Abstract
Data for the chromosomal FISH mapping localization of (AG3T3)3 are compiled for 37 species belonging 27 families; for 24 species and 14 families, this is the first such report. The chromosome number and length ranged from 14–136 and 0.56–14.48 μm, respectively. A total of 23 woody plants presented chromosome length less than 3 μm, thus belonging to the small chromosome group. Telomeric signals were observed at each chromosome terminus in 38 plants (90.5%) and were absent at several chromosome termini in only four woody plants (9.5%). Non-telomeric signals were observed in the chromosomes of 23 plants (54.8%); in particular, abundant non-telomeric (AG3T3)3 was obviously observed in Chimonanthus campanulatus. Telomeric signals outside of the chromosome were observed in 11 woody plants (26.2%). Overall, ten (AG3T3)3 signal pattern types were determined, indicating the complex genome architecture of the 37 considered species. The variation in signal pattern was likely due to chromosome deletion, duplication, inversion, and translocation. In addition, large primary constriction was observed in some species, probably due to or leading to chromosome breakage and the formation of new chromosomes. The presented results will guide further research focused on determining the chromosome number and disclosing chromosome rearrangements of woody plants.
1. Introduction
Most eukaryotes possess chromosomal termini made up of 5–8 bp simple tandem DNA repeats—commonly called telomeric repeats [1,2,3]—which, in all eukaryotes, share similarities. Telomeric DNA repeats typically follow the formula (TxAyGz)n. Furthermore, they are well-conserved, and previous findings have revealed their diversity. There exist at least 17 variable telomeric sequences: TAG3 [4], TA2G2 [5], TA2G3 [5], T2AG2 [6], T2AG3 [7], T3AG3 [8,9,10], AG3T3 [11], T4AG3 [12], A2TG6 [13], T2AT2AG3 [7], T6AG3 [14,15], TCAG2 [16], T2AG2C [17], T2CAG2 [18], T3CAG2 [18], C3TA3 [19], T2N4AG3 [20], and T2ATG3CTCG2 [21]. Undoubtedly, the most widespread telomeric repeat is T2AG3 in animals [22], while that in most plants is T3AG3 [23].
Telomeric repeats are not often localized at chromosomal termini and have also been found in multiple intercalary sites of chromosomes in many species [3,23,24,25]. Interstitial telomeric sequences may represent a significant part of the telomeric DNA [3]. Cytogenetic analysis has found two major types of interstitial telomeric sequences: one is heterochromatic and large, found in centromeric or pericentromeric regions, while the other type is short and distributed at various positions in chromosomes [3]. Unlike telomeric repeats located at termini, interstitial telomeric sequences confer karyotype plasticity. Their unstable and high-length polymorphisms may increase chromosomal fragility and contribute to chromosomal reorganization [3,23,24]. Interstitial telomeric sequences have been shown to cause a change in the chromosome number in Cardamine cordifolia A. Gray [26], as well as in Asteraceae and Brassicaceae [27]. Interstitial telomeric sequences may be derived from centric fusions of Robertsonian translocation [28,29,30], generated through pericentric inversions [31,32], or be the product of equilocal dispersion of telomeric DNA to interstitial regions of the chromosome through transposition or heterologous recombination [33,34].
Many studies have reported on the T3AG3 distribution in woody plants. T3AG3 were observed at each chromosome end in Populus trichocarpa Torr. and A. Gray ex Hook. [35], Picea abies (L.) H. Karst. and Larix decidua Mill. [36], Abies alba Mill. [37,38], Cycas revoluta Thunb. [39], Strobus spp. [40], Aralia elata (Miq.) Seem., Dendropanax morbiferus H. Lév., Eleutherococcus sessiliflorus (Rupr. Et Maxim.) Seem. and Kalopanax septemlobus (Thunb. ex A.Murr.) Koidz [41]. Furthermore, T3AG3 has been observed at the interstitial regions and at all chromosome ends in Pinus spp. [42,43,44,45] and Podocarpus spp. [46]. No T3AG3 signal was found in species of Sessea, Vestia, and Cestrum, but the T3AG3 sequence was dispersed in the Cestrum genome [14,15].
