Plastid Phylogenomics and Plastomic Diversity of the Extant Lycophytes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Taxon Sampling, DNA Extraction, and Sequencing
2.2. Plastome Assembly, and Annotation
2.3. Phylogenetic Analysis
2.4. Comparative Plastomic Analysis
2.5. Divergence Time Estimation and Ancestral State Reconstruction
2.6. Estimation of Substitution Rates
2.7. RNA Editing Analysis
3. Results
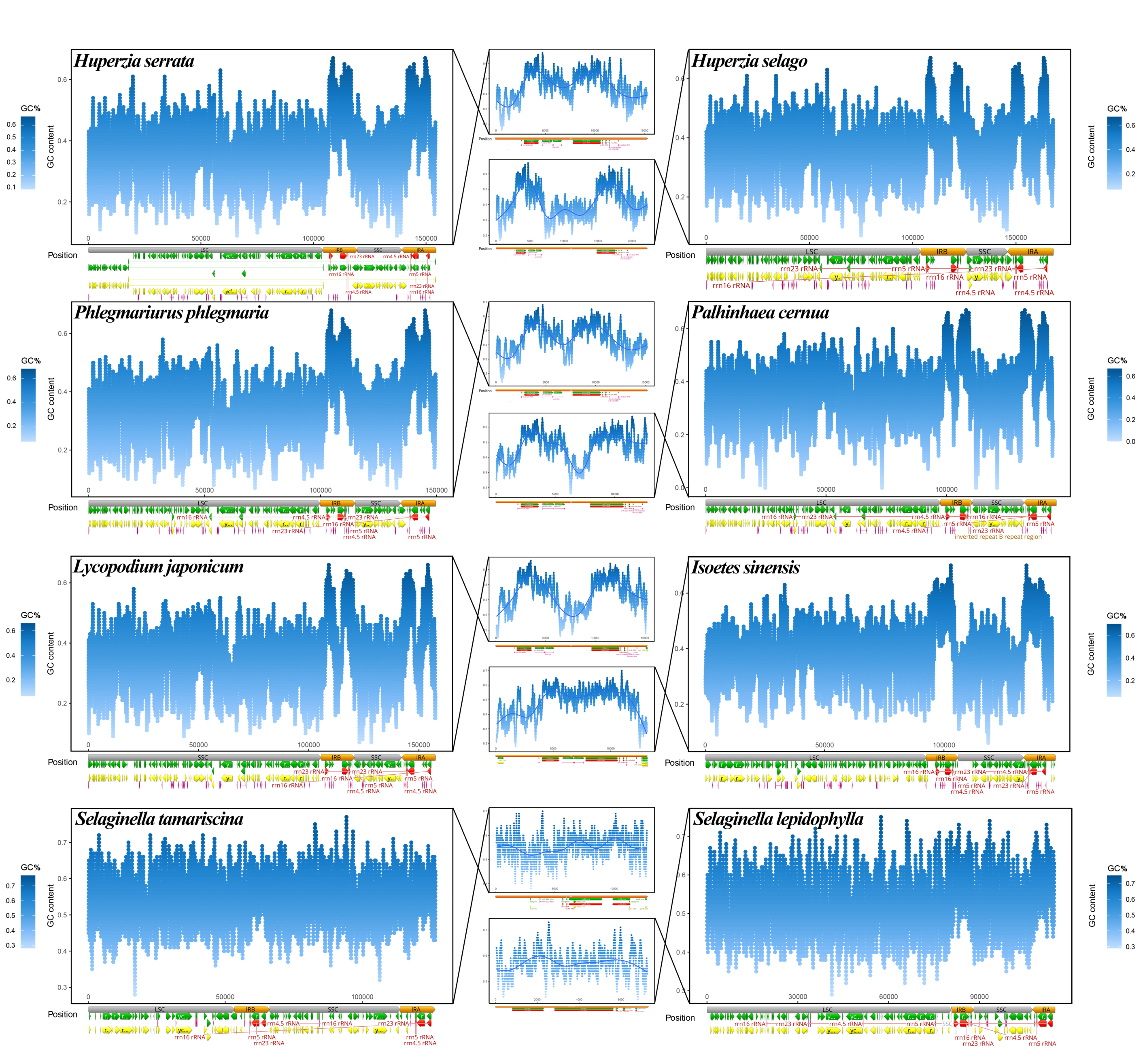
3.1. Features of Lycophyte Plastomes
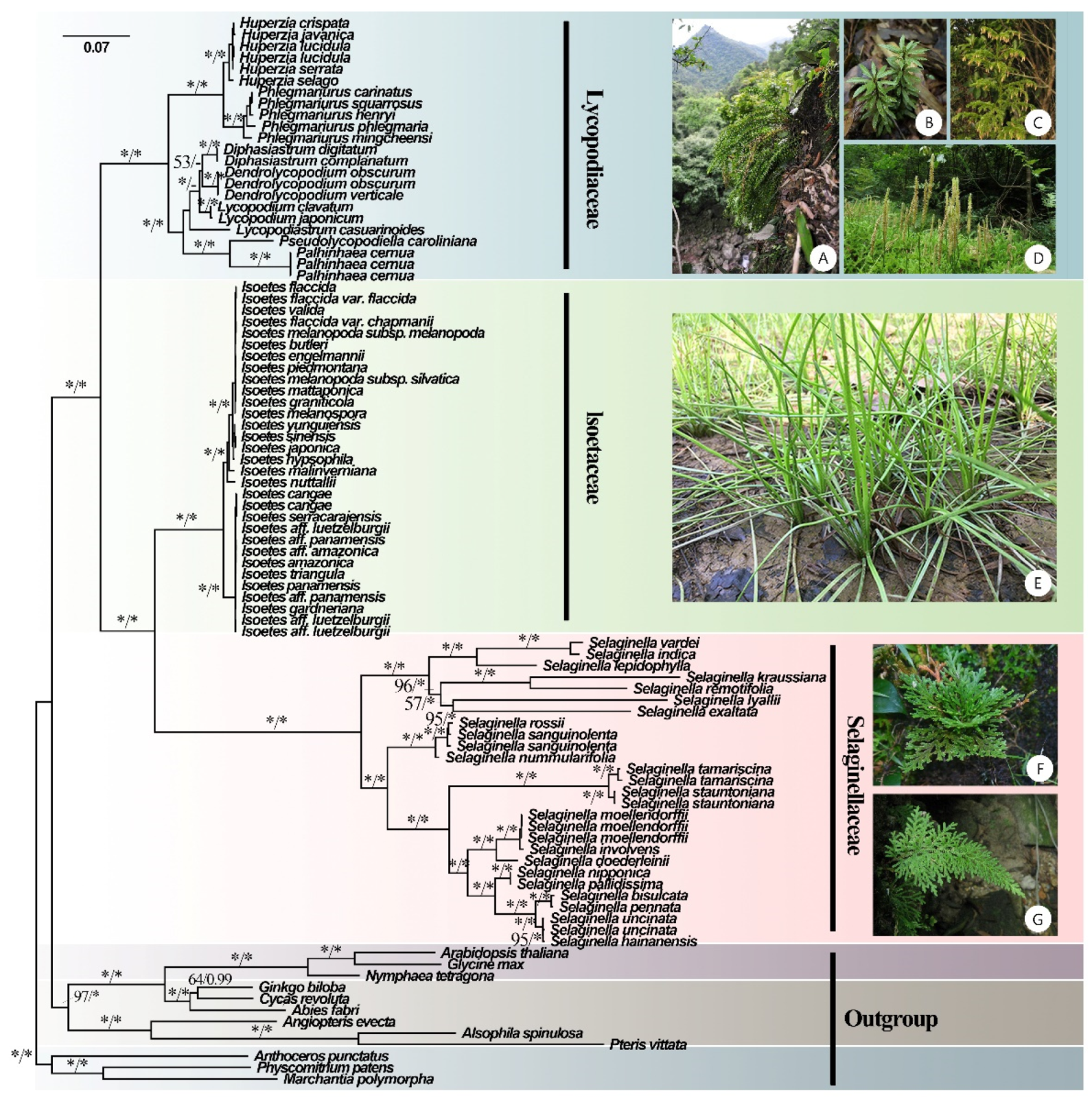
3.2. Phylogenetic Analyses
3.3. GC Content Distribution among Plastomes
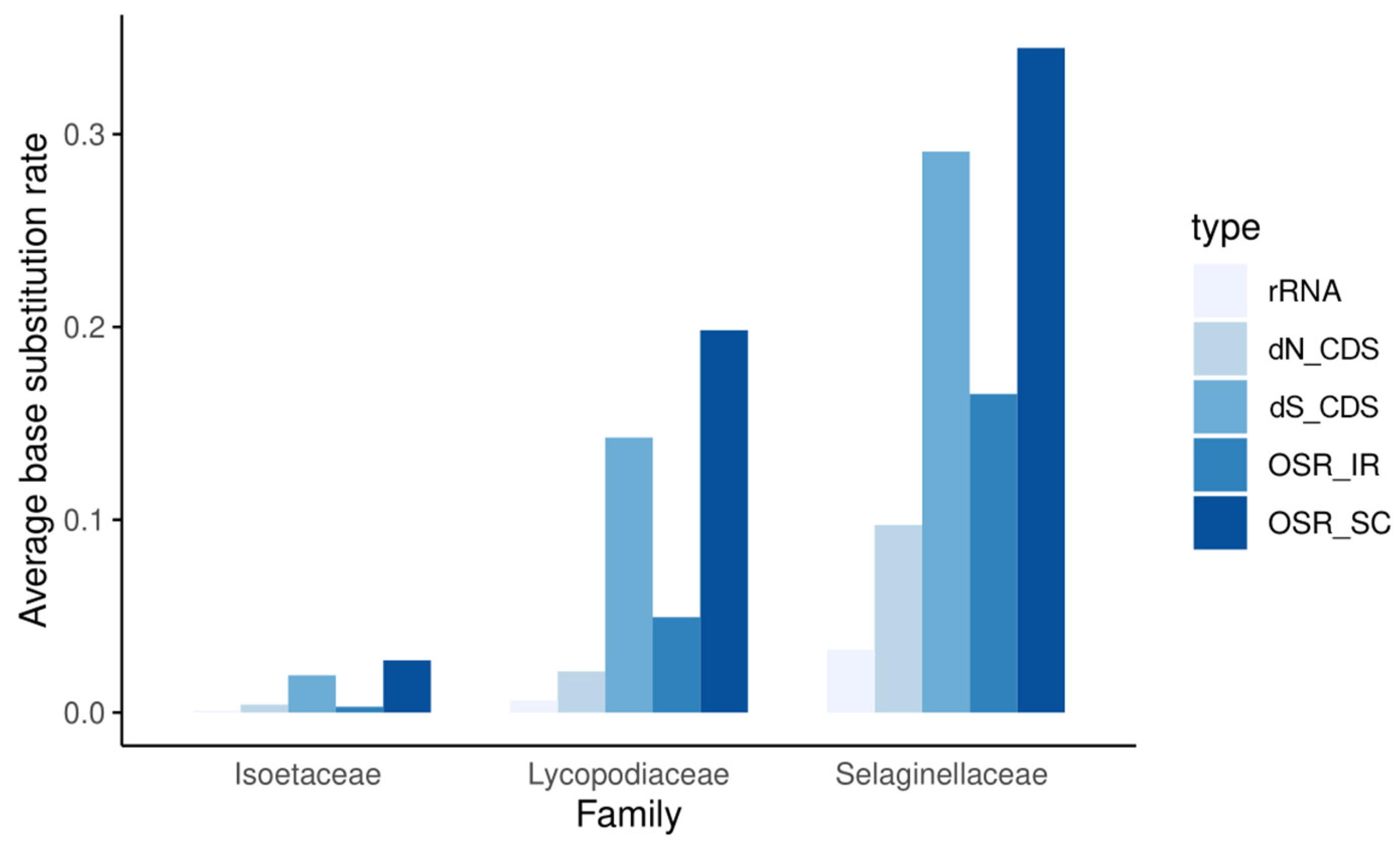
3.4. Base Substitution Rate Estimation
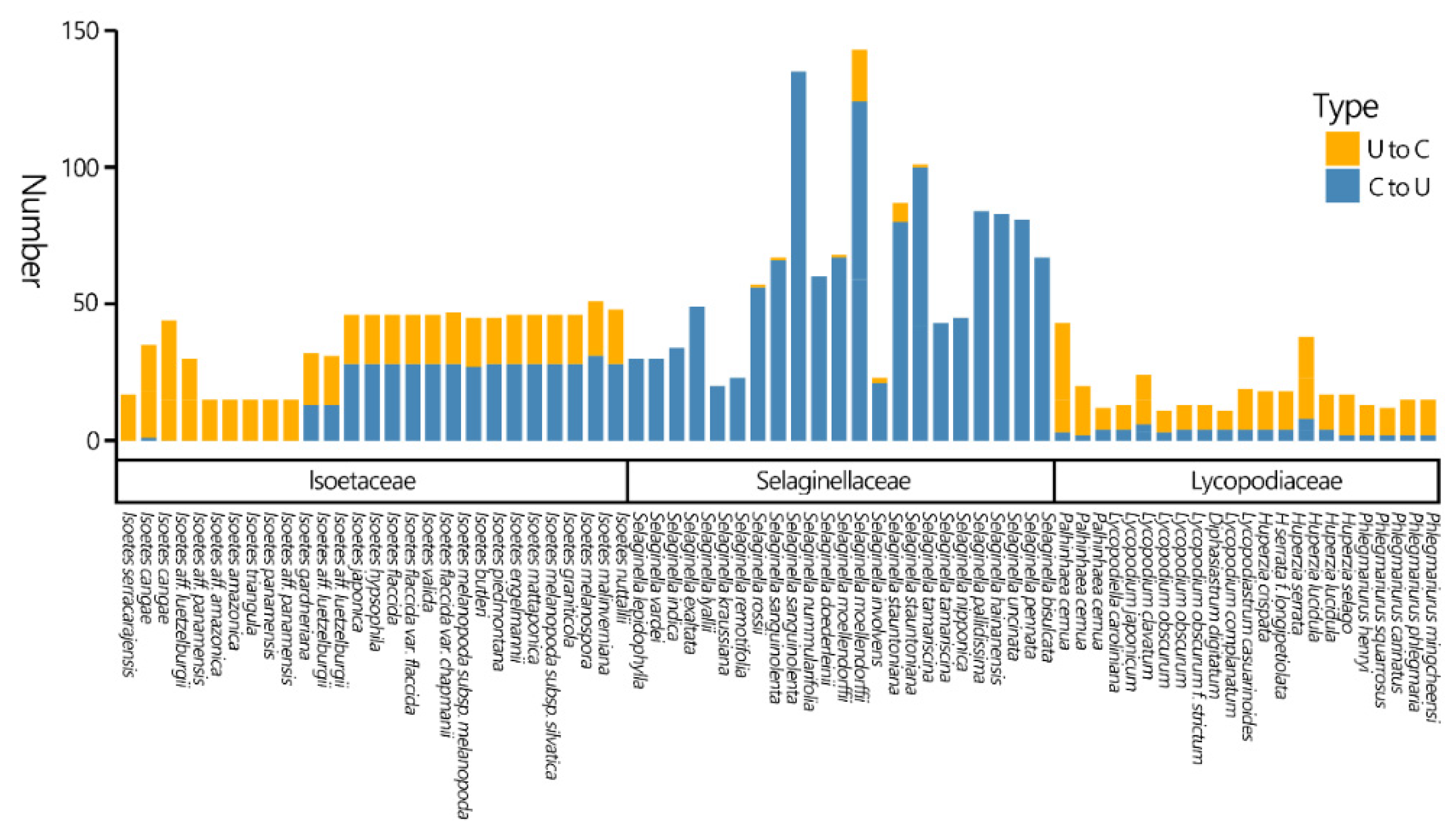
3.5. RNA Editing Analysis
3.6. Divergence Time Inference and Ancestral State Reconstruction
4. Discussion
4.1. Plastomic Diversity of Lycophytes
4.2. Phylogenetic Relationships and Divergence Times
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kenrick, P.; Crane, P.R. The origin and early evolution of plants on land. Nature 1997, 389, 33–39. [Google Scholar] [CrossRef]
- Morris, J.L.; Puttick, M.N.; Clark, J.W.; Edwards, D.; Kenrick, P.; Pressel, S.; Wellman, C.H.; Yang, Z.; Schneider, H.; Donoghue, P.C.J. The timescale of early land plant evolution. Proc. Natl. Acad. Sci. USA 2018, 115, E2274–E2283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bennici, A. Unresolved problems on the origin and early evolution of land plants. Riv. Biol. 2007, 100, 55–67. [Google Scholar] [PubMed]
