Mosaic Evolution of Molecular Pathways for Sex Pheromone Communication in a Butterfly
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insects
2.2. Transcriptome
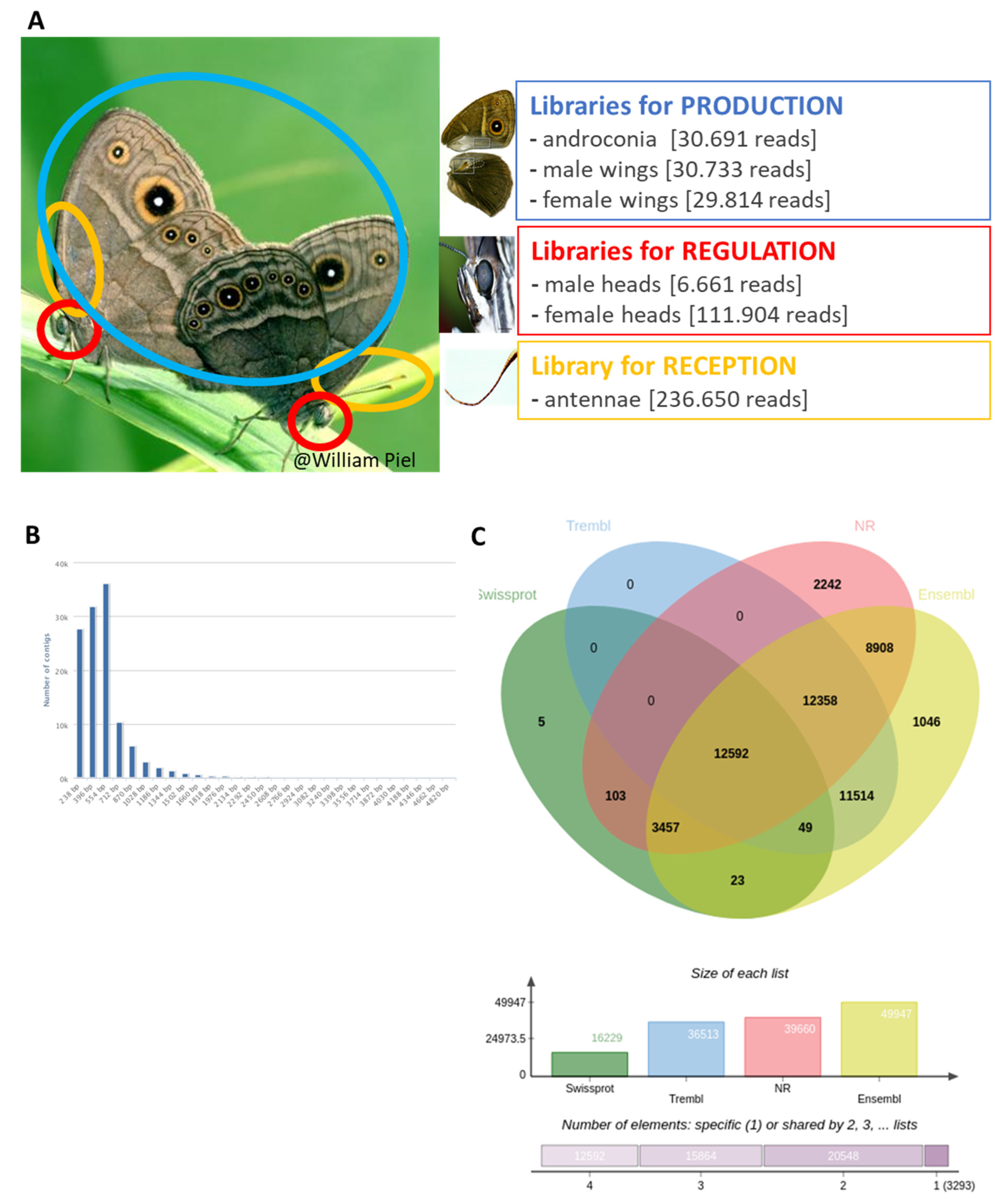
2.2.1. Tissue Collection
2.2.2. RNA Extraction
2.2.3. mRNA Isolation, cDNA Synthesis and Sequencing
2.2.4. Transcriptome Assembly, Quantification, and Annotation
2.2.5. Candidate Gene Identification Using Transcriptome Sequencing
2.2.6. Identification of Specific Gene Families
2.3. Phylogenies
2.4. Real Time Quantitative PCR
2.5. Behavioral Experiments
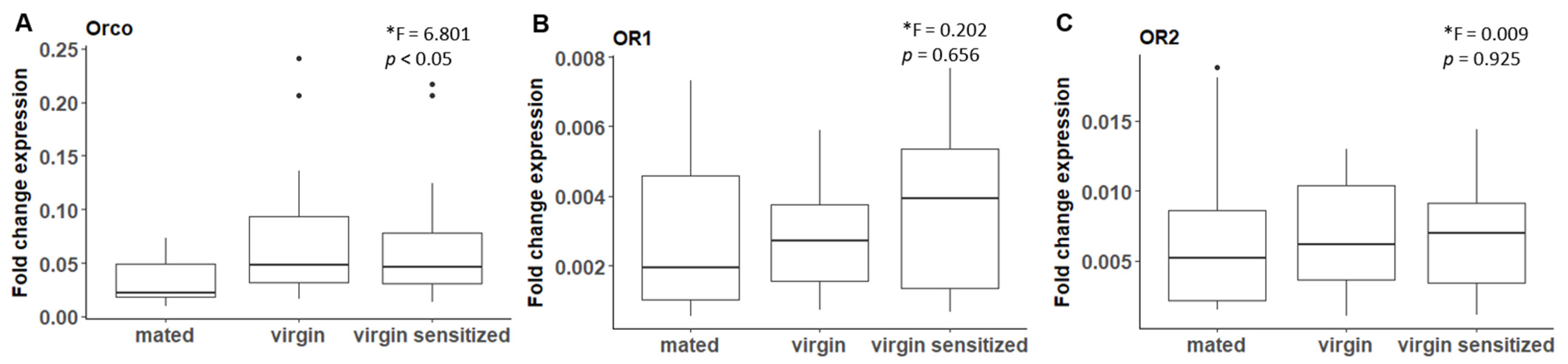
2.5.1. Mating Experiments for Quantifying Odorant Receptor Expression Levels
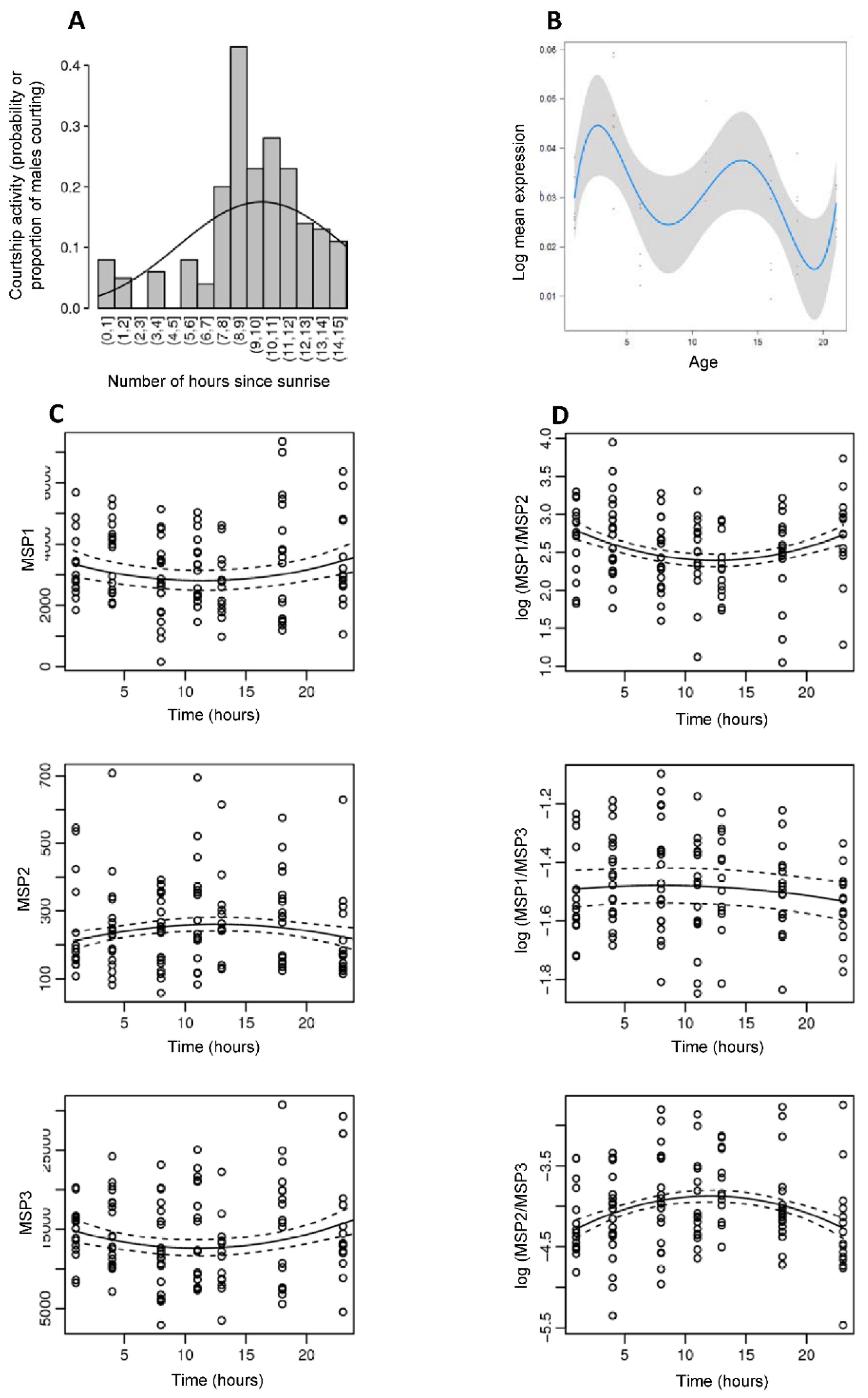
2.5.2. Daily Variation in Courtship Activity
2.5.3. Daily Variation in Male Sex Pheromone Production
2.6. Quantification of Male Sex Pheromone Production
3. Results and Discussion
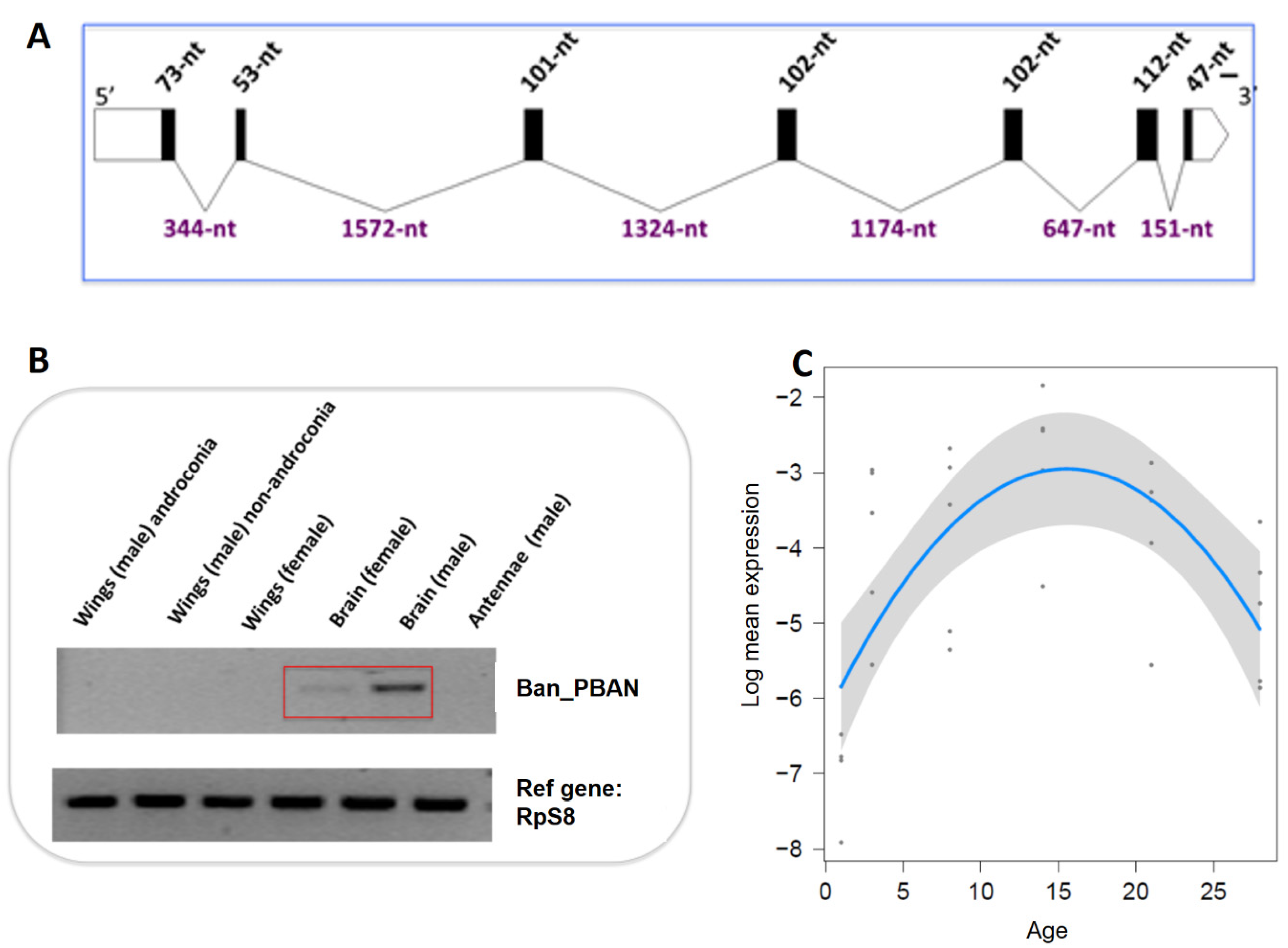
3.1. B. anynana Sex Pheromone Biosynthesis
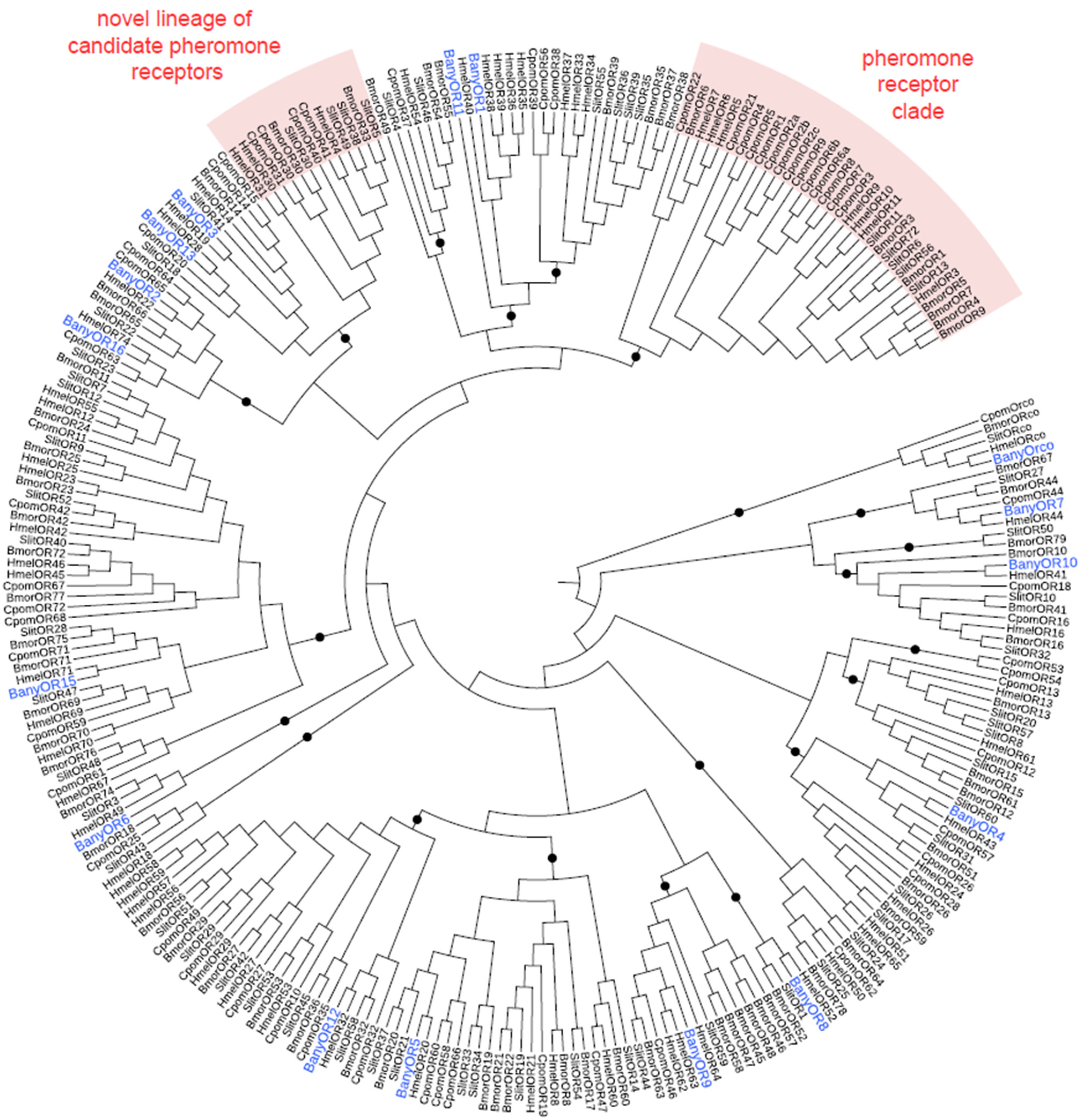
3.2. B. anynana Sex Pheromone Reception
3.3. B. anynana Sex Pheromone Regulation
4. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Cracraft, J. Mandible of Archaeopteryx Provides an Example of Mosaic Evolution. Nature 1970, 226, 1268. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Robles, A.; Hopkins, W.D.; Sherwood, C.C. Modular Structure Facilitates Mosaic Evolution of the Brain in Chimpanzees and Humans. Nat. Commun. 2014, 5, 4469. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Currie, P.; Pittman, M.; Xing, L.; Meng, Q.; Lü, J.; Hu, D.; Yu, C. Mosaic Evolution in an Asymmetrically Feathered Troodontid Dinosaur with Transitional Features. Nat. Commun. 2017, 8, 14972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daeschler, E.B.; Shubin, N.H.; Jenkins, F.A. A Devonian Tetrapod-like Fish and the Evolution of the Tetrapod Body Plan. Nature 2006, 440, 757–763. [Google Scholar] [CrossRef]
- van Gestel, J.; Ackermann, M.; Wagner, A. Microbial Life Cycles Link Global Modularity in Regulation to Mosaic Evolution. Nat. Ecol. Evol. 2019, 3, 1184–1196. [Google Scholar] [CrossRef] [PubMed]
- Fernández, R.; Gabaldón, T. Gene Gain and Loss across the Metazoan Tree of Life. Nat. Ecol. Evol. 2020, 4, 524–533. [Google Scholar] [CrossRef]
- Guijarro-Clarke, C.; Holland, P.W.H.; Paps, J. Widespread Patterns of Gene Loss in the Evolution of the Animal Kingdom. Nat. Ecol. Evol. 2020, 4, 519–523. [Google Scholar] [CrossRef]
- Berner, D.; Salzburger, W. The Genomics of Organismal Diversification Illuminated by Adaptive Radiations. Trends Genet. 2015, 31, 491–499. [Google Scholar] [CrossRef]
- Stryjewski, K.F.; Sorenson, M.D. Mosaic Genome Evolution in a Recent and Rapid Avian Radiation. Nat. Ecol. Evol. 2017, 1, 1912–1922. [Google Scholar] [CrossRef] [PubMed]
