Mechanisms of Caspases 3/7/8/9 in the Degeneration of External Gills of Chinese Giant Salamanders (Andrias davidianus)
Abstract
:1. Introduction
2. Materials and Methods
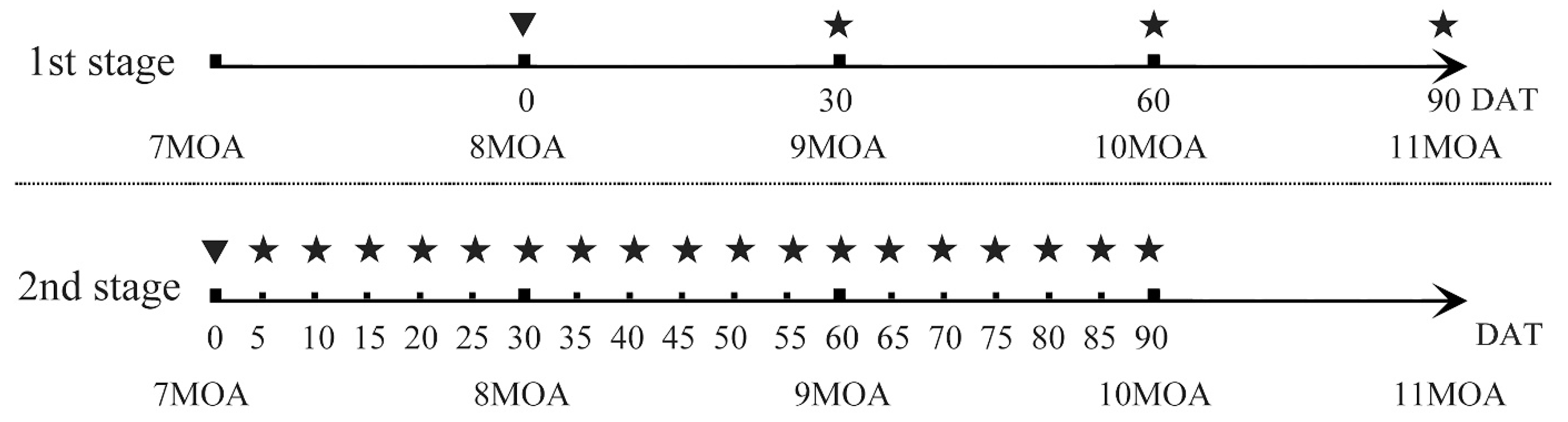
2.1. The TH Treatment Method and Sample Collection
2.2. Cloning of Caspases 3/7/8/9
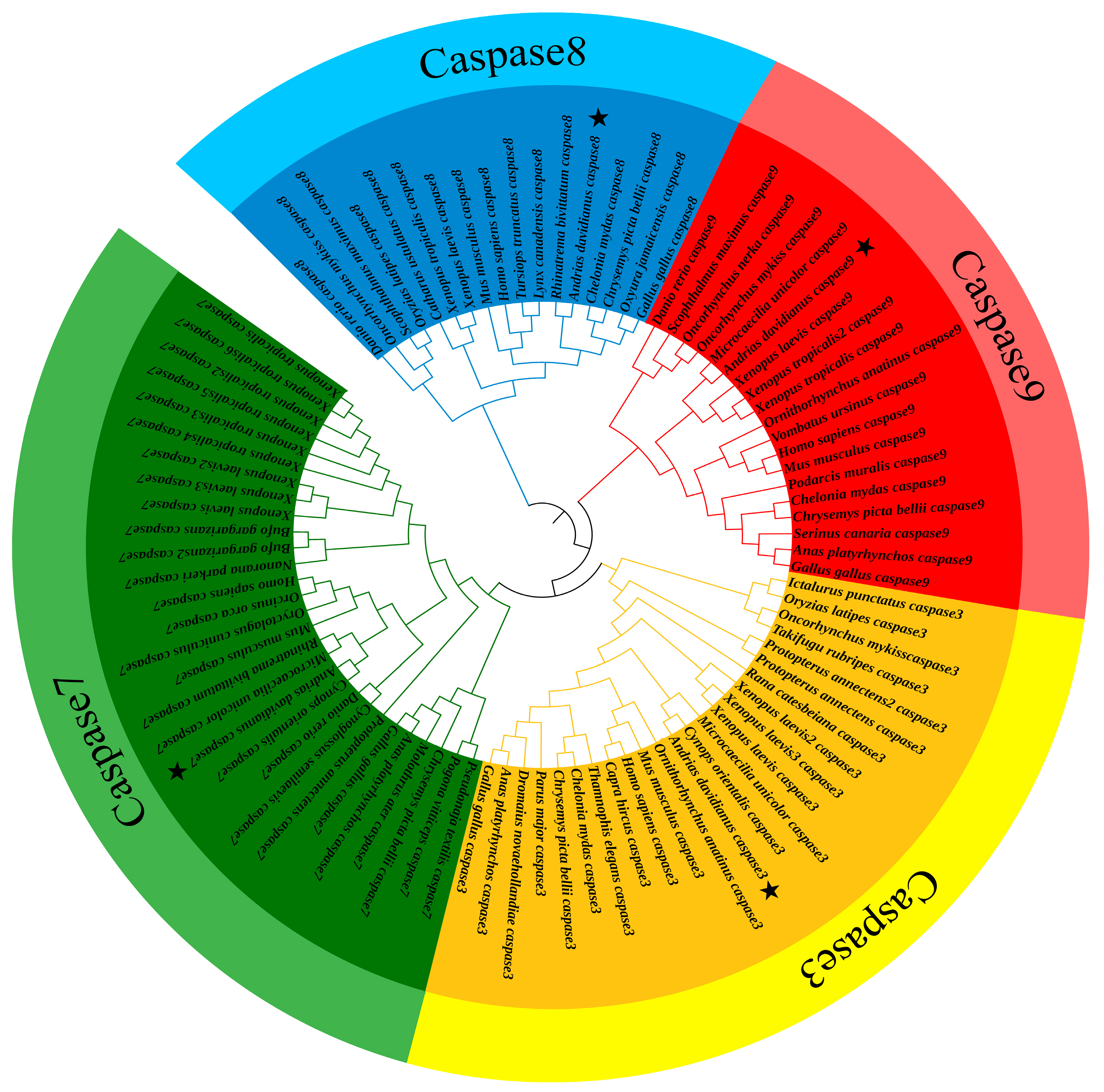
2.3. Sequencing and Phylogenetic Analysis
2.4. Expression Analysis
2.5. TUNEL Assay
3. Results
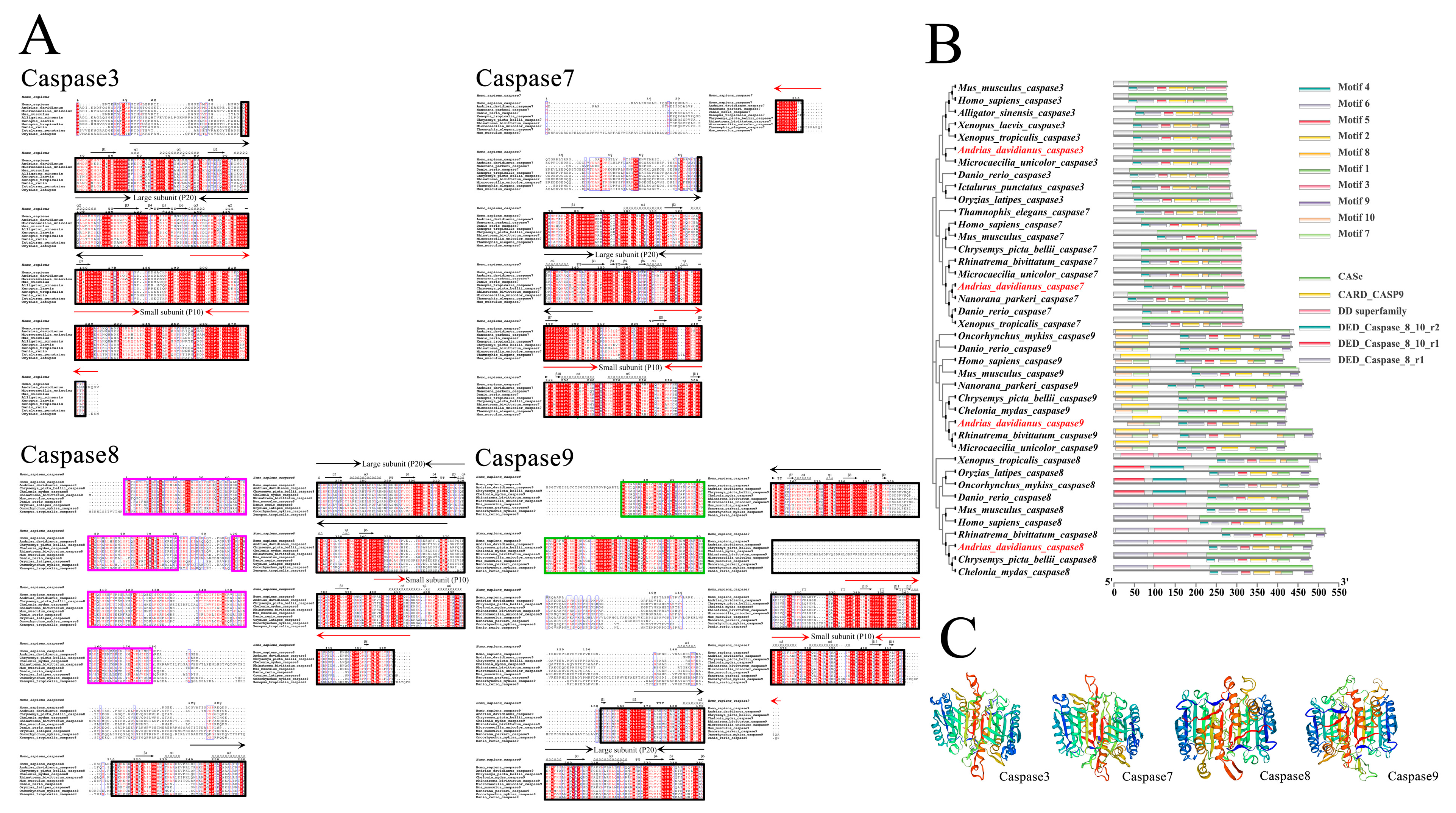
3.1. Characterization and Sequencing: Analysis of caspases 3/7/8/9
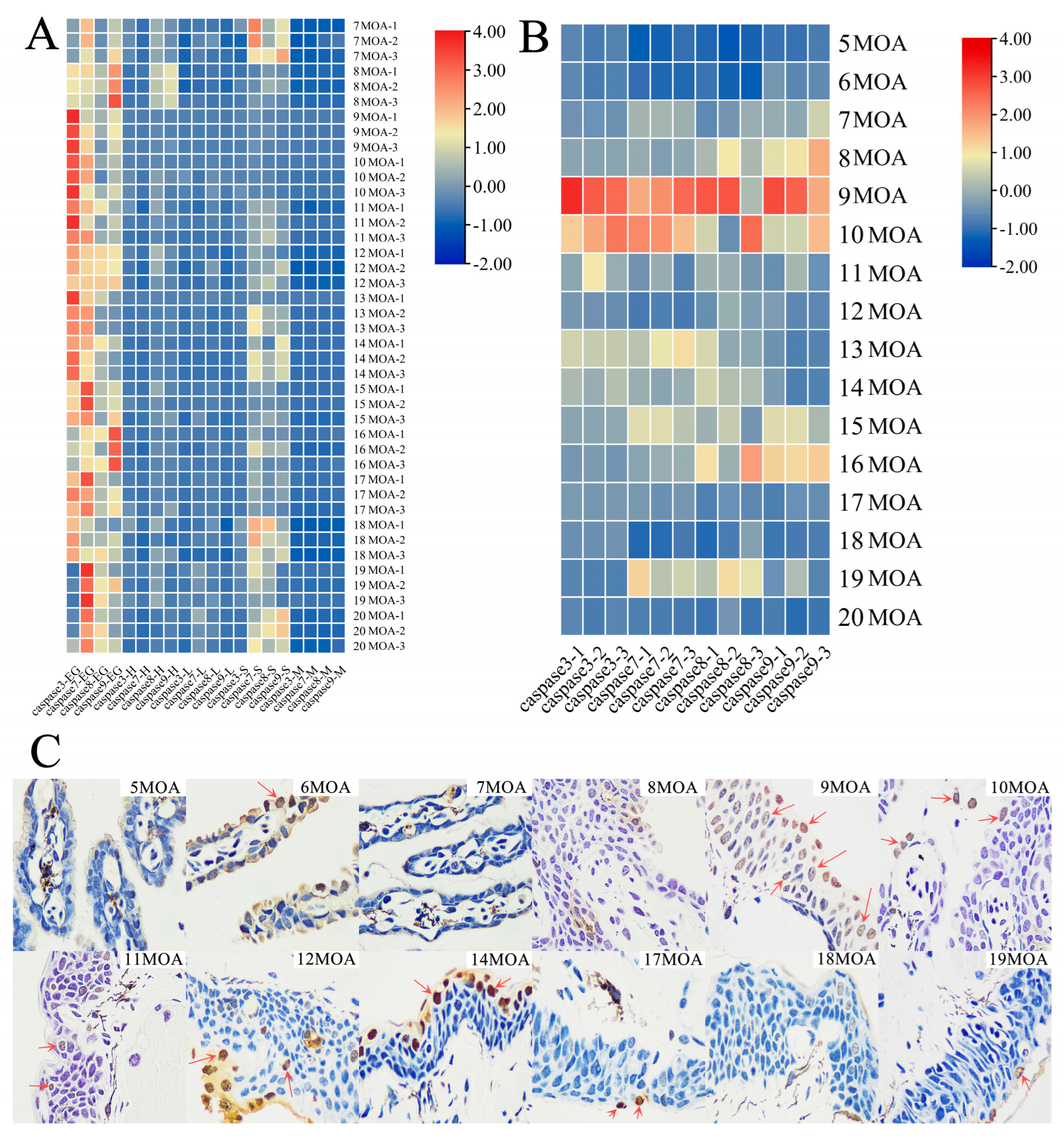
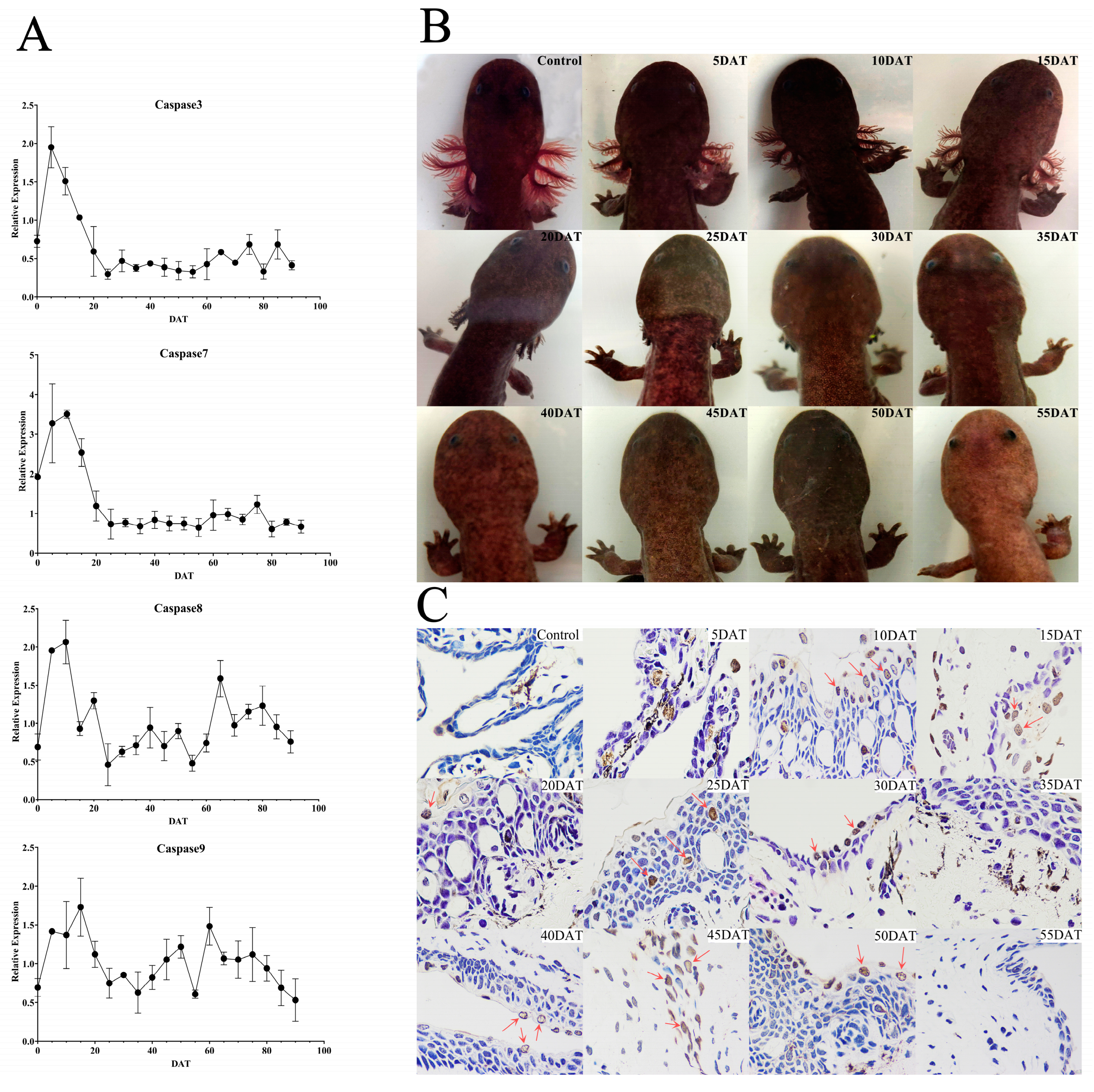
3.2. External Gill Loss and Expression Analysis of Caspases 3/7/8/9 in the Natural Group
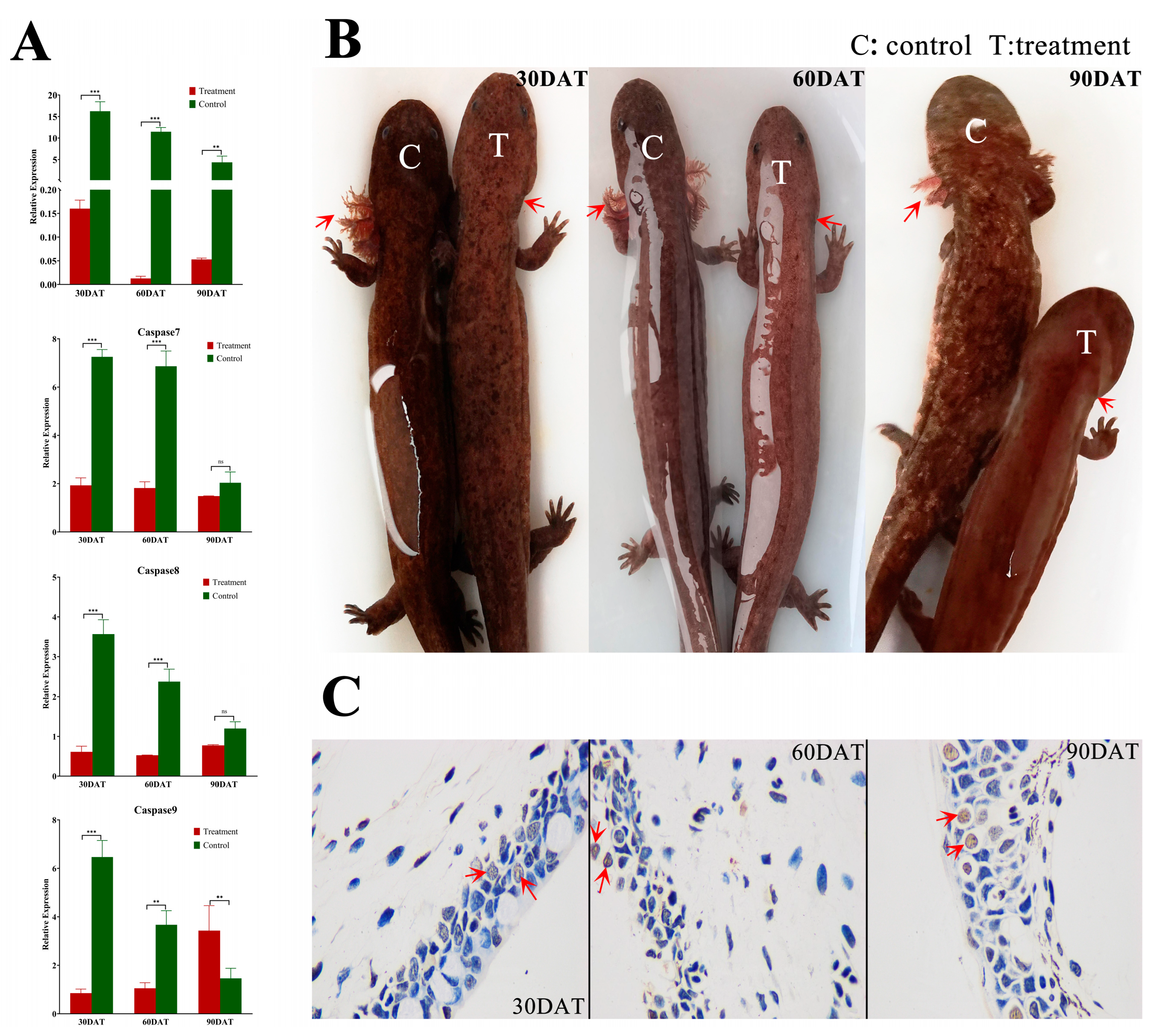
3.3. Analysis of External Gill Loss and Expression of Caspases 3/7/8/9 in the TH Treatment Group
4. Discussion
4.1. Characterization and Phylogeny of Caspases 3/7/8/9
4.2. Natural Metamorphosis and Caspases 3/7/8/9 Expressions
4.3. Acceleration of Metamorphosis through TH Treatment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| TH | Thyroid hormone |
| MOA | Month of age |
| DAT | Day after treatment |
| ORFs | Open reading frames |
References
- Zhao, H.; Deng, J.; Kong, F.; Jiang, W.; Wang, Q.J.; Ma, J.L.; Zhang, H.X. A discussion on the prolongation of the external gill shedding period in giant salamanders. Hebei Fish. 2018, 12, 35–36. [Google Scholar]
- Lewinson, D.; Rosenberg, M.; Warburg, M.R. Ultrastructural and ultracytochemical studies of the gill epithelium in the larvae of Salamandra salamandra (Amphibia, Urodela). Zoomorphology 1987, 107, 17–25. [Google Scholar] [CrossRef]