- Bennici, A. Origin and early evolution of land plants. Commun. Integr. Biol. 2008, 1, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Liu, J.; Zeng, M.; He, J.; Qin, P.; Huang, H.; Xu, L. Identification of WOX family genes in Selaginella kraussiana for studies on stem cells and regeneration in lycophytes. Front. Plant Sci. 2016, 7, 93. [Google Scholar] [CrossRef] [Green Version]
- Kenrick, P.; Strullu-Derrien, C. The origin and early evolution of roots. Plant Physiol. 2014, 166, 570–580. [Google Scholar] [CrossRef] [Green Version]
- White, M.E. The Greening of Gondwana; Reed: Sydney, Australia, 1986. [Google Scholar]
- Schuettpelz, E.; Schneider, H.; Smith, A.R.; Hovenkamp, P.; Prado, J.; Rouhan, G.; Salino, A.; Sundue, M.; Almeida, T.E.; Parris, B.; et al. A community-derived classification for extant lycophytes and ferns. J. Syst. Evol. 2016, 54, 563–603. [Google Scholar] [CrossRef]
- Spencer, V.; Nemec Venza, Z.; Harrison, C.J. What can lycophytes teach us about plant evolution and development? Modern perspectives on an ancient lineage. Evol. Dev. 2020, 23, 174–196. [Google Scholar]
- Wolfe, A.D.; Randle, C.P. Recombination, heteroplasmy, haplotype polymorphism, and paralogy in plastid genes: Implications for plant molecular systematics. Syst. Bot. 2004, 29, 1011–1020. [Google Scholar] [CrossRef]
- Wolfe, K.H.; Li, W.H.; Sharp, P.M. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. USA 1987, 84, 9054–9058. [Google Scholar] [CrossRef] [Green Version]
- Jansen, R.K.; Raubeson, L.A.; Boore, J.L.; Depamphilis, C.W.; Chumley, T.W.; Haberle, R.C.; Wyman, S.K.; Alverson, A.J.; Peery, R.; Herman, S.J.; et al. Methods for obtaining and analyzing whole chloroplast genome sequences. Methods Enzymol. 2005, 395, 348–384. [Google Scholar] [PubMed]
