Abstract
Protein kinases play an essential role in plants’ responses to environmental stress signals. SnRK2 (sucrose non-fermenting 1-related protein kinase 2) is a plant-specific protein kinase that plays a crucial role in abscisic acid and abiotic stress responses in some model plant species. In apple, corn, rice, pepper, grapevine, Arabidopsis thaliana, potato, and tomato, a genome-wide study of the SnRK2 protein family was performed earlier. The genome-wide comprehensive investigation was first revealed to categorize the SnRK2 genes in the Liriodendron chinense (L. chinense). The five SnRK2 genes found in the L. chinense genome were highlighted in this study. The structural gene variants, 3D structure, chromosomal distributions, motif analysis, phylogeny, subcellular localization, cis-regulatory elements, expression profiles in dormant buds, and photoperiod and chilling responses were all investigated in this research. The five SnRK2 genes from L. chinense were grouped into groups (I–IV) based on phylogeny analysis, with three being closely related to other species. Five hormones-, six stress-, two growths and biological process-, and two metabolic-related responsive elements were discovered by studying the cis-elements in the promoters. According to the expression analyses, all five genes were up- and down-regulated in response to abscisic acid (ABA), photoperiod, chilling, and chilling, as well as photoperiod treatments. Our findings gave insight into the SnRK2 family genes in L. chinense and opened up new study options.
1. Introduction
Two species of large deciduous trees make up the genus Liriodendron, which belongs to the Magnoliaceae family. L. chinense is a native of South China, and L. tulipifera comes from eastern North America. The medications produced from the two species varied in America, China, and other countries [1]. The majestic tree L. tulipifera, also known as the tulip tree or yellow poplar, is a particular species found in the eastern United States that may grow to heights of up to 30.48 m [1]. Liriodendron is an excellent decorative tree for landscaping because of its straight trunk, conical crown, unusual leaf shape, and tulip-shaped blooms [2,3]. Liriodendron has also been frequently planted as an industrial timber species because of its quick growth and adaptable wood with superior working characteristics [3]. It is highly prized for its honey production, as a food source for wildlife, and its potential medical benefits [3]. Studies have shown that extracts from Liriodendron leaves have potent cytotoxic effects on tumor cell lines and inhibitory activities against farnesyl protein transferase (FPTase) and tumor cell growth [1]. Bud dormancy, a complicated physiological phenomenon that promotes plant growth, survival, and development, impacts the timing of bud break [4]. Bud dormancy is a crucial characteristic that enables temperate woody perennials to withstand harsh winter conditions. When a plant’s development is halted and its metabolic activity is decreased, it enters a state of dormancy [5,6]. Because it affects the quality of bud break, flowering, and fruiting in the spring, bud dormancy is an important stage in the phenology cycle. A temporary cessation of observable growth is called dormancy [7]. In subtropical trees, including L. chinense, Torreya grandis, Metasequoia glyptostroboides, Cinnamomum chekiangense, and Phoebe chekiangensis, the release of bud dormancy is controlled by photoperiod. The short day had a lower bud burst percentage and delayed the timing of bud burst when compared to the long day [8]. The timing of bud burst displays a continuous phenotypic variation typical of a quantitative attribute for a variety of forest tree species [9]. According to a recent study, only a few woody species are photoperiod sensitive [8,10]. However, a combination of photoperiod, chilling (temperature and duration), and spring or forcing temperatures regulates bud development in temperate [11] and subtropical [8,12] trees. Although bud break and the mechanisms that cause dormancy release are closely related, the underlying molecular pathways are poorly known.
In signal transduction pathways, protein kinases and phosphorylation/dephosphorylation play significant roles in identifying and transmitting stress signals to various cell areas. SnRK (or SNF1-related protein kinase) family members are specific serine/threonine protein kinases found widely in plants and play essential roles in various processes, including growth and development, stress defense, and hormone-mediated signalling [13,14,15,16,17,18,19]. Based on sequence similarity and C-terminal domain structural characteristics, the SnRK family in higher plants is divided into three subfamilies (SnRKl, SnRK2, and SnRK3) [13,20,21]. Many plant genomes have been found to contain the SnRK2 gene family, including in pepper [14], rice [21], cotton [22], Nicotiana tabacum [23], arabidopsis [24], mungbean [25], grapevine [26], maize [27], sugarcane [28], sorghum [29], and apple [30]. Despite this fact, only a tiny portion of the SnRK2 gene has been characterized. Three SnRK2s, SnRK2.2, 2.3, and 2.6, are at the heart of the arabidopsis ABA signaling network, acting as primary positive regulators of ABA signaling in response to water stress as governing seed development and dormancy [26,31,32].
Abscisic acid (ABA) is a key regulator of plant growth and development, including seed germination, maturation and dormancy, cell elongation and division, root and seedling growth, embryo maturation, leaf senescence, and fruit ripening, as well as plant resistance to severe environments [32,33,34,35]. Furthermore, it is necessary for the plant’s reaction to abiotic stressors such as high temperature, cold, drought, and salinity. Increased ABA levels are connected with plant adaptation to these environmental stressors [36]. ABA is required for seed dormancy establishment, maintenance, release, and bud endodormancy. Arabidopsis thaliana (A. thaliana) mutants lacking ABA could not develop seed dormancy [37,38]. Dormancy is influenced not just by ABA levels but also by the ABA signaling pathway. The regulatory component of the ABA receptor (RCAR)/pyrabactin resistance (PYR)/pyrabactin resistance-like (PYL) family of ABA-receptor proteins operate as negative regulators of the protein phosphatase 2c (PP2C, ABI1/ABI2) family of protein phosphatase 2c (PP2C, ABI1/ABI2). The binding of ABA to its RCAR/PYR/PYL receptors and the creation of ABA-receptor PP2C complexes disrupt the interaction between PP2C and SNF1-related protein kinase 2 (SnRK2), which negatively impacts ABA signaling [38,39]. As a result, ABA-responsive genes can be produced by activating downstream transcription factors (such as ABI4 and ABI5) and SnRK2 [38,39]. In the literature, ABA has been suggested to have a role in regulating bud endodormancy, with ABA levels increasing in the autumn and acting as a signal of decreasing day length. Inhibition of cell proliferation and shoot growth, promotion of terminal bud set, and induction of endodormancy are all possible outcomes. In poplar buds, regulators of ABA biosynthesis (NCED3, ABA1, and ABA2), as well as ABA signal transduction components (PP2C, ABI1, and AREB3, among others), were stimulated after 3–4 weeks of short days, and ABA levels in the apex peaked [5,40,41]. However, applying ABA to grapes in the spring had minimal influence on bud break [12], and the impact of freezing on endogenous ABA levels is unknown. Chilling-induced dormancy release in birch was followed by changes in endogenous ABA levels, confirming the involvement of ABA [42,43].
The environmental factors regulate bud dormancy in Liriodendron, particularly describing the role of ABA, chilling, and photoperiod. In this study, we employed genomic approaches and experimental verification to describe and identify ABA SnRK2s receptors, which may be essential signaling regulators in L. chinense response to environmental conditions. There was a total of five LchiSnRK2 genes in the L. chinense genome. Other agriculturally resilient plants with identified SnRK2 genes were studied for evolutionary differences, chromosomal locations, gene structures, and conserved sequence motifs. In addition, the expression profiles of LchiSnRK2s in the dormant bud have been investigated at various phases of development, as well as ABA treatment, photoperiod, and chilling conditions. We also looked into the function of cis-regulatory components identified inside the promoter sequences of ABA-receptors to see how they affect hormone and photoperiod and chilling responses, growth and biological processes, and metabolic responses. L. chinense vegetative dormant buds that had undergone qRT-PCR were used to confirm the transcript expression of LchiSnRK2 genes in response to ABA foliar sprays, photoperiod, and chilling. The results of this study may contribute to a better understanding of the function of ABA receptors in L. chinense and the molecular characterization of LchiSnRK2 genes after the development of genetic material that aids plant stress adaptation.