AG3T3 is similar to but different from T3AG3 and has only been found to be used in 22 woody plants, 14 species, and 11 genera. AG3T3 have been observed at each chromosome end in Hibiscus mutabilis L. [47]; Juglans regia L. ‘Chuanzao1’, J. regia L. ‘Yanyuanzao’, Juglans sigillata Dode ‘Maerkang’, and J. sigillata Dode ‘Muzhilinhe’ [48]; Fraxinus pennsylvanica Marsh., Syringa oblata Ait., Ligustrum lucidum Lindl. and Ligustrum × vicaryi Rehder [49]; and Berberis diaphana Maxim. and Berberis soulieana Schneid [50]. Telomeric AG3T3 ensures integrated and easily countable chromosomes. Furthermore, T3AG3 has been observed in the interstitial regions and at all chromosome ends in Hippophaë rhamnoides ‘Wucifeng’, H. rhamnoides ‘Shenqiuhong’, H. rhamnoides ‘Zhuangyuanhuang’, cultural H. rhamnoides ssp. sinensis, wild H. rhamnoides ssp. sinensis [11], Robinia pseudoacacia L., R. pseudoacacia ‘idaho’, R. pseudoacacia f. decaisneana (Carr.) Voss, Styphnolobium japonicum (L.) Schott, Amorpha fruticose L. [51], and C. campanulatus R.H. Chang and C.S. Ding [52]. Non-telomeric AG3T3 may indicate chromosomal variation. There is still a great need to explore the telomeric and non-telomeric AG3T3 chromosome distribution in other woody plants. To determine the chromosome number and disclose the chromosome rearrangements in woody plants, in this study, we carried out fluorescent in situ hybridization (FISH) mapping to reveal highly abundant (AG3T3)3, established an ideogram to describe the complex genome architecture and, finally, discussed the proposed origin of (AG3T3)3 diversity in woody plants.
2. Materials and Methods
The species chosen for these experiments were firstly considered due to the occurrence of karyotype rearrangements [11,47,48,49,50,51,52,53], and secondly for investigation of species in which (AG3T3)3 has not yet been explored. Zea mays L. was chosen as it possesses the telomeric repetitive unit AGGGTTT conserved in plant chromosome telomeres. It is contained in the sequence named M8-2D, a B chromosome-specific sequence in Z. mays, which has low homology to clones from Z. mays chromosome 4 centromere. M8-2D is localized in B chromosome centromeric and telomeric regions [53]. Hence, we used Z. mays to test the used probe and as a control.
Details of the seeds or seedlings of Z. mays and the 41 woody plants (belonging to 37 species, 27 genera, 18 families) used in the present work are provided in Table 1. All 42 plants were collected from 12 Counties or Districts of Sichuan Province, China.

Table 1.
Details of Z. mays and the 41 woody plants used in this study.
2.1. Probe and Chromosome Preparation
The 21 bp oligo-probe of (AG3T3)3, 5′-AGGGTTTAGGGTTTAGGGTTT-3′, was first reported in Z. mays [53] and has been further applied in Berberidaceae [50], Calycanthaceae [52], Elaeagnaceae [11], Fabaceae [51], Juglandaceae [48], Malvaceae [47], and Oleaceae [49]. This oligo-probe was synthesized by Sangon Biotech Co., Ltd. (Shanghai, China), and tested simultaneously in a single round of FISH. The oligo-probe was 5′-labeled with 6-carboxy-fluorescein (6-FAM; absorption/emission wavelengths 494 nm/518 nm; green).
Collected seeds were germinated in culture dishes with wet filter paper and kept at 25 °C in the daytime and at 18 °C in the night until the roots were ~2 cm in length, which were then cut. The collected seedlings were cultured in soil at room temperature (15–25 °C) until many new roots grew out, which were then cut again. The cut roots were treated with nitrous oxide (N2O) gas for 2–6 h, with treatment time depending on chromosome length and cell wall lignification. Next, the samples were fixed in glacial acetic acid for 5–10 min, then kept in 75% ethyl alcohol. Chromosome preparation was carried out according to the procedure described by Luo et al. [51]. As these techniques have been described elsewhere, they will be detailed briefly here. Approximately 1 mm of the meristematic zone of the root tip was enzymolyzed at 37 °C for 45 min by using cellulase and pectinase (1 mL buffer + 0.04 g cellulase + 0.02 g pectinase, the buffer 50 mL was included 0.5707 g trisodium citrate + 0.4324 g citric acid), which were produced by Yakult Pharmaceutical Ind. Co., Ltd. (Tokyo, Japan) and Kyowa Chemical Products Co., Ltd. (Osaka, Japan), and then mixed into suspension for dropping onto slides. These slides were air dried then examined using an Olympus CX23 microscope (Olympus, Tokyo, Japan).