- Wu, C.S.; Wang, Y.N.; Hsu, C.Y.; Lin, C.P.; Chaw, S.M. Loss of different inverted repeat copies from the chloroplast genomes of pinaceae and cupressophytes and influence of heterotachy on the evaluation of gymnosperm phylogeny. Genome Biol. Evol. 2011, 3, 1284–1295. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.S.; Chaw, S.M. Highly rearranged and size-variable chloroplast genomes in conifers II clade (cupressophytes): Evolution towards shorter intergenic spacers. Plant Biotechnol. J. 2014, 12, 344–353. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, L.A.C.; Da Costa, Z.P.; Callot, C.; Cauet, S.; Zucchi, M.I.; Bergès, H.; Berg, C.V.D.; Vieira, M.L.C. A repertory of rearrangements and the loss of an inverted repeat region in Passiflora chloroplast genomes. Genome Biol. Evol. 2020, 12, 1841–1857. [Google Scholar] [CrossRef] [PubMed]
- Cronk, Q.; Ojeda, I.; Pennington, R.T. Legume comparative genomics: Progress in phylogenetics and phylogenomics. Curr. Opin. Plant Biol. 2006, 9, 99–103. [Google Scholar] [CrossRef]
- Zhang, H.R.; Xiang, Q.P.; Zhang, X.C. The unique evolutionary trajectory and dynamic conformations of DR and IR/DR-coexisting plastomes of the early vascular plant Selaginellaceae (Lycophyte). Genome Biol. Evol. 2019, 11, 1258–1274. [Google Scholar] [CrossRef]
- Mower, J.P.; Ma, P.; Grewe, F.; Taylor, A.; Michael, T.P.; VanBuren, R.; Qiu, Y. Lycophyte plastid genomics: Extreme variation in GC, gene and intron content and multiple inversions between a direct and inverted orientation of the rRNA repeat. New Phytol. 2019, 222, 1061–1075. [Google Scholar] [CrossRef] [Green Version]