2. Materials and Methods
2.1. L. chinense SnRK2 Gene Identification and Characterization
The L. chinense protein database (https://hardwoodgenomics.org/Genome-assembly/2630420, accessed on 5 June 2022) and the NCBI database (https://www.ncbi.nlm.nih.gov/ (accessed on 5 June 2022), PRJNA418360) were used to retrieve the genome sequences for this species [44]. The SnRK2 genes family in L. chinense was discovered using the arabidopsis SnRK2 protein sequences obtained from phytozome v13 (https://phytozome-next.jgi.doe.gov/, accessed on 5 June 2022). The TAIR database (http://www.arabidopsis.org, accessed on 5 June 2022) was used to collect the protein sequences of the well-defined 11 members of the SnRK2 family in A. thaliana: AT3G50500, AT1G78290, AT1G26470, AT5G63650, AT5G66880, AT2G23030, AT4G33950, AT4G40010, AT1G60940, AT5G08590, and AT1G10940. Using A. thaliana SnRK2s as a reference, the protein sequences of the L. chinense SnRK2 family members were found in a specific genome database (https://hardwoodgenomics.org/Genome-assembly/2630420, accessed on 5 June 2022). SnRK2’s protein sequence was analyzed by the Pfam database (http://pfam.xfam.org, accessed on 5 June 2022) and matched with a domain profile (PF00069) [3].
Five (Lchi13910, Lchi00543, Lchi25623, Lchi12999, and Lchi01348) SnRK2 family genes were discovered and confirmed using the L. chinense genome database and NCBI database (https://www.ncbi.nlm.nih.gov/) (accessed on 5 June 2022) [44,45,46].
2.2. SnRK2 Genes’ Chromosomal Distribution in L. chinense
We used https://hardwoodgenomics.org/Genome assembly/2630420, accessed on 5 June 2022, to determine the genome position and protein sequences of all L. chinense SnRK2 genes, and we assessed the distribution positions of SnRK2 genes on scaffold or chromosome. SnRK2 genes were found on the chromosomes of L. chinense using MapGene2Chromosome (MG2C; http://mg2c.iask.in/mg2c v2.0/) (accessed on 5 June 2022) [44,45,46].
2.3. Phylogenetic Tree Construction
Protein sequences of SnRK2 genes from Liriodendron chinense, arabidopsis (A. thaliana), tomato (Solanum lycopersicum), pepper (Capsicum annuum), potato (Solanum tuberosum), grape (Vitis vinifera), corn (Zea mays), rice (Oryza sativa), and apple (Malus domestica) were used to conduct the phylogenetic analysis, and to display the Liriodendron chinense SnRK2 gene family evolution relating to other species. The protein sequences were often aligned using MEGA11 (V 6.06) software (www.megasoftware.net) (accessed on 5 June 2022). The phylogenetic tree was constructed using the neighbor-joining (NJ) method with 1000 bootstrap repetitions. Fig Tree V1.4.4, accessed on 5 June 2022, was used to visualize and edit the phylogenetic tree [2,47,48]. To analyze the evolutionary constraints of each SnRK2 gene pair, the KaKs Calculator 2.0 (https://sourceforge.net/projects/kakscalculator2/, accessed on 5 June 2022) was used to calculate the synonymous (Ks), and non-synonymous (Ka) ratios [49].
2.4. Structure and Significant Motif Analyses of the SnRK2 Family Members
The genome of L. chinense contains five members of the SnRK2 family. Web software that also exhibited the exon/intron arrangements of the SnRK2 genes was used to determine the structural analyses of five SnRK2 genes (http://gsds.cbi.pku.edu.cn, accessed on 5 June 2022). In the protein sequences of the five SnRK2 proteins, the online application MEME v5.4.1, Available online: https://meme-suite.org/meme/tools/glam2scan (accessed on 5 June 2022), revealed more conserved strings or regions [3]. Sequence alphabet DNA, RNA, or protein; site distribution zero or one occurrence per sequence (zoops); motif finding mode classic mode; and 10 motifs were the settings utilized by the application. The TBtools program displayed the MEME results by downloading the supplementary mast file [50,51].
2.5. Analysis of the SnRK2 Family’s Promoter Sequences in L. chinense
We collected 2500 bp upstream sequences of SnRK2 family members using the L. chinense genome assembly database (https://hardwoodgenomics.org/Genome assembly/2630420) (accessed on 5 June 2022). To find cis-regulatory elements (CREs), the obtained sequences were then examined by PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (accessed on 5 June 2022). After calculating the frequency of each CRE motif, we used TBtools to find the most common CREs for the SnRK2 genes [51].
2.6. 3D Structure and Subcellular Localization
We utilized SWISS-MODEL (https://swissmodel.expasy.org/interactive, accessed on 5 June 2022) to estimate the three-dimensional (3D) structure [2]. We utilized Cell-PLoc 2.0 (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/, accessed on 5 June 2022) to predict the subcellular localization of the SnRK2 family of genes [46].
2.7. Plant Material and Environmental Conditions
In order to comprehend the effects of various abscisic acid concentrations (ABA-10 µM and ABA-100 µM), photoperiods (long day and short-day), chilling, and chilling as well as photoperiod during the dormant vegetative bud, tests were carried out at the Zhejiang Agricultural and Forestry University (Hangzhou, China). We transferred two-year-old L. chinense seedlings from the Tianmushan National Forest Station nursery in Hangzhou, China, to our university’s growing chamber to conduct our experiment. The seedlings were relocated from the nursery to the growth chamber for photoperiod and chilling treatments, where they were kept at 20 °C (day/night), 200 mol m−2 s−1 of light intensity, etc., and 50% relative humidity (Figure 1) [2,8].
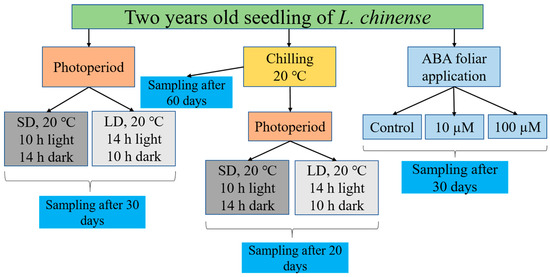
Figure 1.
This schematic flow depicts the experimental setup. The various box designs and color schemes represent different modes of treatment and sampling.
2.8. Extraction of RNA and qRT-PCR Analysis
We used the Total RNAPlant Extraction Kit as directed by the manufacturer (Tiangen, Beijing, China). The cDNA was produced using the TaKaRa PrimeScript 1st Strand cDNA Synthesis Kit (TaKaRa, Dalian, China), following the manufacturer’s instructions. The SYBR Green Real-time PCR Master Mix (TOYOBO, Osaka, Japan) was used to perform the qRT-PCR on an ABI7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) [35].
The transcript profiles of Lchi13910, Lchi00543, Lchi25623, Lchi12999, and Lchi01348 in the dormant bud subjected to various environmental conditions were validated by qRT-PCR analysis (Figure 1). The three technical replicates were used to conduct the qRT-PCR. As a reference gene, the actin gene LtActin97 from Liriodendron was used, along with the sequence provided by Zong et al. [52], to calculate the target genes’ expression levels using the 2−ΔΔCT technique [53]. Primer Premier 5.0 software was used in this investigation to target all employed primers using CDS sequences of SnRK2; more information is available in Supplementary Table S1. GraphPad Prism 9.0.0 software was used to display the outcomes of the qRT-PCR analysis [2].