- Marques, D.; Meier, J.; Seehausen, O. A Combinatorial View on Speciation and Adaptive Radiation. Trends Ecol. Evol. 2019, 34, 531–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shubin, N.; Tabin, C.; Carroll, S. Deep Homology and the Origins of Evolutionary Novelty. Nature 2009, 457, 818–823. [Google Scholar] [CrossRef]
- Shirai, L.T.; Saenko, S.V.; Keller, R.A.; Jeránimo, M.A.; Brakefield, P.M.; Descimon, H.; Wahlberg, N.; Beldade, P. Evolutionary History of the Recruitment of Conserved Developmental Genes in Association to the Formation and Diversification of a Novel Trait. BMC Evol. Biol. 2012, 12, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espeland, M.; Breinholt, J.; Willmott, K.R.; Warren, A.D.; Vila, R.; Toussaint, E.F.A.; Maunsell, S.C.; Aduse-Poku, K.; Talavera, G.; Eastwood, R.; et al. A Comprehensive and Dated Phylogenomic Analysis of Butterflies. Curr. Biol. 2018, 28, 770–778.e5. [Google Scholar] [CrossRef] [Green Version]
- Myers, J. Pheromones and Courtship Behavior in Butterflies. Am. Zool. 1972, 12, 545–551. [Google Scholar] [CrossRef]
- Birch, M.C.; Poppy, G.M.; Baker, T.C. Scents and Eversible Scent Structures of Male Moths. Annu. Rev. Entomol. 1990, 35, 25–58. [Google Scholar] [CrossRef]
- Vane-Wright, R.I.; Boppre, M. Visual and Chemical Signalling in Butterflies: Functional and Phylogenetic Perspectives. Philos. Trans. R. Soc. London B 1993, 340, 197–205. [Google Scholar]
- Nieberding, C.M.; de Vos, H.; Schneider, M.V.; Lassance, J.M.; Estramil, N.; Andersson, J.; Bång, J.; Hedenström, E.; Löfstedt, C.; Brakefield, P.M. The Male Sex Pheromone of the Butterfly Bicyclus Anynana: Towards an Evolutionary Analysis. PLoS ONE 2008, 3, e2751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarto Monteys, V.I.; Quero, C.; Santa-Cruz, M.C.; Rosell, G.; Guerrero, A. Sexual Communication in Day-Flying Lepidoptera with Special Reference to Castniids or “Butterfly-Moths. ” Bull. Entomol. Res. 2016, 106, 421–431. [Google Scholar] [CrossRef] [Green Version]
- Smadja, C.; Butlin, R.K. On the Scent of Speciation: The Chemosensory System and Its Role in Premating Isolation. Heredity (Edinb). 2009, 102, 77–97. [Google Scholar] [CrossRef] [Green Version]
- Wyatt, T.D. Pheromones and Animal Behaviour: Chemical Signals and Signatures, 2nd ed.; Cambridge University Press: Cambridge, UK, 2014. [Google Scholar]
- Moore, P.J.; Reagan-Wallin, N.L.; Haynes, K.F.; Moore, A.J. Odour Conveys Status on Cockroaches. Nature 1997, 389, 25. [Google Scholar] [CrossRef]
- Svensson, M. Sexual Selection in Moths: The Role of Chemical Communication. Biol. Rev. 1996, 71, 113–135. [Google Scholar] [CrossRef]
- Hansson, B.S.; Stensmyr, M.C. Evolution of Insect Olfaction. Neuron 2011, 72, 698–711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tillman, J.A.; Seybold, S.J.; Jurenka, R.A.; Blomquist, G.J. Insect Pheromones—An Overview of Biosynthesis and Endocrine Regulation. Insect Biochem. Mol. Biol. 1999, 29, 481–514. [Google Scholar] [CrossRef]