- Eo, S.H.; Doyle, J.M.; Hale, M.C.; Marra, N.J.; Ruhl, J.D.; DeWoody, J.A. Comparative transcriptomics and gene expression in larval tiger salamander (Ambystoma tigrinum) gill and lung tissues as revealed by pyrosequencing. Gene 2012, 492, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Kerr, J.F.R.; Wyllie, A.H.; Currie, A.R. Apoptosis: A Basic Biological Phenomenon with Wideranging Implications in Tissue Kinetics. Br. J. Cancer 1972, 26, 239–257. [Google Scholar] [CrossRef] [Green Version]
- Meier, P.; Finch, A.; Evan, G. Apoptosis in development. Nature 2000, 407, 796–801. [Google Scholar] [CrossRef]
- Li, M.; Ding, Y.; Mu, Y.; Ao, J.; Chen, X. Molecular cloning and characterization of caspase -3 in large yellow croaker (Pseudosciaena crocea). Fish Shellfish Immunol. 2011, 30, 910–916. [Google Scholar] [CrossRef]
- Grossmann, J. Molecular mechanisms of “detachment–induced apoptosis–Anoikis”. Apoptosis 2002, 7, 247–260. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vitale, I.; Abrams, J.M.; Alnemri, E.S.; Baehrecke, E.H.; Blagosklonny, M.V.; Dawson, T.M.; Dawson, V.L.; El-Deiry, W.S.; Fulda, S.; et al. Molecular definitions of cell death subroutines: Recommendations of the Nomenclature Committee on Cell Death. Cell Death Differ. 2012, 19, 107–120. [Google Scholar] [CrossRef]
- Grütter, M.G. Caspase: Key players in programmed cell death. Curr. Opin. Struct. Biol. 2000, 10, 649–655. [Google Scholar] [CrossRef]
- Kumar, S.; Doumanis, J. The fly caspase. Cell Death Differ. 2000, 7, 1039–1044. [Google Scholar] [CrossRef]
- Shalini, S.; Dorstyn, L.; Dawar, S.; Kumar, S. Old, new and emerging functions of caspase. Cell Death Differ. 2015, 22, 526–539. [Google Scholar] [CrossRef] [Green Version]
- Thomsen, N.D.; Koerber, J.T.; Wells, J.A. Structural snapshots reveal distinct mechanisms of procaspase -3 and -7 activation. Proc. Natl. Acad. Sci. USA 2013, 110, 8477–8482. [Google Scholar] [CrossRef] [Green Version]