2.9. Gene Duplication Analysis
We utilized the Riaz et al. [54] criterion to discover duplicated gene pairs: (1) the nucleotide sequence that we aligned spanned >78% of the longer aligned gene, and (2) the identity between the aligned portions must be exceeded >78%.
2.10. Statistical Analysis
The data were analyzed by Statistix 8.1 software (Analytical Software, Tallahassee, FL, USA) using one-way ANOVA, and the data were presented as the mean ± SD (Standard deviation) of the three replicates. The differences in the mean values of the dormant buds between photoperiod, chilling, chilling, as well as photoperiod, and different treatments (ABA-10 µM and ABA-100 µM) of ABA plants were analyzed using an LSD (least significant difference) test at p < 0.05. The graphs were made using GraphPad Prism 9 statistical software (https://www.graphpad.com/) (accessed on 5 June 2022) [55].
3. Results
3.1. Identification of the SnRK2 Gene Family in L. chinense
Using queries of the well-defined protein sequences of 11 SnRK2s from the A. thaliana genome, we investigated five SnRK2 genes in the L. chinense genome (Table 1 and Table S2). We found a lower number than the previously described SnRK2 genes in other species, e.g., the apple, grapevine, arabidopsis, rice, tomato, potato, corn, and pepper (Table S3). Four proteins (Lchi13910, Lchi25623, Lchi12999, and Lchi01348) were determined to have only one Protein kinase domain, according to domain analysis (PF00069), and for one protein (Lchi00543), we did not find any domain, respectively (Table S4).

Table 1.
Characteristics of the five SnRK2 genes identified in L. chinense. Note: GSL—Genomic sequence length, CDSL—Coding sequence length, PSL—Protein sequence length, SCL—Subcellular localization.
Table 1 contains comprehensive statistics for five SnRK2 genes. All five SnRK2s genes, including Lchi13910, Lchi00543, Lchi25623, Lchi12999, and Lchi01348, were localized on different chromosome/scaffolds (Figure 2; Table 1), respectively. There were explicit mentions of the lengths of the genomic sequence, coding sequence, proteins, gene strands, and exons, respectively (Table 1). According to the results of subcellular localization, all proteins were expected to be found in the nucleus (Table 1).
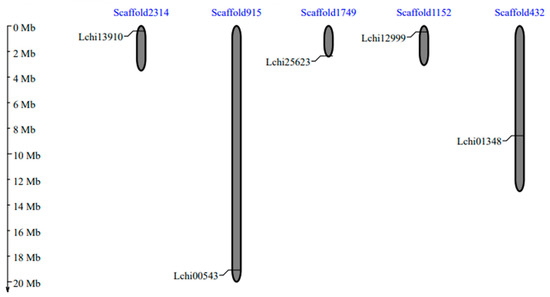
Figure 2.
The SnRK2 gene distribution on the chromosomes of L. chinense serves as a scaffold. The chromosomal/scaffold numbers are found at the top of each chromosome. The names of each LcSnRK2 gene are displayed on the left side of each chromosome. The bars on the scaffold/chromosomes represent the SnRK2 genes.
3.2. SnRK2 Gene Phylogenetic Relationships
Liriodendron chinense predicted SnRK2 proteins were examined using multiple sequence alignment of arabidopsis, tomato, pepper, potato, grape, corn, rice, and apple were shown that a phylogenetic tree in (Table S3) showed four important groups (Group I to IV) (Figure 3). According to the findings, group I consisted of 22 SnRK2 members, Group II included 11 SnRK2 members, Group III contained 20 SnRK2 members, and Group IV comprised 25 SnRK2 members (Figure 3). It is important to note that homologs of LchiSnRK2s genes from different plant species were discovered in each group, with Group III having the highest number of individuals in Groups I, II, and IV (Figure 3). The LchiSnRK2s also share a closer evolutionary connection with the other species within each group.
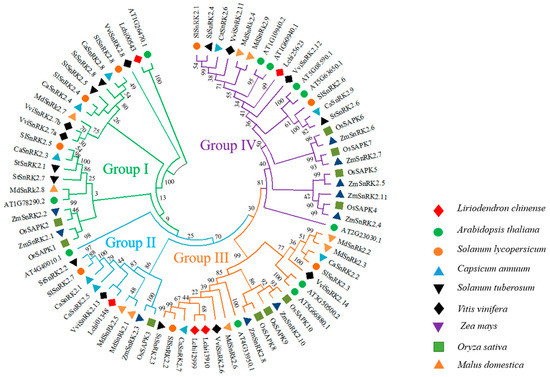
Figure 3.
A phylogenetic analysis of SnRK2 proteins from Liriodendron chinense (5), Arabidopsis thaliana (11), Solanum lycopersicum (8), Capsicum annuum (9), Solanum tuberosum (8), Vitis vinifera (8), Zea mays (10), Oryza sativa (10), and Malus domestica was carried out using the maximum likelihood method (9). There are four groups of SnRK2 proteins: I, II, III, and IV, each of which is represented by a different color.
3.3. Liriodendron chinense SnRK2 Genes Structures and Conserved Motifs Investigation
The gene expansion of the L. chinense family was investigated by examining the exon-intron patterns of the SnRK2 genes (Tables S5 and S6). To further comprehend the structural characteristics of SnRK2 genes, the exon–intron structures and conserved motifs (Figure 4A–C) were examined. The SnRK2s’ exons and introns varied from 1 to 8 and 2 to 9, respectively (Figure 4B). The SnRK2 gene family has a variety of gene architectures, with most SnRK2 genes having five to eight introns; however, certain SnRK2 gene family members, such as Lchi00543, have fewer introns. The maximum number of exons and introns found in Lchi25623 and Lchi01348 was nine. These findings demonstrated that a gene structure that was highly similar to that of their evolutionary relatives was shared by a group of SnRK2 individuals. The SWISS-MODEL program was used to estimate the three-dimensional (3D) structure, and the projected 3D structures showed that the LchiSnRK2 protein has similar conserved structures. Figure 5 provides comprehensive details on the predicted 3D structures.
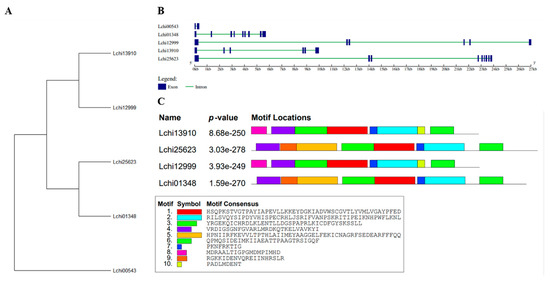
Figure 4.
L. chinense’s SnRK2 family gene structure and motif analysis: (A) Based on phylogenetic relationships and domain identification, the SnRK2s were classified into four groups. Gene structure for SnRK2 (B). The blue horizontal line denotes exon regions, while the green horizontal line denotes intron regions. (C) Conserved motif compositions were found in L. chinense SnRK2s. Various color boxes represent numerous motifs.
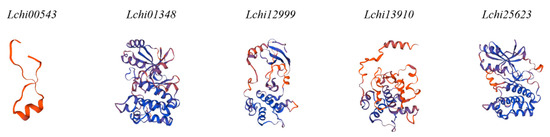
Figure 5.
SnRK2 family 3D structures displaying functional sites.