- Smith, D.R. Updating our view of organelle genome nucleotide landscape. Front. Genet. 2012, 3, 175. [Google Scholar] [CrossRef] [Green Version]
- Du, X.; Lu, J.; Li, D. Advances in the evolution of plastid genome structure in lycophytes and ferns. Biodivers. Sci. 2019, 27, 1172–1183. [Google Scholar]
- Kang, J.S.; Zhang, H.R.; Wang, Y.R.; Liang, S.Q.; Mao, Z.Y.; Zhang, X.C.; Xiang, Q. Distinctive evolutionary pattern of organelle genomes linked to the nuclear genome in Selaginellaceae. Plant J. 2020, 104, 1657–1672. [Google Scholar] [CrossRef]
- Shim, H.; Lee, H.J.; Lee, J.; Lee, H.O.; Kim, J.H.; Yang, T.J.; Kim, N.-S. Plastid genomes of the early vascular plant genus Selaginella have unusual direct repeat structures and drastically reduced gene numbers. Int. J. Mol. Sci. 2021, 22, 641. [Google Scholar] [CrossRef]
- Smith, D.R. Unparalleled GC content in the plastid DNA of Selaginella. Plant Mol. Biol. 2009, 71, 627–639. [Google Scholar] [CrossRef] [PubMed]
- Small, R.L.; Lickey, E.B.; Shaw, J.; Hauk, W.D. Amplification of noncoding chloroplast DNA for phylogenetic studies in lycophytes and monilophytes with a comparative example of relative phylogenetic utility from Ophioglossaceae. Mol. Phylogenet. Evol. 2005, 36, 509–522. [Google Scholar] [CrossRef]
- Zhou, X.M.; Rothfels, C.J.; Zhang, L.; He, Z.R.; Le Péchon, T.; He, H.; Lu, N.T.; Knapp, R.; Lorence, D.; He, X.-J.; et al. A large-scale phylogeny of the lycophyte genus Selaginella (Selaginellaceae: Lycopodiopsida) based on plastid and nuclear loci. Cladistics 2016, 32, 360–389. [Google Scholar] [CrossRef] [PubMed]