- Duan, H.; Orth, K.; Chinnaiyan, A.M.; Poirier, G.G.; Froelich, C.J.; He, W.W.; Dixit, V.M. ICE-LAP6, a novel member of the ICE/Ced-3 gene family, is activated by the cytotoxic T cell protease granzyme B. J. Biol. Chem. 1996, 271, 16720–16724. [Google Scholar] [CrossRef] [Green Version]
- Tata, J.R. Amphibian metamorphosis as a model for the developmental actions of thyroid hormone. Mol. Cell. Endocrinol. 2006, 246, 10–20. [Google Scholar] [CrossRef]
- Thambirajah, A.A.; Koide, E.M.; Imbery, J.J.; Helbing, C.C. Contaminant and Environmental Influences on Thyroid Hormone Action in Amphibian Metamorphosis. Front Endocrinol. 2019, 10, 276. [Google Scholar] [CrossRef] [Green Version]
- Abdollahpour, H.; Falahatkar, B.; Efatpanah, I.; Meknatkhah, B.; Van Der Kraak, G. Hormonal and physiological changes in Sterlet sturgeon Acipenser ruthenus treated with thyroxine. Aquaculture 2019, 507, 293–300. [Google Scholar] [CrossRef]
- Alinezhad, S.; Abdollahpour, H.; Jafari, N.; Falahatkar, B. Effects of thyroxine immersion on Sterlet sturgeon (Acipenser ruthenus) embryos and larvae: Variations in thyroid hormone levels during development. Aquaculture 2020, 519, 734745. [Google Scholar] [CrossRef]
- Brown, D.D.; Cai, L. Amphibian metamorphosis. Dev. Biol. 2007, 306, 20–33. [Google Scholar] [CrossRef] [Green Version]
- Larras-Regard, E.; Taurog, A.; Dorris, M. Plasma T4 and T3 levels in Ambystoma tigrinum at various stages of metamorphosis. Gen. Comp. Endocrinol. 1981, 43, 443–450. [Google Scholar] [CrossRef]
- Alberch, P.; Gale, E.A.; Larsen, P.R. Plasma T4 and T3 levels in naturally metamorphosing Eurycea bislineata(Amphibia: Plethodontidae). Gen. Comp. Endocrinol. 1986, 61, 153–163. [Google Scholar] [CrossRef]