2.2. FISH Hybridization
Slides with well-spread samples were used for hybridization. Chromosomes were first subjected to a series of fixation (10 min, 4% paraformaldehyde, room temperature), dehydrated (5 min, 75%, 95%, 100% ethanol, room temperature), denatured (2 min, deionized formamide, 80 °C), and a second dehydration (5 min, 75%, 95%, 100% ethanol, −20 °C), and then hybridized (10 μL hybridization mixture of 0.375 μL of (AG3T3)3, 4.675 μL of 2× SSC, and 4.95 μL of ddH2O) for 1–2 h at 37 °C. Subsequently, hybridized chromosomes were washed with 2× SSC and ddH2O twice for 5 min at room temperature, and air-dried. Finally, they were counterstained with 4,6-diamidino-2-phenylindole (DAPI, Vector Laboratories, Inc., Burlingame, CA, USA) for 5 min, referring to the procedure described by Luo et al. [51]. Slides were checked using an Olympus BX-63 microscope (Olympus Corporation, Tokyo, Japan), and FISH images were captured by a DP-70 CCD camera connected to the microscope.
2.3. Karyotype Analysis
Raw images were processed using the DP Manager (Olympus Corporation, Tokyo, Japan) and Photoshop CC 2015 (Adobe Systems Incorporated, San Jose, CA, USA) software. At least ten slides of each plant were observed, and at least fifteen cells with good chromosome spread were used for chromosome counting and length measurement. All chromosomes were aligned from longest to shortest. The chromosome ratio was determined by comparing the length of the longest chromosome to that of the shortest chromosome. Further karyotype analysis could not be carried out due to the small chromosome size and unclear centromere location of many of the species.
3. Results
3.1. Karyotype Analysis Revealed Differences among 37 Species
For 24 species and 14 families, this is the first time that (AG3T3)3 testing has been reported. To visualize the chromosomal distribution of (AG3T3)3 in Z. mays and the 41 woody plants, we performed FISH analysis, as shown in Figure 1(A1–A11), Figure 2(B1–B16), and Figure 3(C1–C15). To clearly display each chromosome distribution of the (AG3T3)3, we cut those in Figure 1, Figure 2 and Figure 3 and aligned them, as shown in Figure 4(A1–A11), Figure 5(B1–B16), and Figure 6(C1–C15).
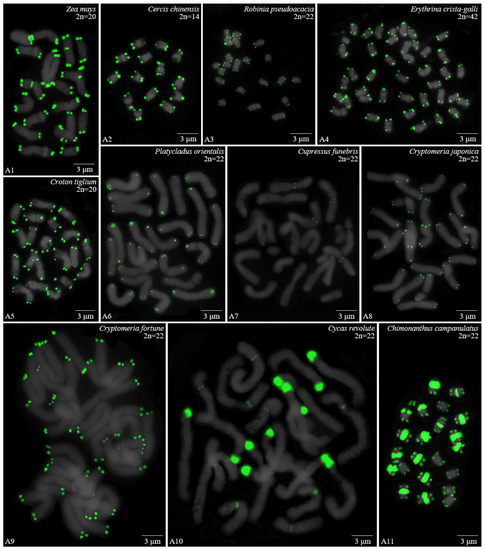
Figure 1.
Oligo-FISH depicting the (AG3T3)3 present in Z. mays and ten woody plants. Chromosomes in A1–A8 and A11 are from metaphase, while the chromosomes in A9–A10 are from prometaphase. Oligo-probe (AG3T3)3 is exhibited by green signals: (A1) Z. mays, 2n = 20; (A2) C. chinensis, 2n = 14; (A3) R. pseudoacacia, 2n = 22; (A4) E. crista-galli, 2n = 42; (A5) C. tiglium, 2n = 20; (A6) P. orientalis, 2n = 22; (A7) C. funebris, 2n = 22; (A8) C. japonica, 2n = 22; (A9) C. fortunei, 2n = 22; (A10) C. revoluta, 2n = 22; and (A11) C. campanulatus, 2n = 22. Bar: 3 μm.