- Pereira, J.B.S.; Labiak, P.H.; Stützel, T.; Schulz, C. Origin and biogeography of the ancient genus Isoëtes with focus on the Neotropics. Bot. J. Linn. Soc. 2017, 185, 253–271. [Google Scholar] [CrossRef]
- Bauret, L.; Field, A.R.; Gaudeul, M.; Selosse, M.A.; Rouhan, G. First insights on the biogeographical history of Phlegmariurus (Lycopodiaceae), with a focus on Madagascar. Mol. Phylogenet. Evol. 2018, 127, 488–501. [Google Scholar] [CrossRef]
- Testo, W.L.; Sessa, E.; Barrington, D.S. The rise of the Andes promoted rapid diversification in Neotropical Phlegmariurus (Lycopodiaceae). New Phytol. 2019, 222, 604–613. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Zhou, X.; Rothfels, C.J.; Shepherd, L.D.; Knapp, R.; Zhang, L.; Lu, N.T.; Fan, X.; Wan, X.; Gao, X.; et al. A global phylogeny of Lycopodiaceae (Lycopodiales; lycophytes) with the description of a new genus, Brownseya, from Oceania. Taxon 2022, 71, 25–51. [Google Scholar] [CrossRef]
- Wolf, P.G.; Karol, K.G.; Mandoli, D.F.; Kuehl, J.; Arumuganathan, K.; Ellis, M.W.; Mishler, B.D.; Kelch, D.G.; Olmstead, R.G.; Boore, J.L. The first complete chloroplast genome sequence of a lycophyte, Huperzia lucidula (Lycopodiaceae). Gene 2005, 350, 117–128. [Google Scholar] [CrossRef] [Green Version]