- Sterling, J.; Fu, L.; Matsuura, K.; Shi, Y.B. Cytological and morphological analyses reveal distinct features of intestinal development during Xenopus tropicalis metamorphosis. PLoS ONE 2012, 7, e47407. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Zhu, W.; Liu, J.; Wang, S.; Jiang, J. Identification and differential regulation of microRNAs during thyroid hormone-dependent metamorphosis in Microhyla fissipes. BMC Genom. 2018, 19, 507. [Google Scholar] [CrossRef]
- Trudeau, V.L.; Thomson, P.; Zhang, W.S.; Reynaud, S.; Navarro-Martin, L.; Langlois, V.S. Agrochemicals disrupt multiple endocrine axes in amphibians. Mol. Cell. Endocrinol. 2020, 513, 110861. [Google Scholar] [CrossRef]
- Brown, A.K.; Wong, C.S. Distribution and fate of pharmaceuticals and their metabolite conjugates in a municipal wastewater treatment plant. Water Res. 2018, 144, 774–783. [Google Scholar] [CrossRef]
- Hu, Q.M.; Xiao, H.B.; Tian, H.F.; Meng, Y. Characterization and expression of cyp19a gene in the Chinese giant salamander Andrias davidianus. Comp Biochem Phys B 2016, 192, 21–29. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.Y.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; He, Y.; Xia, R. TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv 2018, 289660. [Google Scholar] [CrossRef]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef] [Green Version]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.J.; Han, L.H.; Cong, R.S.; Liang, J. Caspase family proteases and apoptosis. Acta Biochim. Biophys. Sin. 2005, 37, 719–727. [Google Scholar] [CrossRef] [Green Version]