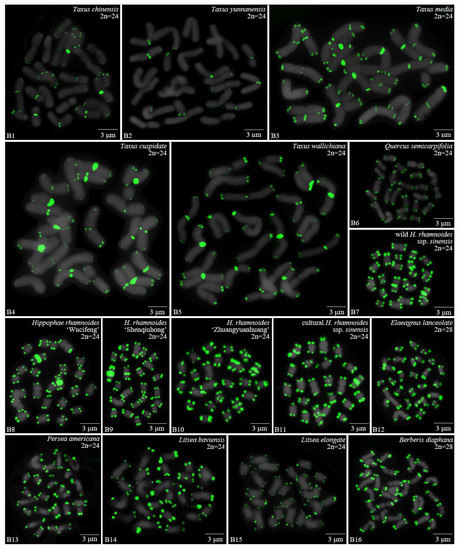
Figure 2.
Oligo-FISH depicting the (AG3T3)3 present in 16 woody plants. All chromosomes in B1–B16 are from metaphase. Oligo-probe (AG3T3)3 is exhibited by green signals: (B1) T. chinensis, 2n = 24; (B2) T. yunnanensis, 2n = 24; (B3) T. media, 2n = 24; (B4) T. cuspidata, 2n = 24; (B5) T. wallichiana, 2n = 24; (B6) Q. semecarpifolia, 2n = 24; (B7) wild H. rhamnoides ssp. sinensis, 2n = 24; (B8) H. rhamnoides ‘Wucifeng’, 2n = 24; (B9) H. rhamnoides ‘Shenqiuhong’, 2n = 24; (B10) H. rhamnoides ‘Zhuangyuanhuang’, 2n = 24; (B11) cultural H. rhamnoides ssp. sinensis, 2n = 24; (B12) E. lanceolata, 2n = 28; (B13) P. americana, 2n = 24; (B14) L. baviensis, 2n = 24; (B15) L. elongate, 2n = 24; and (B16) B. diaphana, 2n = 28. Bar: 3 μm.
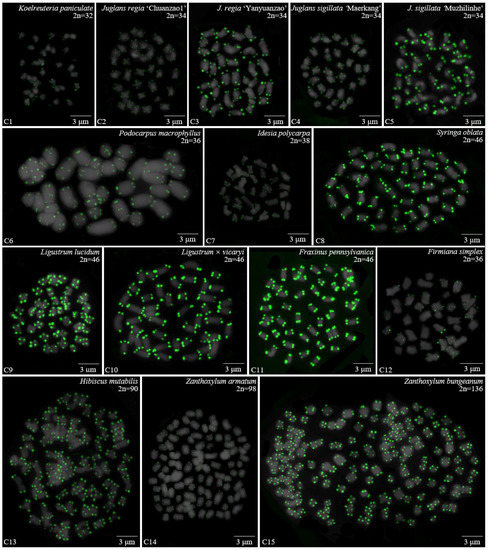
Figure 3.
Oligo-FISH depicting the (AG3T3)3 present in 15 woody plants. All chromosomes in C1–C15 are from metaphase. Oligo-probe (AG3T3)3 is exhibited by green signals: (C1) K. paniculata, 2n = 32; (C2) J. regia ‘Chuanzao1’, 2n = 34; (C3) J. regia ‘Yanyuanzao’, 2n = 34; (C4) J. sigillata ‘Maerkang’, 2n = 34; (C5) J. sigillata ‘Muzhilinhe’, 2n = 34; (C6) P. macrophyllus, 2n = 36; (C7) I. polycarpa, 2n = 38; (C8) S. oblata, 2n = 46; (C9) L. lucidum, 2n = 46; (C10) L. × vicaryi, 2n = 46; (C11) F. pennsylvanica, 2n = 46; (C12) F. simplex, 2n = 36; (C13) H. mutabilis, 2n = 90; (C14) Z. armatum, 2n = 98; and (C15) Z. bungeanum, 2n = 136. Bar: 3 μm.