- Jurenka, R. Insect Pheromone Biosynthesis. In The Chemistry of Pheromones and Other Semiochemicals I. Topics in Current Chemistry; Schulz, S., Ed.; Springer: Berlin/Heidelberg, Germany, 2004; Volume 239, pp. 97–132. [Google Scholar]
- Blomquist, G.; Jurenka, R.; Schal, C.; Tittiger, C. 12—Pheromone Production: Biochemistry and Molecular Biology. In Insect Endocrinology; Gilbert, L., Ed.; Academic Press: London, UK, 2012; pp. 523–567. [Google Scholar]
- Leal, W.S. Odorant Reception in Insects: Roles of Receptors, Binding Proteins, and Degrading Enzymes. Annu. Rev. Entomol. 2013, 58, 373–391. [Google Scholar] [CrossRef]
- Rafaeli, A. Revelations on the Regulatory Mechanisms in Moth Sex-Pheromone Signals. In Management of Insect Pests to Agriculture: Lessons Learned from Deciphering their Genome, Transcriptome and Proteome; Czosnek, H., Ghanim, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 115–129. ISBN 9783319240497. [Google Scholar]
- Zhang, J.; Walker, W.B.; Wang, G. Pheromone Reception in Moths: From Molecules to Behaviors. In Progress in Molecular Biology and Translational Science; Glatz, R., Ed.; Academic Press: London, UK, 2015; Volume 130, pp. 109–128. [Google Scholar]
- Yew, J.Y.; Chung, H. Insect Pheromones: An Overview of Function, Form, and Discovery. Prog. Lipid Res. 2015, 59, 88–105. [Google Scholar] [CrossRef]
- Helmkampf, M.; Cash, E.; Gadau, J. Evolution of the Insect Desaturase Gene Family with an Emphasis on Social Hymenoptera. Mol. Biol. Evol. 2015, 32, 456–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelosi, P.; Zhou, J.J.; Ban, L.P.; Calvello, M. Soluble Proteins in Insect Chemical Communication. Cell. Mol. Life Sci. 2006, 63, 1658–1676. [Google Scholar] [CrossRef] [PubMed]
- Vieira, F.G.; Sánchez-Gracia, A.; Rozas, J. Comparative genomic analysis of the odorant-binding protein family in 12 Drosophila genomes: Purifying selection and birth-and-death evolution. Genome Biol. 2007, 8, R235. [Google Scholar] [CrossRef] [Green Version]
- de Fouchier, A.; Montagné, N.; Mirabeau, O.; Jacquin-Joly, E. Current Views on the Function and Evolution of Olfactory Receptors in Lepidoptera. Short Views Insect Biochem. Mol. Biol. S 2014, 2, 385–408. [Google Scholar]
- Bastin-Héline, L.; de Fouchier, A.; Cao, S.; Koutroumpa, F.; Caballero-Vidal, G.; Robakiewicz, S.; Monsempes, C.; François, M.C.; Ribeyre, T.; Maria, A.; et al. A Novel Lineage of Candidate Pheromone Receptors for Sex Communication in Moths. eLife 2019, 8, e49826. [Google Scholar] [CrossRef]
- Liénard, M.A.; Wang, H.L.; Lassance, J.M.; Löfstedt, C. Sex Pheromone Biosynthetic Pathways Are Conserved between Moths and the Butterfly Bicyclus Anynana. Nat. Commun. 2014, 5, 3957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costanzo, K.; Monteiro, A. The Use of Chemical and Visual Cues in Female Choice in the Butterfly Bicyclus Anynana. Proc. R. Soc. B Biol. Sci. 2007, 274, 845–851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bacquet, P.M.B.; Brattström, O.; Wang, H.L.; Allen, C.E.; Löfstedt, C.; Brakefield, P.M.; Nieberding, C.M. Selection on Male Sex Pheromone Composition Contributes to Butterfly Reproductive Isolation. Proc. R. Soc. B Biol. Sci. 2015, 282, 20142734. [Google Scholar] [CrossRef] [Green Version]