- Palacios-Rodriguez, Y.; Garcia-Lainez, G.; Sancho, M.; Gortat, A.; Orzaez, M.; Perez-Paya, E. Polypeptide modulators of caspase recruitment domain (CARD)-CARD-mediated protein-protein interactions. J. Biol. Chem. 2011, 286, 44457–44466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Twiddy, D.; Cain, K. Caspase-9 cleavage, do you need it? Biochem. J. 2007, 405, e1–e2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desroches, A.; Boucher, D.; Denault, J.B. Caspase Family. In Encyclopedia of Signaling Molecules; Springer: New York, NY, USA, 2016; pp. 1–20. [Google Scholar]
- Dai, Z.; Li, S.R.; Zhu, P.F.; Liu, L.; Wang, B.; Liu, Y.P.; Luo, X.D.; Zhao, X.D. Isocostunolide inhibited glioma stem cell by suppression proliferation and inducing caspase dependent apoptosis. Bioorg. Med. Chem. Lett. 2017, 27, 2863–2867. [Google Scholar] [CrossRef] [PubMed]
- Sakamaki, K.; Satou, Y. Caspase: Evolutionary aspects of their functions in vertebrates. J. Fish Biol. 2009, 74, 727–753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hime, P.M.; Lemmon, A.R.; Lemmon, E.C.M.; Prendini, E.; Brown, J.M.; Thomson, R.C.; Kratovil, J.D.; Noonan, B.P.; Pyron, R.A.; Peloso, P.L.V.; et al. Phylogenomics Reveals Ancient Gene Tree Discordance in the Amphibian Tree of Life. Syst. Biol. 2021, 70, 49–66. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, K.; Takahashi, A.; Yaoita, Y. Structure, Expression, and Function of the Xenopus laevis Caspase Family. J. Biol. Chem. 2000, 275, 10484–10491. [Google Scholar] [CrossRef] [Green Version]
- Das, B.; Cai, L.; Carter, M.G.; Piao, Y.L.; Sharov, A.A.; Ko, M.S.; Brown, D.D. Gene expression changes at metamorphosis induced by thyroid hormone in Xenopus laevis tadpoles. Dev. Biol. 2006, 291, 342–355. [Google Scholar] [CrossRef] [Green Version]