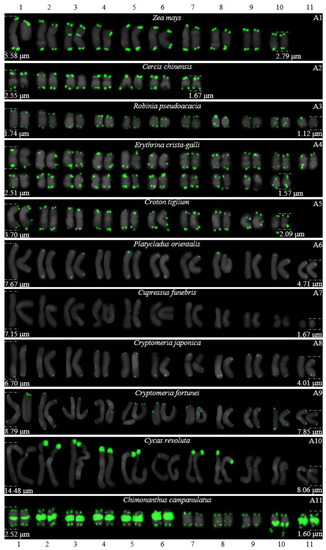
Figure 4.
FISH karyograms of Z. mays and the ten woody plants from Figure 1. Oligo-probe (AG3T3)3 is exhibited by green signals. The upper/lower numbers (1–11) represent the chromosome pairs. The first/last chromosome of each plant were used to calculate the chromosome length: (A1) Z. mays, 2.79–5.58 μm; (A2) C. chinensis, 1.67–2.55 μm; (A3) R. pseudoacacia, 1.12–1.74 μm; (A4) E. crista-galli, 1.57–2.51 μm; (A5) C. tiglium, 2.09–3.70 μm; (A6) P. orientalis, 4.71–7.67 μm; (A7) C. funebris, 1.67–7.15 μm; (A8) C. japonica, 4.01–6.70 μm; (A9) C. fortunei, 7.85–8.79 μm; (A10) C. revoluta, 8.06–14.48 μm; and (A11) C. campanulatus, 1.60–2.52 μm.
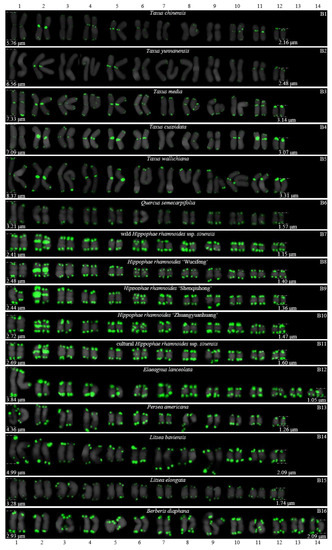
Figure 5.
FISH karyograms of the 16 woody plants cutting from Figure 2. Oligo-probe (AG3T3)3 is exhibited by green signals. The upper/lower numbers (1–14) represent the chromosome pairs. The first/last chromosome of each plant were used to calculate the chromosome length: (B1) T. chinensis, 2.16–5.76 μm; (B2) T. yunnanensis, 2.48–6.56 μm; (B3) T. media, 3.14–7.33 μm; (B4) T. cuspidata, 3.07–7.09 μm; (B5) T. wallichiana, 3.31–8.37 μm; (B6) Q. semecarpifolia, 1.57–3.21 μm; (B7) wild H. rhamnoides ssp. sinensis, 1.15–2.41 μm; (B8) H. rhamnoides ‘Wucifeng’, 1.40–2.48 μm; (B9) H. rhamnoides ‘Shenqiuhong’, 1.36–2.44 μm; (B10) H. rhamnoides ‘Zhuangyuanhuang’, 1.47–2.72 μm; (B11) cultural H. rhamnoides ssp. sinensis, 1.60–2.69 μm; (B12) E. lanceolata, 1.05–3.84 μm; (B13) P. americana, 1.26–4.36 μm; (B14) L. baviensis, 2.09–4.99 μm; (B15) L. elongate, 1.74–3.28 μm; and (B16) B. diaphana, 2.09–2.93 μm.

Figure 6.