- Andersson, M. Sexual Selection; Princeton University Press: Princeton, NJ, USA, 1994. [Google Scholar]
- Johansson, B.G.; Jones, T.M. The Role of Chemical Communication in Mate Choice. Biol. Rev. 2007, 82, 265–289. [Google Scholar] [CrossRef] [PubMed]
- Nieberding, C.M.; Fischer, K.; Saastamoinen, M.; Allen, C.E.; Wallin, E.A.; Hedenström, E.; Brakefield, P.M. Cracking the Olfactory Code of a Butterfly: The Scent of Ageing. Ecol. Lett. 2012, 15, 415–424. [Google Scholar] [CrossRef] [PubMed]
- van Bergen, E.; Brakefield, P.M.; Heuskin, S.; Zwaan, B.J.; Nieberding, C.M. The Scent of Inbreeding: A Male Sex Pheromone Betrays Inbred Males. Proc. R. Soc. B Biol. Sci. 2013, 280, 20130102. [Google Scholar] [CrossRef] [Green Version]
- Heuskin, S.; Vanderplanck, M.; Bacquet, P.; Holveck, M.-J.; Kaltenpoth, M.; Engl, T.; Pels, C.; Taverne, C.; Lognay, G.; Nieberding, C.M. The Composition of Cuticular Compounds Indicates Body Parts, Sex and Age in the Model Butterfly Bicyclus Anynana (Lepidoptera). Front. Ecol. Evol. 2014, 2, 37. [Google Scholar] [CrossRef] [Green Version]
- Darragh, K.; Vanjari, S.; Mann, F.; Gonzalez-Rojas, M.F.; Morrison, C.R.; Salazar, C.; Pardo-Diaz, C.; Merrill, R.M.; McMillan, W.O.; Schulz, S.; et al. Male Sex Pheromone Components in Heliconius Butterflies Released by the Androconia Affect Female Choice. PeerJ 2017, 5, e3953. [Google Scholar] [CrossRef] [Green Version]
- Brakefield, P.M.; Filali, E.E.; Van Der Laan, R.; Brueker, C.J.; Saccheri, I.J.; Zwaan, B. Effective Population Size, Reproductive Success and Sperm Precedence in the Butterfly, Bicyclus Anynana, in Captivity. J. Evol. Biol. 2001, 14, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Brakefield, P.; Beldade, P.; Zwaan, B. Dissection of Larval and Pupal Wings from the African Butterfly Bicyclus Anynana. Cold Spring Harb. Protoc. 2009, 2009. [Google Scholar] [CrossRef]
- Jérôme, M.; Noirot, C.; Klopp, C. Assessment of Replicate Bias in 454 Pyrosequencing and a Multi-Purpose Read-Filtering Tool. BMC Res. Notes 2011, 4, 2–5. [Google Scholar] [CrossRef] [Green Version]
- Briscoe, A.D.; Macias-Muñoz, A.; Kozak, K.M.; Walters, J.R.; Yuan, F.; Jamie, G.A.; Martin, S.H.; Dasmahapatra, K.K.; Ferguson, L.C.; Mallet, J.; et al. Female Behaviour Drives Expression and Evolution of Gustatory Receptors in Butterflies. PLoS Genet. 2013, 9, e1003620. [Google Scholar] [CrossRef] [PubMed]