- Rowe, I.; Coen, L.; Le Blay, K.; Le Mevel, S.; Demeneix, B.A. Autonomous regulation of muscle fibre fate during metamorphosis in Xenopus tropicalis. Dev. Dyn. 2002, 224, 381–390. [Google Scholar] [CrossRef]
- Rowe, I.; Le Blay, K.; Du Pasquier, D.; Palmier, K.; Levi, G.; Demeneix, B.; Coen, L. Apoptosis of tail muscle during amphibian metamorphosis involves a caspase 9-dependent mechanism. Dev. Dyn. 2005, 233, 76–87. [Google Scholar] [CrossRef]
- McDonald, P.C.; Nagel, J.M.; Dedhar, S. Anastasis, Recovery from the Brink of Death as a Mechanism of Drug Resistance. In Biological Mechanisms and the Advancing Approaches to Overcoming Cancer Drug Resistance; Elsevier: Amsterdam, The Netherlands, 2021; pp. 251–260. [Google Scholar] [CrossRef]
- Matalova, E.; Lesot, H.; Svandova, E.; Vanden Berghe, T.; Sharpe, P.T.; Healy, C.; Vandenabeele, P.; Tucker, A.S. Caspase-7 participates in differentiation of cells forming dental hard tissues. Dev. Growth Differ. 2013, 55, 615–621. [Google Scholar] [CrossRef]
- Chaudhary, S.; Madhukrishna, B.; Adhya, A.K.; Keshari, S.; Mishra, S.K. Overexpression of caspase 7 is ERalpha dependent to affect proliferation and cell growth in breast cancer cells by targeting p21(Cip). Oncogenesis 2016, 5, e219. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Zoysa, M.D.; Whang, I.; Lee, S.; Kim, Y.; Oh, C.; Choi, C.Y.; Yeo, S.-Y.; Lee, J. Molluscan death effector domain (DED)-containing caspase-8 gene from disk abalone (Haliotis discus discus): Molecular characterization and expression analysis. Fish Shellfish. Immunol. 2011, 30, 480–487. [Google Scholar] [CrossRef]
- Djordjevic, A.; Djordjevic, J.; Elakovic, I.; Adzic, M.; Matic, G.; Radojcic, M.B. Fluoxetine affects hippocampal plasticity, apoptosis and depressive-like behavior of chronically isolated rats. Prog. Neuropsychopharmacol. Biol. Psychiatry 2012, 36, 92–100. [Google Scholar] [CrossRef]