FISH karyogram of 15 woody plants cutting from Figure 3. Oligo-probe (AG3T3)3 is exhibited by green signals. The upper/lower numbers (1–19) represent the chromosome pairs. The first/last chromosome of each plant were used to calculate the chromosome length: (C1) K. paniculata, 0.56–1.12 μm; (C2) J. regia ‘Chuanzao1’, 0.63–2.60 μm; (C3) J. regia ‘Yanyuanzao’, 1.01–2.55 μm; (C4) J. sigillata ‘Maerkang’, 0.66–1.88 μm; (C5) J. sigillata ‘Muzhilinhe‘, 0.87–2.79 μm; (C6) P. macrophyllus, 1.92–7.01 μm; (C7) I. polycarpa, 0.94–2.23 μm; (C8) S. oblata, 1.64–2.20 μm; (C9) L. lucidum, 0.98–1.36 μm; (C10) L. × vicaryi, 1.37–3.07 μm; (C11) F. pennsylvanica, 0.98–2.06 μm; (C12) F. simplex, 1.08–2.37 μm; (C13) H. mutabilis, 1.22–2.90 μm; (C14) Z. armatum, 0.98–1.99 μm; and (C15) Z. bungeanum, 0.94–2.62 μm.
The chromosome number and length for the considered species were sorted in Table 2. The chromosome number in the 42 plants ranged from 14 (C. chinensis, A3) to 136 (Z. bungeanum, C15). A total of 14 woody plants possessed 24 chromosomes (one third), whereas seven woody plants possessed 22 chromosomes (one sixth). The longest chromosome length of each plant ranged from 1.12 μm (K. paniculata, C1) to 14.48 μm (C. revoluta, A10), while the shortest chromosome length of each plant ranged from 0.56 μm (K. paniculate, C1) to 8.06 μm (C. revoluta, A10). A total of 23 woody plants (nearly one third) had chromosome length less than 3 μm, thus falling into the small chromosome category. Due to the indistinct location of centromeres and small size of chromosomes in many of the considered woody plants, further karyotype analysis—such as long/short arm length and karyotype formula—was not carried out. Karyotype asymmetry was assessed using the ratio of longest to shortest chromosome length. The largest ratio was 4.28 in C. funebris (A7), while the smallest ratio was 1.12 in C. fortunei (A9). The ratio for 17 plants ranged from 1 to 2 (40.5%), while that of 19 plants ranged from 2 to 3 (45.2%). The ratio was greater than 3 for six plants: C. funebris (A7), E. lanceolata (B12), P. americana (B13), J. regia ‘Chuanzao1’ (C2), J. sigillata ‘Muzhilinhe’ (C5), and P. macrophyllus (C6). These results indicated that abundant differences exist among 37 of the considered species.

Table 2.
Chromosome number and length of the 42 plants used in this study.
3.2. The Diverse Signal Patterns of (AG3T3)3 Reveal the Complex Genome Architecture