- Banks, J.A.; Nishiyama, T.; Hasebe, M.; Bowman, J.L.; Gribskov, M.; DePamphilis, C.; Albert, V.A.; Aono, N.; Aoyama, T.; Ambrose, B.A.; et al. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 2011, 332, 960–963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.; Xin, T.; Bartels, D.; Li, Y.; Gu, W.; Yao, H.; Liu, S.; Yu, H.; Pu, X.; Zhou, J.; et al. Genome analysis of the ancient tracheophyte Selaginella tamariscina reveals evolutionary features relevant to the acquisition of desiccation tolerance. Mol. Plant 2018, 11, 983–994. [Google Scholar] [CrossRef] [Green Version]
- Wickell, D.; Kuo, L.-Y.; Yang, H.-P.; Ashok, A.D.; Irisarri, I.; Dadras, A.; de Vries, S.; de Vries, J.; Huang, Y.-M.; Li, Z.; et al. Underwater CAM photosynthesis elucidated by Isoëtes genome. Nat. Commun. 2021, 12, 6348. [Google Scholar] [CrossRef]
- Nunes, G.; Oliveira, R.R.M.; Guimarães, J.T.F.; Giulietti, A.M.; Caldeira, C.; Vasconcelos, S.; Pires, E.; Dias, M.; Watanabe, M.; Pereira, J.; et al. Quillworts from the Amazon: A multidisciplinary populational study on Isoëtes serracarajensis and Isoëtes cangae. PLoS ONE 2018, 13, e0201417. [Google Scholar] [CrossRef]
- Schafran, P.W.; Zimmer, E.A.; Taylor, W.C.; Musselman, L.J. a whole chloroplast genome phylogeny of diploid species of Isoëtes (Isoëtaceae, Lycopodiophyta) in the southeastern United States. Castanea 2018, 83, 224–235. [Google Scholar] [CrossRef]
- Pereira, J.B.S.; Giulietti, A.M.; Prado, J.; Vasconcelos, S.; Watanabe, M.T.C.; Pinangé, D.S.B.; Oliveira, R.R.M.; Pires, E.D.; Caldeira, C.F.; Oliveira, G. Plastome-based phylogenomics elucidate relationships in rare Isoëtes species groups from the Neotropics. Mol. Phylogenet. Evol. 2021, 161, 107177. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 21 April 2020).