- van Schooten, B.; Jiggins, C.D.; Briscoe, A.D.; Papa, R. Genome-Wide Analysis of Ionotropic Receptors Provides Insight into Their Evolution in Heliconius Butterflies. BMC Genomics 2016, 17, 254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ernst, D.A.; Westerman, E.L. Stage- and Sex-Specific Transcriptome Analyses Reveal Distinctive Sensory Gene Expression Patterns in a Butterfly. BMC Genomics 2021, 22, 584. [Google Scholar] [CrossRef] [PubMed]
- Nowell, R.W.; Elsworth, B.; Oostra, V.; Zwaan, B.J.; Wheat, C.W.; Saastamoinen, M.; Saccheri, I.J.; van’t Hof, A.E.; Wasik, B.R.; Connahs, H.; et al. A High-Coverage Draft Genome of the Mycalesine Butterfly Bicyclus Anynana. Gigascience 2017, 6, gix035. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.L.; He, P.; Zhang, L.; Fang, S.Q.; Dong, S.L.; Zhang, Y.J.; Li, F. Large-Scale Identification of Odorant-Binding Proteins and Chemosensory Proteins from Expressed Sequence Tags in Insects. BMC Genomics 2009, 10, 632. [Google Scholar] [CrossRef] [Green Version]
- Dasmahapatra, K.K.; Walters, J.R.; Briscoe, A.D.; Davey, J.W.; Whibley, A.; Nadeau, N.J.; Zimin, A.V.; Salazar, C.; Ferguson, L.C.; Martin, S.H.; et al. Butterfly Genome Reveals Promiscuous Exchange of Mimicry Adaptations among Species. Nature 2012, 487, 94–98. [Google Scholar]
- Walker, W.B.; Gonzalez, F.; Garczynski, S.F.; Witzgall, P. The Chemosensory Receptors of Codling Moth Cydia Pomonella-Expression in Larvae and Adults. Sci. Rep. 2016, 6, 23518. [Google Scholar] [CrossRef] [Green Version]
- Walker, W.B.; Roy, A.; Anderson, P.; Schlyter, F.; Hansson, B.S.; Larsson, M.C. Transcriptome Analysis of Gene Families Involved in Chemosensory Function in Spodoptera Littoralis (Lepidoptera: Noctuidae). BMC Genomics 2019, 20, 428. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [Green Version]
- Anisimova, M.; Gascuel, O. Approximate Likelihood-Ratio Test for Branches: A Fast, Accurate, and Powerful Alternative. Syst. Biol. 2006, 55, 539–552. [Google Scholar] [CrossRef] [PubMed]
- Poivet, E.; Gallot, A.; Montagné, N.; Glaser, N.; Legeai, F.; Jacquin-Joly, E. A Comparison of the Olfactory Gene Repertoires of Adults and Larvae in the Noctuid Moth Spodoptera Littoralis. PLoS ONE 2013, 8, e60263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogt, R.G.; Große-Wilde, E.; Zhou, J.J. The Lepidoptera Odorant Binding Protein Gene Family: Gene Gain and Loss within the GOBP/PBP Complex of Moths and Butterflies. Insect Biochem. Mol. Biol. 2015, 62, 142–153. [Google Scholar] [CrossRef] [PubMed]
- Arun, A.; Baumlé, V.; Amelot, G.; Nieberding, C.M. Selection and Validation of Reference Genes for QRT-PCR Expression Analysis of Candidate Genes Involved in Olfactory Communication in the Butterfly Bicyclus Anynana. PLoS ONE 2015, 10, e0120401. [Google Scholar] [CrossRef] [PubMed]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Legendre, P.; Legendre, L. Numerical Ecology, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 1998. [Google Scholar]