- Ishizuya-Oka, A.; Hasebe, T.; Shi, Y.B. Apoptosis in amphibian organs during metamorphosis. Apoptosis 2010, 15, 350–364. [Google Scholar] [CrossRef] [Green Version]
- Shibata, Y.; Tanizaki, Y.; Zhang, H.; Lee, H.; Dasso, M.; Shi, Y.B. Thyroid Hormone Receptor Is Essential for Larval Epithelial Apoptosis and Adult Epithelial Stem Cell Development but Not Adult Intestinal Morphogenesis during Xenopus tropicalis Metamorphosis. Cells 2021, 10, 536. [Google Scholar] [CrossRef]
- Laudet, V. The origins and evolution of vertebrate metamorphosis. Curr. Biol. 2011, 21, R726–R737. [Google Scholar] [CrossRef] [Green Version]
- Buchholz, D.R. More similar than you think: Frog metamorphosis as a model of human perinatal endocrinology. Dev. Biol. 2015, 408, 188–195. [Google Scholar] [CrossRef] [Green Version]
- Thampi, P.; Liu, J.; Zeng, Z.; MacLeod, J.N. Changes in the appendicular skeleton during metamorphosis in the axolotl salamander (Ambystoma mexicanum). J. Anat. 2018, 233, 468–477. [Google Scholar] [CrossRef]
| Primer Name | Primer Sequence | Primer Purpose |
|---|---|---|
| Caspase 3-F | 5′-GAGGCAGCGAGGACTATTGT-3′ | Caspase 3 amplification |
| Caspase 3-R | 5′-TGGTGGCTCATTGTTCTTGTT-3′ | |
| Caspase 7-F | 5′-TTTTACCCGCCACCTCCTATCC-3′ | Caspase 7 amplification |
| Caspase 7-R | 5′-ACAACAGTAACACAGTTCCCCC-3′ | |
| Caspase 8-F | 5′-GATGACAAACCCCATGTAAGG-3′ | Caspase 8 amplification |
| Caspase 8-R | 5′-TCTCCCAAATGAAGGTGCTC-3′ | |
| Caspase 9-F | 5′-CTCATGTCCGGTACGGTAGA-3′ | Caspase 9 amplification |
| Caspase 9-R | 5′-CAGAGGTTTGTGACCGTATGC-3′ | |
| M13-F | 5′-CGCCAGGGTTTTCCCAGTCACGAC-3′ | Universal primer |
