation Mutagenesis. Genes. 2022; 13(8):1412. https://doi.org/10.3390/genes13081412
Chicago/Turabian StyleSobitan, Adebiyi, William Edwards, Md Shah Jalal, Ayanfeoluwa Kolawole, Hemayet Ullah, Atanu Duttaroy, Jiang Li, and Shaolei Teng. 2022. "Prediction of the Effects of Missense Mutations on Human Myeloperoxidase Protein Stability Using In Silico Saturation Mutagenesis" Genes 13, no. 8: 1412. https://doi.org/10.3390/genes13081412
APA StyleSobitan, A., Edwards, W., Jalal, M. S., Kolawole, A., Ullah, H., Duttaroy, A., Li, J., & Teng, S. (2022). Prediction of the Effects of Missense Mutations on Human Myeloperoxidase Protein Stability Using In Silico Saturation Mutagenesis. Genes, 13(8), 1412. https://doi.org/10.3390/genes13081412