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; Depamphilis, C.W.; Yi, T.S.; Li, D.-Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef] [Green Version]
- Qu, X.J.; Moore, M.J.; Li, D.Z.; Yi, T.S. PGA: A software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 2019, 15, 50. [Google Scholar] [CrossRef] [Green Version]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Wang, Y.H.; Jin, J.J.; Stull, G.W.; Bruneau, A.; Cardoso, D.; De Queiroz, L.P.; Moore, M.J.; Zhang, S.-D.; Chen, S.-Y.; et al. Exploration of plastid phylogenomic conflict yields new insights into the deep relationships of Leguminosae. Syst. Biol. 2020, 69, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; Gao, F.; Jakovlic, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [Green Version]
- Lanfear, R.; Calcott, B.; Kainer, D.; Mayer, C.; Stamatakis, A. Selecting optimal partitioning schemes for phylogenomic datasets. BMC Evol. Biol. 2014, 14, 82. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [Green Version]
- Darling, A.E.; Mau, B.; Perna, N.T. Progressivemauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puttick, M.N. MCMCtreeR: Functions to prepare MCMCtree analyses and visualize posterior ages on trees. Bioinformatics 2019, 35, 5321–5322. [Google Scholar] [CrossRef] [PubMed]
- Hao, S.; Xue, J.; Wang, Q.; Liu, Z. Yuguangia ordinata gen. et sp. nov., a new lycopsid from the Middle Devonian (Late Givetian) of Yunnan, China, and its phylogenetic implications. Int. J. Plant Sci. 2007, 168, 1161–1175. [Google Scholar] [CrossRef]
- Banks, H.P.; Bonamo, P.M.; Grierson, J.D. Leclercqia complexa gen. et sp. nov., a new Lycopod from the late middle devonian of eastern New York. Rev. Palaeobot. Palynol. 1972, 14, 19–40. [Google Scholar] [CrossRef]
- Bonamo, P.M. Rellimia thomsonii (Progymnospermopsida) from the middle devonian of New York State. Am. J. Bot. 1977, 64, 1272. [Google Scholar] [CrossRef]
- Strother, P.K.; Traverse, A. Plant microfossils from llandoverian and wenlockian rocks of Pennsylvania. Palynology 1979, 3, 1–21. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [Green Version]
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Lam, T.T.Y.; Zhu, H.; Guan, Y. Two methods for mapping and visualizing associated data on phylogeny using GGTree. Mol. Biol. Evol. 2018, 35, 3041–3043. [Google Scholar] [CrossRef]
- Louca, S.; Doebeli, M. Efficient comparative phylogenetics on large trees. Bioinformatics 2018, 34, 1053–1055. [Google Scholar] [CrossRef]
- Robison, T.A.; Wolf, P.G. ReFernment: An R package for annotating RNA editing in plastid genomes. Appl. Plant Sci. 2019, 7, e01216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, Y.; Yu, J.; Shang, H.; Liu, B.; Yan, Y. Complete chloroplast genome of Isoëtes hypsophila (Isoëtaceae), the Endangered quillwort in China. Mitochondrial DNA Part B Resour. 2021, 6, 2908–2909. [Google Scholar] [CrossRef]
- Palmer, J.D.; Thompson, W.F. Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell 1982, 29, 537–550. [Google Scholar] [CrossRef]
- Hecht, J.; Grewe, F.; Knoop, V. Extreme RNA editing in coding islands and abundant microsatellites in repeat sequences of Selaginella moellendorffii mitochondria: The root of frequent plant mtDNA recombination in early tracheophytes. Genome Biol. Evol. 2011, 3, 344–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monroe, J.G.; Srikant, T.; Carbonell-Bejerano, P.; Becker, C.; Lensink, M.; Exposito-Alonso, M.; Klein, M.; Hildebrandt, J.; Neumann, M.; Kliebenstein, D.; et al. Mutation bias reflects natural selection in Arabidopsis thaliana. Nature 2022, 602, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Browne, P.D.; Nielsen, T.K.; Kot, W.; Aggerholm, A.; Gilbert, M.T.P.; Puetz, L.; Rasmussen, M.; Zervas, A.; Hansen, L.H. GC bias affects genomic and metagenomic reconstructions, underrepresenting GC-poor organisms. Gigascience 2020, 9, giaa008. [Google Scholar] [CrossRef]