Additionally, we used the MEME online servers to clarify the conserved motifs of the SnRK2 genes. Furthermore, five SnRK2 genes included 10 conserved motifs (Figure 4C). Due to the prediction, motifs one, two, three, four, six, and seven in most SnRK2 proteins. Motifs one, two, three, four, six, and seven were recognized in Lchi13910, Lchi25623, Lchi12999, and Lchi01348. Motifs five and nine were identified in Lchi25623, and Lchi01348, while motifs eight and 10 were recognized in Lchi13910, and Lchi12999, respectively. SnRK2 proteins exhibited three highly conserved motifs (one, two, and five) with 50 amino acids; one motif (three) with 40 amino acids, while two motifs (four and six) with 29 amino acids, motif nine with 20, motif eight with 19, and motifs seven and 10 with nine amino acids (Figure S1). These findings suggest that the gene structure and amino acid sequence of members of the same subfamily of LchiSnRK share a significant degree of similarity. LchiSnRK proteins’ three-dimensional (3D) structures show that three subfamily proteins have similar 3D structures at their N terminals but differ at their C terminals (Figure 5).
3.4. Identification of Cis-Regulatory Elements in the Promoters of Five SnRK2 Genes
Using a 2500 bp region from each gene’s TAS (transcriptional activation site) to analyze the PlantCARE database to determine the cis-regulatory elements and functions of the genes. In the chosen region of the SnRK2 genes’ promoters in L. chinense, we identified possible cis-elements (Table S7). In SnRK2s, several cis-regulatory components are displayed in Figure 6A,B. On the items that were found, there is extensive information in Table S8. The SnRK2s gene family comprises 191 distinct CREs, including elements sensitive to hormones, stress, growth and biological processes, and metabolism (Figure 6B). Identifying five hormone-related responsive elements—ABA, auxin, GA, MeJA, and SA—represents and suggests the right targets for research into how hormones work in stressful situations (Figure 6; Table S8). The majority of hormone-related responsive regions are specific to a few genes, as seen in Figure 5. Five genes include multiple copies of the responsive elements for auxin, ABA, SA, MeJA, and GA, illustrating their critical roles in phytohormone-related responses in plants (Figure 7A–D). These SnRK2s genes may respond to stress-related stimuli as evidenced by the prediction of six stress-related response components (anoxic specific inducibility, circadian regulation, defense and stress, drought, light, and low temperature) (Figure 7A–D; Table S8). In addition, 80 light-responsive components were discovered, suggesting that SnRK2s are essential for responding to light stress. All SnRK2 genes had one defensive and stress-response element, two anoxic specific inducibility, three circadian control, eight drought response elements, and two low-temperature response elements (Figure 7; Table S5). Most of the promoters of the SnRK2 genes contained CREs responsive to biological processes and growth, including meristem expression (two) and cell cycle regulation (three). In addition, two sensitive metabolic elements—one each for the control of zein metabolism and anaerobic induction—were found (Figure 7A–D; Table S8).
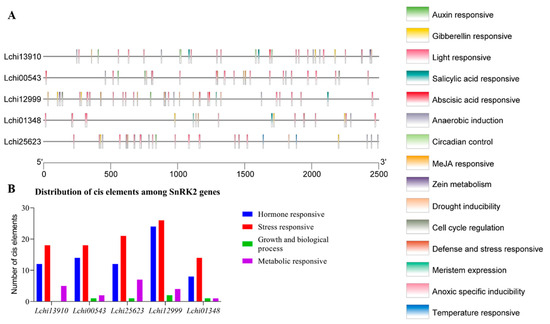
Figure 6.
Cis-regulatory elements (CREs) found in the promoters of SnRK2 genes. (A) The positional distribution of the projected CREs on the SnRK2 promoters is shown by vertical bars. Five SnRK2 genes’ promoter sequences (2500 bp) were examined using PlantCARE. In this legend, each cis-color element is represented: (B) Hormones, stress, growth and biological processes, and sensitive metabolic components are linked to the distribution of cis-elements in the promoters of SnRK2 genes. The detected cis-elements are displayed in colored boxes.
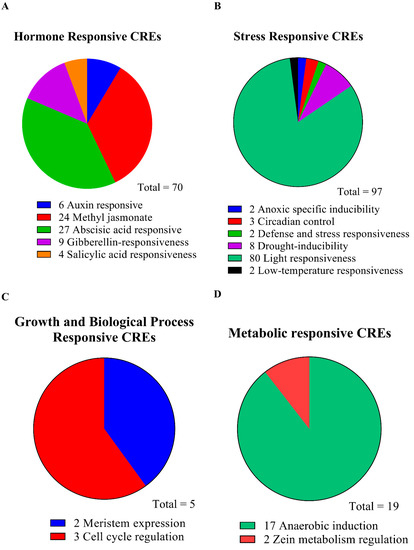
Figure 7.
The distribution of cis-regulatory elements (CREs) in the promoters of SnRK2 genes is based on their hypothesized roles. The four different types of CREs are (A) hormone-responsive CREs, (B) stress-responsive CREs, (C) growth and biological process-responsive CREs, and (D) metabolic-responsive CREs.
3.5. SnRK2 Gene Expression Analysis Using Real-Time qRT-PCR in Response to Dormant Bud in L. chinense
Five LchiSnRK2 genes were subjected to qRT-PCR-based expression profiling to look into their expression profiles under contrasting photoperiods (long day (P-LD) and short-day (P-SD)), chilling, various abscisic acid concentrations (ABA-10 µM and ABA-100 µM), and chilling as well as photoperiod (C + P-LD, C + P-SD) conditions (Figure 8). The expression level of the Lchi13910 gene significantly increased under ABA-10 µM and photoperiod long day (P-LD) and significantly decreased under chilling as well as photoperiod long day (C + P-LD), according to the RT-qPCR data. In other conditions, including ABA-100 µM, P-SD, and C + P-SD, Lchi13910 expression level displayed a non-significant difference. Similarly, the expression of the Lchi00543 gene significantly decreased under all environmental circumstances, including chilling, photoperiod, and various ABA concentrations. In P-SD, chilling, and C + P-LD conditions, the expression level of Lchi25623 was significantly up-regulated, whereas, in ABA-100 µM conditions, it was significantly downregulated. In the ABA-10 µM and P-LD conditions, Lchi25623 displayed non-significant differences. The Lchi12999 gene was significantly up-regulated in the chilling condition, whereas it was down-regulated in the ABA-100 µM, P-LD, and C + P-LD conditions. In the ABA-10 µM, P-SD, and C + P-SD conditions, the expression level of Lchi12999 did not display any significant differences. The Lchi01348 transcript expression was also highly up-regulated in P-SD but significantly down-regulated in all other environmental conditions (Figure 8).
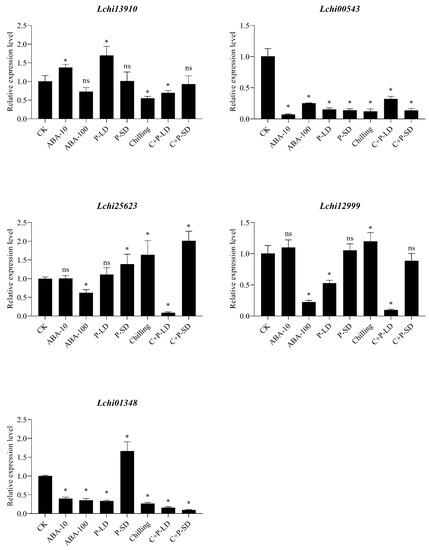
Figure 8.
The expression of LchiSnRK2s by qRT-PCR in dormant buds of L. chinense at the seedling stage under different photoperiods (long day (P-LD) and short-day (P-SD)), chilling, various abscisic acid concentrations (ABA-10 µM and ABA-100 µM), and chilling as well as photoperiod (C + P-LD, C + P-SD). The Least Significant Difference (LSD) test indicates a significant (p ≤ 0.05) difference between the control and all conditions. * = Showed significant differences and ns = Showed non-significant differences.