- Nieberding, C.M.; San Martin, G.; Saenko, S.; Allen, C.E.; Brakefield, P.M.; Visser, B. Sexual Selection Contributes to Partial Restoration of Phenotypic Robustness in a Butterfly. Sci. Rep. 2018, 8, 14315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tupec, M.; Buček, A.; Valterová, I.; Pichová, I. Biotechnological Potential of Insect Fatty Acid-Modifying Enzymes. Zeitschrift für Naturforschung C 2017, 72, 387–403. [Google Scholar] [CrossRef]
- Tupec, M.; Buček, A.; Janoušek, V.; Vogel, H.; Prchalová, D.; Kindl, J.; Pavlíčková, T.; Wenzelová, P.; Jahn, U.; Valterová, I.; et al. Expansion of the Fatty Acyl Reductase Gene Family Shaped Pheromone Communication in Hymenoptera. eLife 2019, 8, e39231. [Google Scholar] [CrossRef]
- Zhang, Y.N.; Xia, Y.H.; Zhu, J.Y.; Li, S.Y.; Dong, S.L. Putative Pathway of Sex Pheromone Biosynthesis and Degradation by Expression Patterns of Genes Identified from Female Pheromone Gland and Adult Antenna of Sesamia Inferens (Walker). J. Chem. Ecol. 2014, 40, 439–451. [Google Scholar] [CrossRef]
- Gu, S.H.; Wu, K.M.; Guo, Y.Y.; Pickett, J.A.; Field, L.M.; Zhou, J.J.; Zhang, Y.J. Identification of Genes Expressed in the Sex Pheromone Gland of the Black Cutworm Agrotis Ipsilon with Putative Roles in Sex Pheromone Biosynthesis and Transport. BMC Genomics 2013, 14, 636. [Google Scholar] [CrossRef] [Green Version]
- Moto, K.; Yoshiga, T.; Yamamoto, M.; Takahashi, S.; Okano, K.; Ando, T.; Nakata, T.; Matsumoto, S. Pheromone Gland-Specific Fatty-Acyl Reductase of the Silkmoth, Bombyx Mori. Proc. Natl. Acad. Sci. USA 2003, 100, 9156–9161. [Google Scholar] [CrossRef] [Green Version]
- Ando, T.; Inomata, S.; Yamamoto, M. Lepidopteran Sex Pheromones. In The Chemistry of Pheromones and Other Semiochemicals I. Topics in Current Chemistry; Schulz, S., Ed.; Springer: Berlin/Heidelberg, Germany, 2004. [Google Scholar]
- Guo, H.; Del Corso, A.; Huang, L.Q.; Mura, U.; Pelosi, P.; Wang, C.Z. Aldehyde Reductase Activity in the Antennae of Helicoverpa Armigera. Insect Mol. Biol. 2014, 23, 330–340. [Google Scholar] [PubMed]
- Yamamoto, K.; Higashiura, A.; Suzuki, M.; Shiotsuki, T.; Sugahara, R.; Fujii, T.; Nakagawa, A. Structural Characterization of an Aldo-Keto Reductase (AKR2E5) from the Silkworm Bombyx Mori. Biochem. Biophys. Res. Commun. 2016, 474, 104–110. [Google Scholar] [CrossRef] [Green Version]
- Zhan, S.; Merlin, C.; Boore, J.L.; Reppert, S.M. The Monarch Butterfly Genome Yields Insights into Long-Distance Migration. Cell 2011, 147, 1171–1185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakurai, T.; Nakagawa, T.; Mitsuno, H.; Mori, H.; Endo, Y.; Tanoue, S.; Yasukochi, Y.; Touhara, K.; Nishioka, T. Identification and Functional Characterization of a Sex Pheromone Receptor in the Silkmoth Bombyx Mori. Proc. Natl. Acad. Sci. USA 2004, 101, 16653–16658. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.D.; Löfstedt, C. Moth Pheromone Receptors: Gene Sequences, Function, and Evolution. Front. Ecol. Evol. 2015, 3, 105. [Google Scholar] [CrossRef] [Green Version]