- Pace, N.R. Structure and synthesis of the ribosomal ribonucleic acid of prokaryotes. Bacteriol. Rev. 1973, 37, 562–603. [Google Scholar] [CrossRef]
- Guo, W.; Grewe, F.; Mower, J.P. Variable frequency of plastid RNA editing among ferns and repeated loss of uridine-to-cytidine editing from vascular plants. PLoS ONE 2015, 10, e0117075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oldenkott, B.; Yamaguchi, K.; Tsuji-Tsukinoki, S.; Knie, N.; Knoop, V. Chloroplast RNA editing going extreme: More than 3400 events of C-to-U editing in the chloroplast transcriptome of the lycophyte Selaginella uncinata. Rna 2014, 20, 1499–1506. [Google Scholar] [CrossRef] [Green Version]
- Du, X.Y.; Lu, J.M.; Li, D.Z. Extreme plastid RNA editing may confound phylogenetic reconstruction: A case study of Selaginella lycophytes. Plant Divers. 2020, 42, 356–361. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Morton, B.R.; Maier, U.G. The evolution of chloroplast RNA editing. Mol. Biol. Evol. 2006, 23, 1912–1921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, D.R. Unparalleled variation in RNA editing among selaginella plastomes. Plant Physiol. 2020, 182, 12–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rüdinger, M.; Volkmar, U.; Lenz, H.; Groth-Malonek, M.; Knoop, V. Nuclear DYW-type PPR gene families diversify with increasing RNA editing frequencies in liverwort and moss mitochondria. J. Mol. Evol. 2012, 74, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Kidston, R.; Lang, W.H. On Old Red Sandstone plants showing structure, from the Rhynie Chert Bed, Aberdeenshire. Part II. Additional notes on Rhynia gwynne-vaughani, Kidston and Lang: With descriptions of Rhynia major, N. Sp., and Hornea lignieri, N.G.N. sp. Trans. R. Soc. Edinb. 1920, 52, 603–627. [Google Scholar] [CrossRef] [Green Version]
- Rickards, R.B. The age of the earliest club mosses: The Silurian Baragwanathia flora in Victoria, Australia. Geol. Mag. 2000, 137, 207–209. [Google Scholar] [CrossRef]
- Evkaikina, A.I.; Berke, L.; Romanova, M.A.; Proux-Wéra, E.; Ivanova, A.N.; Rydin, C.; Pawlowski, K.; Voitsekhovskaja, O.V. The Huperzia selago shoot tip transcriptome sheds new light on the evolution of leaves. Genome Biol. Evol. 2017, 9, 2444–2460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Field, A.R.; Testo, W.; Bostock, P.D.; Holtum, J.A.M.; Waycott, M. Molecular phylogenetics and the morphology of the Lycopodiaceae subfamily Huperzioideae supports three genera: Huperzia, Phlegmariurus and Phylloglossum. Mol. Phylogenet. Evol. 2016, 94, 635–657. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Wang, T.; Shu, J.; Xiang, Q.; Yang, T.; Zhang, X.; Yan, Y. Plastid Phylogenomics and Plastomic Diversity of the Extant Lycophytes. Genes 2022, 13, 1280. https://doi.org/10.3390/genes13071280
Chen S, Wang T, Shu J, Xiang Q, Yang T, Zhang X, Yan Y. Plastid Phylogenomics and Plastomic Diversity of the Extant Lycophytes. Genes. 2022; 13(7):1280. https://doi.org/10.3390/genes13071280
Chicago/Turabian StyleChen, Sisi, Ting Wang, Jiangping Shu, Qiaoping Xiang, Tuo Yang, Xianchun Zhang, and Yuehong Yan. 2022. "Plastid Phylogenomics and Plastomic Diversity of the Extant Lycophytes" Genes 13, no. 7: 1280. https://doi.org/10.3390/genes13071280
APA StyleChen, S., Wang, T., Shu, J., Xiang, Q., Yang, T., Zhang, X., & Yan, Y. (2022). Plastid Phylogenomics and Plastomic Diversity of the Extant Lycophytes. Genes, 13(7), 1280. https://doi.org/10.3390/genes13071280