4. Discussion
Only two species of the genus Liriodendron exist in nature, L. chinense and L. tulipifera. It is an extinct plant. These are commonly planted for landscaping and timber production in China and the US due to their great material qualities and ornamental value [44,46]. China’s central-western and southern regions are home to L. chinense (Hemsl.) Sarg. (L. chinense), which is typically found in the mountains at altitudes between 450 and 1800 m [56]. Researchers studying the disjunctive distribution of flowering plants between eastern Asia and eastern North America have shown that Liriodendron is an ideal natural resource [57]. Numerous items, including furniture and agricultural machinery, are made from Liriodendron wood [1]. Several studies have demonstrated the powerful cytotoxic effects of extracts from Liriodendron leaves on tumor cell lines, as well as their inhibitory activity against farnesyl protein transferase (FPTase) and tumor cell development. [1,56] Despite Liriodendron’s high economic worth and L. chinense’s threatened status, little is known about their population’s genetic makeup and geographic variation [56]. ABA appears crucial for bud dormancy induction and maintenance but not for dormancy release. Bud dormancy-related gene expression in stratified peach seeds was studied with measurements of ABA levels to look into the relationship between seed and bud dormancy [58]. In numerous plant species, ABA has been shown to perform a notable role in inducing seed and bud dormancy and controlling stress responses [59]. For perennial plants to survive, the transition from bud dormancy is an essential developmental process [59]. Diverse endogenous genetic variables and environmental cues control the process; however, the underlying mechanisms are still poorly understood. According to other studies, ABA is only required for bud growth and maintenance, not for the release of dormancy [59]. In Liriodendron twigs, photoperiod substantially impacted the budburst percentage (BB%) and interacted with chilling [8]. The BB% in Liriodendron twigs responded strongly to photoperiod. In this species, the BB% under LD conditions was consistently higher than 50%, but under SD conditions, it was lower than 20%. This was true regardless of the length of the chilling period [8]. The high sensitivity to photoperiod would delay or prevent bud bursting during warm winters (high sensitivity of days to budburst, Liriodendron twigs, Phoebe, and Torreya seedlings), lowering the risk of frost damage brought on by false springs [8].
There are numerous forms of unfavorable environmental conditions to which plants are subjected. Plants can mitigate the negative impacts of extreme environmental conditions by utilizing various morphological, physiological, and molecular defense mechanisms [25,60]. According to Gillebert et al. [61], protein-kinase-mediated phosphorylation is crucial for reducing ecological pressures, and the SnRK2 genes, as well-known members of the serine-threonine protein kinase subfamily, are essential for combating environmental challenges such as drought [62,63]. SnRK2s mostly regulate the development of abscisic acid-dependent plants and their responses to environmental stressors. SnRK2s are either not activated or weakly activated in plants in response to ABA regulating abiotic stress responses. Furthermore, SnRK2 genes are essential controllers of the ABA signal-transduction pathway [64,65,66]. Fortunately, L. chinense whole-genome sequencing data are now accessible [44], which presents a fantastic chance to investigate this gene family at the genome level.
According to numerous research, each SnRK2 gene uniquely responds to multiple environmental conditions. The majority of SnRK2s in model plant species such as rice [21], arabidopsis [24], and maize [27] have been described to date. The first SnRK2 gene was discovered from an ABA-treated wheat embryo cDNA library [67]. Many plant species have been found to contain SnRK2 genes. However, the SnRK2 gene family in L. chinense had not yet been identified. Five distinct SnRK2 genes were detected in the L. chinense genome by genome-wide analysis in the current study. This is fewer than the number of SnRK2 genes found in other plant species, such as 20 in cotton [22], 22 in N. tabacum [23], 10 in arabidopsis [24], 10 in rice [21], eight in mungbean [25], eight in grapevine [26], 11 in maize [27], 10 in sugarcane [28], 10 in sorghum [29], nine in pepper [13], and 12 in apple [30]. According to published research, 8 to 22 SnRK2 genes have been reported for several plant species.
The gene structure of LchiSnRK2s was analyzed and revealed two patterns. Two genes had nine exons and eight introns, while two other genes had six exons and five introns, and only one gene had two exons and one intron. The former gene structure pattern is consistent with those of cotton [22], N. tabacum [23], arabidopsis [24], rice [21], mungbean [25], grapevine [26], maize [27], sugarcane [28], sorghum [29], pepper [13], and apple [30]. The SnRK2 family in plants has undergone very little evolutionary change.
A thorough phylogenetic analysis was carried out by building a phylogenetic tree to explore the evolutionary relationship between L. chinense, A. thaliana, Solanum lycopersicum, Capsicum annuum, Solanum tuberosum, Vitis vinifera, Zea mays, Oryza sativa, and Malus domestica. The phylogenetic analysis showed that the LchiSnRK2 genes were grouped into the four previously described groups (I, II, III, and IV) [24,26,66]. Lchi00543 was the only member of the group I, and Lchi01348 was the only member of group II. Lchi13910 and Lchi12999 were the two members of group III, and Lchi25623 was in Group IV. These findings demonstrate a potential evolutionary pattern within the SnRK2 gene family [25].
The cis-regulatory motifs and elements present in the promoter sequences of the LchiSnRK2 genes will help us better understand how these genes react to different environmental situations. Our findings revealed four types of cis-elements, including those that responded to hormones, stress, growth and biological processes, and metabolism (Figure 6 and Figure 7; Table S8). Light, ABA, methyl jasmonate, anaerobic induction, gibberellin, drought, auxin, salicylic acid, circadian control, cell cycle regulation, defenses and stress, low temperature, meristem, and other extreme cis-elements were among them. Previous reviews show cis-elements promote plant stress responses [22,23,24]. SnRK2 genes were shown to have a significant role in specific agricultural plants under various environmental conditions by others who also reported comparable findings in these same plants [2]. These findings may improve our understanding of LchiSnRK2 genes in diverse contexts and conditions. The possible functionalities of various LchiSnRK2 were also characterized using 3D structural modeling. The LchiSnRK2 box, which forms a single helix and is packed parallel to the -helix in the N-terminal lobe, is a notable feature of LchiSnRK2. For kinase activity, the LchiSnRK2 box-C contact has been essential [68]. Understanding the biological functions of LchiSnRK2 proteins is possible by their modeled 3D structures.
Numerous investigations have shown that SnRK2s have a role in various abiotic stress responses. This study looked at how LchiSnRK2 expression changed in response to ABA, chilling, and photoperiod treatments. ABA also increased the expression of numerous SnRK2 genes from maize [27]. Numerous SnRK2 genes are differently controlled by ABA in various organs of the rice plant [21]. Wheat TaSnRK2.4, TaSnRK2.7, and TaSnRK2.8 expression levels are increased in response to various stressors [62]. SnRK2s, such as TaSnRK2.3, TaSnRK2.4, TaSnKR2.7, and TaSnRK2.8 from wheat [69,70,71]; and ZmSnRK2.3 and ZmSnRK2.7 from maize, have been demonstrated to Huai et al. [27]. Additionally, LchiSnRK2 genes may be effective genetic modifiers for chilling, photoperiod, and various ABA levels tolerance in plants and crops.