- Shen, S.; Cao, S.; Zhang, Z.; Kong, X.; Liu, F.; Wang, G.; Zhang, S. Evolution of Sex Pheromone Receptors in Dendrolimus Punctatus Walker (Lepidoptera: Lasiocampidae) Is Divergent from Other Moth Species. Insect Biochem. Mol. Biol. 2020, 122, 103375. [Google Scholar] [CrossRef] [PubMed]
- Yuvaraj, J.K.; Corcoran, J.A.; Andersson, M.N.; Newcomb, R.D.; Anderbrant, O.; Löfstedt, C. Characterization of Odorant Receptors from a Non-Ditrysian Moth, Eriocrania Semipurpurella Sheds Light on the Origin of Sex Pheromone Receptors in Lepidoptera. Mol. Biol. Evol. 2017, 34, 2733–2746. [Google Scholar] [CrossRef] [Green Version]
- Soques, S.; Vásquez, G.M.; Grozinger, C.M.; Gould, F. Age and Mating Status Do Not Affect Transcript Levels of Odorant Receptor Genes in Male Antennae of Heliothis Virescens and Heliothis Subflexa. J. Chem. Ecol. 2010, 36, 1226–1233. [Google Scholar] [CrossRef]
- Saveer, A.M.; Kromann, S.H.; Birgersson, G.; Bengtsson, M.; Lindblom, T.; Balkenius, A.; Hansson, B.S.; Witzgall, P.; Becher, P.G.; Ignell, R. Floral to Green: Mating Switches Moth Olfactory Coding and Preference. Proc. R. Soc. B Biol. Sci. 2012, 279, 2314–2322. [Google Scholar] [CrossRef]
- Zeng, F.F.; Sun, X.; Dong, H.B.; Wang, M.Q. Analysis of a CDNA Library from the Antenna of Cnaphalocrocis Medinalis and the Expression Pattern of Olfactory Genes. Biochem. Biophys. Res. Commun. 2013, 433, 463–469. [Google Scholar] [CrossRef]
- Immonen, E.; Ritchie, M.G. The Genomic Response to Courtship Song Stimulation in Female Drosophila Melanogaster. Proc. R. Soc. B Biol. Sci. 2012, 279, 1359–1365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Latorre-Estivalis, J.M.; Omondi, B.A.; DeSouza, O.; Oliveira, I.H.R.; Ignell, R.; Lorenzo, M.G. Molecular Basis of Peripheral Olfactory Plasticity in Rhodnius Prolixus, a Chagas Disease Vector. Front. Ecol. Evol. 2015, 3, 74. [Google Scholar] [CrossRef] [Green Version]
- Pelosi, P.; Iovinella, I.; Felicioli, A.; Dani, F.R. Soluble Proteins of Chemical Communication: An Overview across Arthropods. Front. Physiol. 2014, 5, 320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelosi, P.; Zhu, J.; Knoll, W. Odorant-Binding Proteins as Sensing Elements for Odour Monitoring. Sensors 2018, 18, 3248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venthur, H.; Zhou, J.J. Odorant Receptors and Odorant-Binding Proteins as Insect Pest Control Targets: A Comparative Analysis. Front. Physiol. 2018, 9, 1163. [Google Scholar] [CrossRef]
- Sánchez-Gracia, A.; Vieira, F.G.; Rozas, J. Molecular Evolution of the Major Chemosensory Gene Families in Insects. Heredity (Edinb). 2009, 103, 208–216. [Google Scholar] [CrossRef]
- Vieira, F.G.; Rozas, J. Comparative Genomics of the Odorant-Binding and Chemosensory Protein Gene Families across the Arthropoda: Origin and Evolutionary History of the Chemosensory System. Genome Biol. Evol. 2011, 3, 476–490. [Google Scholar] [CrossRef]