| M13-R | 5′-CAGCGGATAACAATTTCACACAGG-3′ | |
| β-actin-F | 5′-GCCGTGACCTGACAGACTACCT-3′ | RT-qPCR |
| β-actin-R | 5′-AGTCCAGGGCGACATAGCAGAG-3′ | |
| GAPDH-F | 5′-GACCACTGTCCACGCAGTCAC-3′ | |
| GAPDH-R | 5′-GATGTTCTGGTTGGCACCTCT-3′ | |
| Q Caspase 3-F | 5′-GGACATTGAGGCAAAGCCAGAA-3′ | |
| Q Caspase 3-R | 5′-TGAGGTTTCCAGCATCCACATC-3′ | |
| Q Caspase 7-F | 5′-GCAGATCCTCACCAGGGTCAAC-3′ | |
| Q Caspase 7-R | 5′-CGTCAGCATGGACACCACACAA-3′ | |
| Q Caspase 8-F | 5′-CAGACGGCAGATGTCCAACG-3′ | |
| Q Caspase 8-R | 5′-TATCATCACCTCTCGGGCAGC-3′ | |
| Q Caspase 9-F | 5′-TGGGCACCACTGTCCAACTC-3′ | |
| Q Caspase 9-R | 5′-ATCTCCGCTGTCCATTACCGA-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, S.; Tan, C.; Sun, X.; Tang, X.; Huang, X.; Yan, F.; Zhu, G.; Wang, Q. Mechanisms of Caspases 3/7/8/9 in the Degeneration of External Gills of Chinese Giant Salamanders (Andrias davidianus). Genes 2022, 13, 1360. https://doi.org/10.3390/genes13081360
Yang S, Tan C, Sun X, Tang X, Huang X, Yan F, Zhu G, Wang Q. Mechanisms of Caspases 3/7/8/9 in the Degeneration of External Gills of Chinese Giant Salamanders (Andrias davidianus). Genes. 2022; 13(8):1360. https://doi.org/10.3390/genes13081360
Chicago/Turabian StyleYang, Shijun, Caixia Tan, Xuerong Sun, Xiong Tang, Xiao Huang, Fan Yan, Guangxiang Zhu, and Qin Wang. 2022. "Mechanisms of Caspases 3/7/8/9 in the Degeneration of External Gills of Chinese Giant Salamanders (Andrias davidianus)" Genes 13, no. 8: 1360. https://doi.org/10.3390/genes13081360
APA StyleYang, S., Tan, C., Sun, X., Tang, X., Huang, X., Yan, F., Zhu, G., & Wang, Q. (2022). Mechanisms of Caspases 3/7/8/9 in the Degeneration of External Gills of Chinese Giant Salamanders (Andrias davidianus). Genes, 13(8), 1360. https://doi.org/10.3390/genes13081360