5. Conclusions
In conclusion, five LchiSnRK2 genes were identified during genome-wide analysis of L. chinense SnRK2 genes. The SnRK2 gene family in L. chinense was subjected to a genome-wide investigation, and the expression patterns in dormant vegetative buds under ABA foliar sprays, photoperiod, and chilling conditions were studied. Through molecular breeding, it may be possible to increase the photoperiod, chilling tolerance, and bud dormancy release of L. chinense and other crops. Our understanding of the mechanism behind L. chinense stress tolerance may be aided by the molecular elucidation of these genes.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes13081305/s1, Figure S1. The details of the 10 motifs found in LchiSnRK2 proteins. Table S1. Details about the primers used in this study’s qRT-PCR gene expression investigation. Table S2. Arabidopsis protein detected by a blast in L. chinense. Table S3. Information on the protein sequences of the nine species’ known SnRK2 members. Table S4. List of identified domains. Table S5. Information of CDS sequence of SnRK2 family in Liriodendron chinense. Table S6. Genomic sequence of SnRK2 family in L. chinense. Table S7. Promoter sequence of SnRK2 family in Liriodendron chinense. Table S8. Information of cis-elements detected in the promoter’s regions of SnRK2s family.
Author Contributions
Conceptualization, Q.H.; methodology, Q.H., M.Z. and W.C.; software, Q.H. and M.F.A.; formal analysis, Q.H. and M.W.R.; writing—original draft preparation, Q.H.; writing—review and editing, Q.H., R.K., M.A., M.S.A. and M.S.E.; supervision, R.Z. and J.W.; funding acquisition, R.Z. and J.W. All authors have read and agreed to the published version of the manuscript.
Funding
The study was financed by the Youth Elite Science Sponsorship Program of CAST (YESS, 2020QNRC001), National Forestry and Grassland Technological Innovation Program for Young TopNotch Talents (2020132604), Chinese National Natural Science Foundation (32171832), Breeding program for T. grandis (2021C02066-11), and Overseas Expertise Introduction Project for Discipline Innovation (111 Project D18008). The authors extend their appreciation to the Researchers supporting project number (RSP-2021/173), King Saud University, Riyadh, Saudi Arabia.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chen, J.H.; Yang, G.X.; Ding, Q.; Xia, T.S.; Shi, J.; Jia, A.Q. In vitro tumor cytotoxic activities of extracts from three Liriodendron plants. Pak. J. Pharm. Sci. 2013, 26, 233–237. [Google Scholar]
- Hussain, Q.; Zheng, M.; Furqan, M.; Khan, R.; Yasir, M.; Farooq, S.; Zhang, R.; Wu, J. Genome-wide identification, characterization and expression analysis of the ABA receptor PYL gene family in response to ABA, photoperiod, and chilling in vegetative buds of Liriodendron chinense. Sci. Hortic. 2022, 303, 111200. [Google Scholar] [CrossRef]
- Sheng, Y.; Hao, Z.; Peng, Y.; Liu, S.; Hu, L.; Shen, Y.; Shi, J.; Chen, J. Morphological, phenological, and transcriptional analyses provide insight into the diverse flowering traits of a mutant of the relic woody plant Liriodendron chinense. Hortic. Res. 2021, 8, 174. [Google Scholar] [CrossRef]
- Yang, Q.; Gao, Y.; Wu, X.; Moriguchi, T.; Bai, S.; Teng, Y. Bud endodormancy in deciduous fruit trees: Advances and prospects. Hortic. Res. 2021, 8, 139. [Google Scholar] [CrossRef]
- Arora, R.; Rowland, L.J.; Tanino, K. Induction and release of bud dormancy in woody perennials: A science comes of age. HortScience 2003, 38, 911–921. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Sherif, S.M. Hormonal Orchestration of Bud Dormancy Cycle in Deciduous Woody Perennials. Front. Plant Sci. 2019, 10, 1136. [Google Scholar] [CrossRef]
- Beauvieux, R.; Wenden, B.; Dirlewanger, E. Bud dormancy in perennial fruit tree species: A pivotal role for oxidative cues. Front. Plant Sci. 2018, 9, 657. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, F.; Zheng, J.; Lin, J.; Hänninen, H.; Wu, J. Chilling accumulation and photoperiod regulate rest break and bud burst in five subtropical tree species. For. Ecol. Manag. 2021, 485, 118183. [Google Scholar] [CrossRef]
- Derory, J.; Léger, P.; Garcia, V.; Schaeffer, J.; Hauser, M.; Salin, F.; Luschnig, C.; Plomion, C.; Glössl, J.; Kremer, A.; et al. Transcriptome analysis of bud burst in sessile oak (Quercus petraea). New Phytol. 2006, 170, 723–738. [Google Scholar] [CrossRef]
- Zohner, C.M.; Benito, B.M.; Svenning, J.C.; Renner, S.S. Day length unlikely to constrain climate-driven shifts in leaf-out times of northern woody plants. Nat. Clim. Chang. 2016, 6, 1120–1123. [Google Scholar] [CrossRef]
- Pletsers, A.; Caffarra, A.; Kelleher, C.T.; Donnelly, A. Chilling temperature and photoperiod influence the timing of bud burst in juvenile Betula pubescens Ehrh. and Populus tremula L. trees. Ann. For. Sci. 2015, 72, 941–953. [Google Scholar] [CrossRef] [Green Version]
- Jewaria, P.K.; Hänninen, H.; Li, X.; Bhalerao, R.P.; Zhang, R. A hundred years after: Endodormancy and the chilling requirement in subtropical trees. New Phytol. 2021, 231, 565–570. [Google Scholar] [CrossRef]
- Wu, Z.; Cheng, J.; Hu, F.; Qin, C.; Xu, X.; Hu, K. The SnRK2 family in pepper (Capsicum annuum L.): Genome-wide identification and expression analyses during fruit development and under abiotic stress. Genes Genomics 2020, 42, 1117–1130. [Google Scholar] [CrossRef]
- Wang, X.; Wang, L.; Wang, Y.; Liu, H.; Hu, D.; Zhang, N.; Zhang, S.; Cao, H.; Cao, Q.; Zhang, Z.; et al. Arabidopsis PCaP2 plays an important role in chilling tolerance and ABA response by activating CBF- and SnRK2-mediated transcriptional regulatory network. Front. Plant Sci. 2018, 9, 215. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Zhang, Z.; Gao, J.; Wang, P.; Hu, T.; Wang, Z.; Hou, Y.J.; Wan, Y.; Liu, W.; Xie, S.; et al. Arabidopsis duodecuple mutant of PYL ABA receptors reveals PYL repression of ABA-independent SnRK2 activity. Cell Rep. 2018, 23, 3340–3351. [Google Scholar] [CrossRef]