To better investigate diversity of (AG3T3)3, different types of ideograms for the 42 plants were drawn based on the FISH karyograms shown in Figure 4, Figure 5 and Figure 6, which are illustrated in Figure 7, Figure 8 and Figure 9. Telomeric signals were observed at each chromosome terminus in 38 plants (90.5%, the first class): Z. mays (A1), C. chinensis (A2), R. pseudoacacia (A3), E. crista-galli (A4), C. tiglium (A5), P. orientalis (A6), C. japonica (A8), C. fortune (A9), C. campanulatus (A11), T. chinensis (B1), T. media (B3), T. cuspidata (B4), T. wallichiana (B5), Q. semecarpifolia (B6), wild H. rhamnoides ssp. sinensis (B7), H. rhamnoides ‘Wucifeng’ (B8), H. rhamnoides ‘Shenqiuhong’ (B9), H. rhamnoides ‘Zhuangyuanhuang’ (B10), cultural H. rhamnoides ssp. sinensis (B11), E. lanceolata (B12), P. americana (B13), L. baviensis (B14), L. elongate (B15), B. diaphana (B16), J. regia ‘Chuanzao1’ (C2), J. regia ‘Yanyuanzao’ (C3), J. sigillata ‘Maerkang’ (C4), J. sigillata ‘Muzhilinhe’ (C5), P. macrophyllus (C6), I. polycarpa (C7), S. oblata (C8), L. lucidum (C9), L. × vicaryi (C10), F. pennsylvanica (C11), F. simplex (C12), H. mutabilis (C13), Z. armatum (C14), and Z. bungeanum (C15). Meanwhile, telomeric signals were absent at several chromosome termini in only four woody plants (9.5%, the fourth class): C. funebris (A7), C. revoluta (A10), T. yunnanensis (B2), and K. paniculata (C1). Non-telomeric signals were observed at chromosome termini in 23 plants (54.8%, the second class): Z. mays (A1), R. pseudoacacia (A3), C. funebris (A7), C. revoluta (A10), and C. campanulatus (A11), T. chinensis (B1), T. yunnanensis (B2), T. media (B3), T. cuspidata (B4), T. wallichiana (B5), wild H. rhamnoides ssp. sinensis (B7), H. rhamnoides ‘Wucifeng’ (B8), H. rhamnoides ‘Shenqiuhong’ (B9), H. rhamnoides ‘Zhuangyuanhuang’ (B10), cultural H. rhamnoides ssp. sinensis (B11), J. sigillata ‘Muzhilinhe’ (C5), P. macrophyllus (C6), L. lucidum (C9), F. pennsylvanica (C11), F. simplex (C12), H. mutabilis (C13), Z. armatum (C14), and Z. bungeanum (C15). Telomeric signals outside of the chromosome were observed in 11 woody plants (26.2%, the third class): E. crista-galli (A4), C. tiglium (A5), C. campanulatus (A11), P. americana (B13), L. baviensis (B14), K. paniculata (C1), J. sigillata ‘Muzhilinhe’ (C5), S. oblata (C8), L. × vicaryi (C10), F. pennsylvanica (C11), and F. simplex (C12).
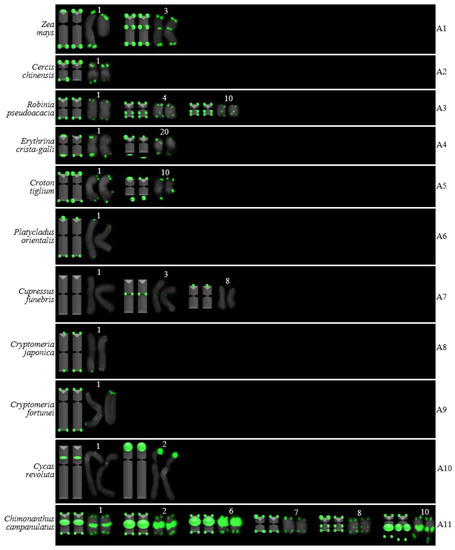
Figure 7.
Ideograms of Z. mays and ten woody plants based on the signal patterns in the FISH karyograms shown in Figure 4: (A1) Z. mays; (A2) C. chinensis; (A3) R. pseudoacacia; (A4) E. crista-galli; (A5) C. tiglium; (A6) P. orientalis; (A7) C. funebris; (A8) C. japonica; (A9) C. fortunei; (A10) C. revoluta; and (A11) C. campanulatus. The numbers (1–4, 6–8, 10, 20) above the karyograms are the numbers of the chromosome pairs. Oligo-probe (AG3T3)3 is exhibited by green signals. Telomeric signals were observed at each chromosome terminus in (A1–A6,A8,A9,A11), while telomeric signals were absent at several chromosome termini in (A7,A10). Non-telomeric signals were observed at several chromosome termini in (A1,A3,A7,A10,A11). Telomeric signals deviated from the chromosome in (A4,A5,A11).
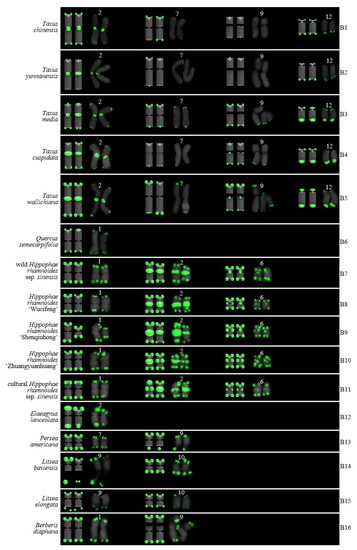
Figure 8.