- Bloch, G.; Hazan, E.; Rafaeli, A. Circadian Rhythms and Endocrine Functions in Adult Insects. J. Insect Physiol. 2013, 59, 56–69. [Google Scholar] [CrossRef]
- Bober, R.; Rafaeli, A. Gene-Silencing Reveals the Functional Significance of Pheromone Biosynthesis Activating Neuropeptide Receptor (PBAN-R) in a Male Moth. Proc. Natl. Acad. Sci. USA 2010, 107, 16858–16862. [Google Scholar] [CrossRef] [Green Version]
- Rosén, W.Q. Endogenous Control of Circadian Rhythms of Pheromone Production in the Turnip Moth, Agrotis Segetum. Arch. Insect Biochem. Physiol. 2002, 50, 21–30. [Google Scholar] [CrossRef]
- Levi-Zada, A.; David, M.; Fefer, D.; Seplyarsky, V.; Sadowsky, A.; Dobrinin, S.; Ticuchinski, T.; Harari, D.; Blumberg, D.; Dunkelblum, E. Circadian Release of Male-Specific Components of the Greater Date Moth, Aphomia (Arenipses) Sabella, Using Sequential SPME/GC/MS Analysis. J. Chem. Ecol. 2014, 40, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Luo, L.; Jiang, X.; Zhang, L.; Niu, C. Expression of Pheromone Biosynthesis Activating Neuropeptide and Its Receptor (PBANR) MRNA in Adult Female Spodoptera Exigua (Lepidoptera: Noctuidae). Arch. Insect Biochem. Physiol. 2010, 75, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Cong, Q.; Shen, J.; Zhang, J.; Hallwachs, W.; Janzen, D.H.; Grishin, N.V. Genomes of Skipper Butterflies Reveal Extensive Convergence of Wing Patterns. Proc. Natl. Acad. Sci. USA 2019, 116, 6232–6237. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Cong, Q.; Shen, J.; Opler, P.; Grishin, N. Genomics of a Complete Butterfly Continent. bioRxiv 2019, 845, 829887. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nieberding, C.M.; Beldade, P.; Baumlé, V.; San Martin, G.; Arun, A.; Lognay, G.; Montagné, N.; Bastin-Héline, L.; Jacquin-Joly, E.; Noirot, C.; et al. Mosaic Evolution of Molecular Pathways for Sex Pheromone Communication in a Butterfly. Genes 2022, 13, 1372. https://doi.org/10.3390/genes13081372
Nieberding CM, Beldade P, Baumlé V, San Martin G, Arun A, Lognay G, Montagné N, Bastin-Héline L, Jacquin-Joly E, Noirot C, et al. Mosaic Evolution of Molecular Pathways for Sex Pheromone Communication in a Butterfly. Genes. 2022; 13(8):1372. https://doi.org/10.3390/genes13081372
Chicago/Turabian StyleNieberding, Caroline M., Patrícia Beldade, Véronique Baumlé, Gilles San Martin, Alok Arun, Georges Lognay, Nicolas Montagné, Lucie Bastin-Héline, Emmanuelle Jacquin-Joly, Céline Noirot, and et al. 2022. "Mosaic Evolution of Molecular Pathways for Sex Pheromone Communication in a Butterfly" Genes 13, no. 8: 1372. https://doi.org/10.3390/genes13081372
APA StyleNieberding, C. M., Beldade, P., Baumlé, V., San Martin, G., Arun, A., Lognay, G., Montagné, N., Bastin-Héline, L., Jacquin-Joly, E., Noirot, C., Klopp, C., & Visser, B. (2022). Mosaic Evolution of Molecular Pathways for Sex Pheromone Communication in a Butterfly. Genes, 13(8), 1372. https://doi.org/10.3390/genes13081372