- Lin, Q.; Wu, F.; Sheng, P.; Zhang, Z.; Zhang, X.; Guo, X.; Wang, J.; Cheng, Z.; Wang, J.; Wang, H.; et al. The SnRK2-APC/C TE regulatory module mediates the antagonistic action of gibberellic acid and abscisic acid pathways. Nat. Commun. 2015, 6, 7981. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Li, W.; Mao, X.; Jing, R.; Jia, H. Differential activation of the wheat SnRK2 family by abiotic stresses. Front. Plant Sci. 2016, 7, 420. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Ji, W.; Gao, P.; Li, Y.; Cai, H.; Bai, X.; Chen, Q.; Zhu, Y. GsAPK, an ABA-activated and calcium-independent SnRK2-type kinase from G. soja, mediates the regulation of plant tolerance to salinity and ABA stress. PLoS ONE 2012, 7, e33838. [Google Scholar] [CrossRef] [Green Version]
- Coello, P.; Hirano, E.; Hey, S.J.; Muttucumaru, N.; Martinez-Barajas, E.; Parry, M.A.J.; Halford, N.G. Evidence that abscisic acid promotes degradation of SNF1-related protein kinase (SnRK) 1 in wheat and activation of a putative calcium-dependent SnRK2. J. Exp. Bot. 2012, 63, 913–924. [Google Scholar] [CrossRef] [Green Version]
- Hrabak, E.M.; Chan, C.W.M.; Gribskov, M.; Harper, J.F.; Choi, J.H.; Halford, N.; Kudla, J.; Luan, S.; Nimmo, H.G.; Sussman, M.R.; et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003, 132, 666–680. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, Y.; Yamamoto, S.; Minami, H.; Kagaya, Y.; Hattori, T. Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell 2004, 16, 1163–1177. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Ge, X.; Yang, Z.; Zhang, C.; Zhao, G.; Chen, E.; Liu, J.; Zhang, X.; Li, F. Genome-wide identification and characterization of SnRK2 gene family in cotton (Gossypium hirsutum L.). BMC Genet. 2017, 18, 54. [Google Scholar] [CrossRef]
- Li, J.; Song, J.; Li, C.; Ma, J.; Liu, J.; Zhu, X.; Li, J.; He, F.; Yang, C. Genome-Wide Identification and Expression Profile Analysis of the SnRK2 Gene Family in Nicotiana tabacum. Biochem. Genet. 2022, 1–16. [Google Scholar] [CrossRef]
- Saha, J.; Chatterjee, C.; Sengupta, A.; Gupta, K.; Gupta, B. Genome-wide analysis and evolutionary study of sucrose non-fermenting 1-related protein kinase 2 (SnRK2) gene family members in Arabidopsis and Oryza. Comput. Biol. Chem. 2014, 49, 59–70. [Google Scholar] [CrossRef]
- Fatima, A.; Khan, M.J.; Awan, H.M.; Akhtar, M.N.; Bibi, N.; Sughra, K.; Khan, M.R.; Ahmad, R.; Ibrahim, M.; Hussain, J.; et al. Genome-wide identification and expression analysis of SnRK2 gene family in mungbean (Vigna radiata) in response to drought stress. Crop Pasture Sci. 2020, 71, 469–476. [Google Scholar] [CrossRef]
- Liu, J.Y.; Chen, N.N.; Cheng, Z.M.; Xiong, J.S. Genome-wide identification, annotation and expression profile analysis of SnRK2 gene family in grapevine. Aust. J. Grape Wine Res. 2016, 22, 478–488. [Google Scholar] [CrossRef]
- Huai, J.; Wang, M.; He, J.; Zheng, J.; Dong, Z.; Lv, H.; Zhao, J.; Wang, G. Cloning and characterization of the SnRK2 gene family from Zea mays. Plant Cell Rep. 2008, 27, 1861–1868. [Google Scholar] [CrossRef]
- Li, C.; Nong, Q.; Xie, J.; Wang, Z.; Liang, Q.; Solanki, M.K.; Malviya, M.K.; Liu, X.; Li, Y.; Htun, R.; et al. Molecular Characterization and Co-expression Analysis of the SnRK2 Gene Family in Sugarcane (Saccharum officinarum L.). Sci. Rep. 2017, 7, 17659. [Google Scholar] [CrossRef] [Green Version]
- Li, L.-B.; Zhang, Y.-R.; Liu, K.-C.; NI, Z.-F.; Fang, Z.-J.; Sun, Q.-X.; Gao, J.-W. Identification and Bioinformatics Analysis of SnRK2 and CIPK Family Genes in Sorghum. Agric. Sci. China 2010, 9, 19–30. [Google Scholar] [CrossRef]
- Shao, Y.; Qin, Y.; Zou, Y.; Ma, F. Genome-wide identification and expression profiling of the SnRK2 gene family in Malus prunifolia. Gene 2014, 552, 87–97. [Google Scholar] [CrossRef]
- Fujita, Y.; Nakashima, K.; Yoshida, T.; Katagiri, T.; Kidokoro, S.; Kanamori, N.; Umezawa, T.; Fujita, M.; Maruyama, K.; Ishiyama, K.; et al. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in arabidopsis. Plant Cell Physiol. 2009, 50, 2123–2132. [Google Scholar] [CrossRef] [Green Version]
- Fujita, Y.; Yoshida, T.; Yamaguchi-Shinozaki, K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant. 2013, 147, 15–27. [Google Scholar] [CrossRef]
- Hussain, Q.; Asim, M.; Zhang, R.; Khan, R.; Farooq, S.; Wu, J. Transcription factors interact with ABA through gene expression and signaling pathways to mitigate drought and salinity stress. Biomolecules 2021, 11, 1159. [Google Scholar] [CrossRef]
- Hussain, Q.; Shi, J.; Scheben, A.; Zhan, J.; Wang, X.; Liu, G.; Yan, G.; King, G.J.; Edwards, D.; Wang, H. Genetic and signalling pathways of dry fruit size: Targets for genome editing-based crop improvement. Plant Biotechnol. J. 2020, 18, 1124–1140. [Google Scholar] [CrossRef]
- Hussain, Q.; Zhan, J.; Liang, H.; Wang, X.; Liu, G.; Shi, J.; Wang, H. Key genes and mechanisms underlying natural variation of silique length in oilseed rape (Brassica napus L.) germplasm. Crop J. 2022, 10, 617–626. [Google Scholar] [CrossRef]
- Boneh, U.; Biton, I.; Schwartz, A.; Ben-Ari, G. Characterization of the ABA signal transduction pathway in Vitis vinifera. Plant Sci. 2012, 187, 89–96. [Google Scholar] [CrossRef]
- Karssen, C.M.; Brinkhorst-van der Swan, D.L.C.; Breekland, A.E.; Koornneef, M. Induction of dormancy during seed development by endogenous abscisic acid: Studies on abscisic acid deficient genotypes of Arabidopsis thaliana (L.) Heynh. Planta 1983, 157, 158–165. [Google Scholar] [CrossRef]
- Li, J.; Xu, Y.; Niu, Q.; He, L.; Teng, Y.; Bai, S. Abscisic acid (ABA) promotes the induction and maintenance of pear (Pyrus pyrifolia white pear group) flower bud endodormancy. Int. J. Mol. Sci. 2018, 19, 310. [Google Scholar] [CrossRef] [Green Version]
- Hubbard, K.E.; Nishimura, N.; Hitomi, K.; Getzoff, E.D.; Schroeder, J.I. Early abscisic acid signal transduction mechanisms: Newly discovered components and newly emerging questions. Genes Dev. 2010, 24, 1695–1708. [Google Scholar] [CrossRef] [Green Version]
- Rombauts, S.; Ruttink, T.; Arend, M.; Morreel, K.; Bhalerao, R.P.; Boerjan, W.; Rohde, A. A Molecular Timetable for Apical Bud Formation and Dormancy Induction in Poplar. Plant Cell 2007, 19, 2370–2390. [Google Scholar] [CrossRef] [Green Version]
- Rohde, A.; Bhalerao, R.P. Plant dormancy in the perennial context. Trends Plant Sci. 2007, 12, 217–223. [Google Scholar] [CrossRef]
- Zheng, C.; Halaly, T.; Acheampong, A.K.; Takebayashi, Y.; Jikumaru, Y. Abscisic acid (ABA) regulates grape bud dormancy, and dormancy release stimuli may act through modification of ABA metabolism. J. Exp. Bot. 2015, 66, 1527–1542. [Google Scholar] [CrossRef]
- Li, C.; Junttila, O.; Heino, P.; Palva, E.T. Low temperature sensing in silver birch (Betula pendula Roth) ecotypes. Plant Sci. 2004, 167, 165–171. [Google Scholar] [CrossRef]
- Chen, J.; Hao, Z.; Guang, X.; Zhao, C.; Wang, P.; Xue, L.; Zhu, Q.; Yang, L.; Sheng, Y.; Zhou, Y.; et al. Liriodendron genome sheds light on angiosperm phylogeny and species—Pair differentiation. Nat. Plants 2019, 5, 18–25. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Liu, H.; Hao, Z.; Zong, Y.; Xia, H.; Shen, Y.; Li, H. Genome-wide identification and expression analysis of r2r3-myb family genes associated with petal pigment synthesis in Liriodendron. Int. J. Mol. Sci. 2021, 22, 11291. [Google Scholar] [CrossRef]
- Hu, L.; Wang, P.; Hao, Z.; Lu, Y.; Xue, G.; Cao, Z.; Qu, H.; Cheng, T.; Shi, J.; Chen, J. Gibberellin oxidase gene family in L. Chinense: Genome-wide identification and gene expression analysis. Int. J. Mol. Sci. 2021, 22, 7167. [Google Scholar] [CrossRef]
- Yadav, S.K.; Santosh Kumar, V.V.; Verma, R.K.; Yadav, P.; Saroha, A.; Wankhede, D.P.; Chaudhary, B.; Chinnusamy, V. Genome-wide identification and characterization of ABA receptor PYL gene family in rice. BMC Genomics 2020, 21, 676. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Raza, A.; Wei, S.; Ang, G.; Mehmood, S.S.; Hussain, M.A.; Nie, W.; Yan, L.; Zou, X.; Zhang, X. Catalase (Cat) gene family in rapeseed (Brassica napus L.): Genome-wide analysis, identification and expression pattern in response to multiple hormones and abiotic stress conditions. Int. J. Mol. Sci. 2021, 22, 4281. [Google Scholar] [CrossRef]
- Su, W.; Raza, A.; Gao, A.; Jia, Z.; Zhang, Y.; Hussain, M.A.; Mehmood, S.S.; Cheng, Y.; Lv, Y.; Zou, X. Genome-wide analysis and expression profile of superoxide dismutase (Sod) gene family in rapeseed (Brassica napus L.) under different hormones and abiotic stress conditions. Antioxidants 2021, 10, 1182. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Zong, Y.; Hao, Z.; Tu, Z.; Shen, Y.; Zhang, C.; Wen, S.; Yang, L.; Ma, J.; Li, H. Genome-wide survey and identification of AP2/ERF genes involved in shoot and leaf development in Liriodendron chinense. BMC Genomics 2021, 22, 807. [Google Scholar] [CrossRef]
- Liu, H.; Yang, L.; Tu, Z.; Zhu, S.; Zhang, C.; Li, H. Genome-wide identification of MIKC-type genes related to stamen and gynoecium development in Liriodendron. Sci. Rep. 2021, 11, 6585. [Google Scholar] [CrossRef]
- Riaz, M.W.; Lu, J.; Shah, L.; Yang, L.; Chen, C.; Mei, X.D.; Xue, L.; Manzoor, M.A.; Abdullah, M.; Rehman, S.; et al. Expansion and Molecular Characterization of AP2/ERF Gene Family in Wheat (Triticum aestivum L.). Front. Genet. 2021, 12, 632155. [Google Scholar] [CrossRef]
- Khan, R.; Ma, X.; Zhang, J.; Wu, X.; Iqbal, A.; Wu, Y.; Zhou, L.; Wang, S. Circular drought-hardening confers drought tolerance via modulation of the antioxidant defense system, osmoregulation, and gene expression in tobacco. Physiol. Plant. 2021, 172, 1073–1088. [Google Scholar] [CrossRef]
- Long, X.; Weng, Y.; Liu, S.; Hao, Z.; Sheng, Y.; Guan, L.; Shi, J.; Chen, J. Genetic diversity and differentiation of relict plant Liriodendron populations based on 29 Novel EST-SSR markers. Forests 2019, 10, 334. [Google Scholar] [CrossRef] [Green Version]
- Xiang, Q.Y.; Soltis, D.E.; Soltis, P.S. The Eastern Asian and Eastern and Western North American Floristic Disjunction: Congruent Phylogenetic Patterns in Seven Diverse Genera. Mol. Phylogenet. Evol. 1998, 10, 178–190. [Google Scholar] [CrossRef] [Green Version]
- Leida, C.; Conesa, A.; Llácer, G.; Badenes, M.L.; Ríos, G. Histone modifications and expression of DAM6 gene in peach are modulated during bud dormancy release in a cultivar-dependent manner. New Phytol. 2012, 193, 67–80. [Google Scholar] [CrossRef]
- Ionescu, I.A.; Møller, B.L.; Sánchez-Pérez, R. Chemical control of flowering time. J. Exp. Bot. 2017, 68, 369–382. [Google Scholar] [CrossRef]
- Fujita, M.; Fujita, Y.; Noutoshi, Y.; Takahashi, F.; Narusaka, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Crosstalk between abiotic and biotic stress responses: A current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 2006, 9, 436–442. [Google Scholar] [CrossRef]
- Ghillebert, R.; Swinnen, E.; Wen, J.; Vandesteene, L.; Ramon, M.; Norga, K.; Rolland, F.; Winderickx, J. The AMPK/SNF1/SnRK1 fuel gauge and energy regulator: Structure, function and regulation. FEBS J. 2011, 278, 3978–3990. [Google Scholar] [CrossRef]
- Kulik, A.; Wawer, I.; Krzywińska, E.; Bucholc, M.; Dobrowolska, G. SnRK2 protein Kinases—Key regulators of plant response to abiotic stresses. OMI A J. Integr. Biol. 2011, 15, 859–872. [Google Scholar] [CrossRef]
- Fujii, H.; Zhu, J.K. Osmotic stress signaling via protein kinases. Cell Mol. Life Sci. 2012, 69, 3165–3173. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef]
- Park, S.Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.F.F.; et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [Green Version]
- Umezawa, T.; Sugiyama, N.; Mizoguchi, M.; Hayashi, S.; Myouga, F.; Yamaguchi-Shinozaki, K.; Ishihama, Y.; Hirayama, T.; Shinozaki, K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 17588–17593. [Google Scholar] [CrossRef] [Green Version]
- Robert, J.A.; Walker-Simmon, M.K. Isolation of a wheat cDNA clone for an abscisic acid-inducible transcript with homology to protein kinases. Proc. Natl. Acad. Sci. USA 2015, 89, 10183–10187. [Google Scholar]
- Ng, L.M.; Soon, F.F.; Zhou, X.E.; West, G.M.; Kovach, A.; Suino-Powell, K.M.; Chalmers, M.J.; Li, J.; Yong, E.L.; Zhu, J.K.; et al. Structural basis for basal activity and autoactivation of abscisic acid (ABA) signaling SnRK2 kinases. Proc. Natl. Acad. Sci. USA 2011, 108, 21259–21264. [Google Scholar] [CrossRef] [Green Version]
- Tian, S.; Mao, X.; Zhang, H.; Chen, S.; Zhai, C.; Yang, S.; Jing, R. Cloning and characterization of TaSnRK2.3, a novel SnRK2 gene in common wheat. J. Exp. Bot. 2013, 64, 2063–2080. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Mao, X.; Wang, C.; Jing, R. Overexpression of a common wheat gene TASnRK2.8 enhances tolerance to drought, salt and low temperature in Arabidopsis. PLoS ONE 2010, 5, 2–13. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Mao, X.; Jing, R.; Chang, X.; Xie, H. Characterization of a common wheat (Triticum aestivum L.) TaSnRK2.7 gene involved in abiotic stress responses. J. Exp. Bot. 2011, 62, 975–988. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).