Ideograms of 16 woody plants based on the signal patterns in the FISH karyograms shown in Figure 5: (B1) T. chinensis; (B2) T. yunnanensis; (B3) T. media; (B4) T. cuspidata; (B5) T. wallichiana; (B6) Q. semecarpifolia; (B7) wild H. rhamnoides ssp. sinensis; (B8) H. rhamnoides ‘Wucifeng’; (B9) H. rhamnoides ‘Shenqiuhong’; (B10) H. rhamnoides ‘Zhuangyuanhuang’; (B11) cultural H. rhamnoides ssp. sinensis; (B12) E. lanceolata; (B13) P. americana; (B14) L. baviensis; (B15) L. elongate; and (B16) B. diaphana. The numbers (1, 2, 6, 7, 9, 10, 12) above the karyograms are the numbers of the chromosome pairs. Oligo-probe (AG3T3)3 is exhibited by green signals. Telomeric signals were observed at each chromosome terminus in (B1,B3–B16), while telomeric signals were absent at several chromosome termini in (B2). Non-telomeric signals were observed at several chromosome termini in (B1–B5,B7–B11). Telomeric signals deviated from the chromosome in (B13,B14).
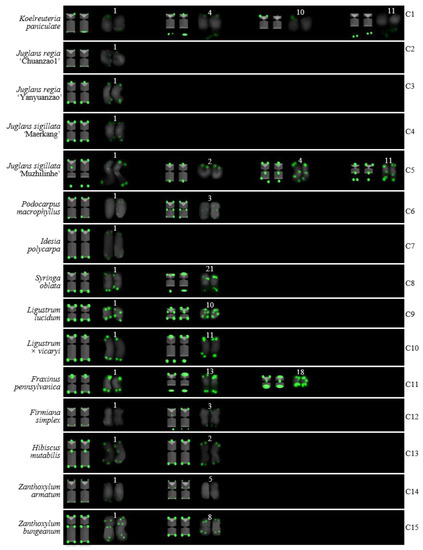
Figure 9.
Ideograms of 15 woody plants based on the signal patterns in the FISH karyograms shown in Figure 6: (C1) K. paniculata; (C2) J. regia ‘Chuanzao1’; (C3) J. regia ‘Yanyuanzao’; (C4) J. sigillata ‘Maerkang’; (C5) J. sigillata ‘Muzhilinhe‘; (C6) P. macrophyllus; (C7) I. polycarpa; (C8) S. oblata; (C9) L. lucidum; (C10) L. × vicaryi; (C11) F. pennsylvanica; (C12) F. simplex; (C13) H. mutabilis; (C14) Z. armatum; and (C15) Z. bungeanum. The numbers (1–5, 8, 10, 11, 13, 18, 21) above the karyograms are the numbers of the chromosome pairs. Oligo-probe (AG3T3)3 is exhibited by green signals. Telomeric signals were observed at each chromosome terminus in (C2–C15), while telomeric signals were absent at several chromosome termini in C1. Non-telomeric signals were observed at several chromosome termini in (C5,C6,C9,C11–C15). Telomeric signals deviated from the chromosome in (C1,C5,C8,C10–C12).
Furthermore, we summarized the results shown in Figure 7, Figure 8 and Figure 9 in order to produce the (AG3T3)3 signal pattern presented in Figure 10. The results for Z. mays and 41 woody plants, belonging to 18 families, are shown, including six plants in Elaeagnaceae (yellow), five in Taxaceae (red), four in Cupressaceae (dark blue), four in Juglandaceae (light green), four in Oleaceae (orange), three in Fabaceae (pink), three in Lauraceae (light blue), two in Malvaceae (dark green), two in Rutaceae (grey), and one in each of Berberidaceae, Calycanthaceae, Cycadaceae, Euphorbiaceae, Fagaceae, Poaceae, Podocarpaceae, Salicaceae, and Sapindaceae, respectively. Except for Lauraceae and Rutaceae, which each presented a single signal pattern type, the other families (Elaeagnaceae, Cupressaceae, Juglandaceae, Oleaceae, Lauraceae, and Malvaceae) all presented at least two signal pattern types.

Figure 10.
Signal pattern of (AG3T3)3 in Z. mays and 41 woody plants. Signal patterns type I–X are summarized based on Figure 7, Figure 8 and Figure 9. The number between the signal pattern and the species represents the type/type combination, including the species number. The number after the species represents the ratio of longest to shortest chromosome length. Elaeagnaceae includes six woody plants (yellow), Taxaceae includes five woody plants (red), Cupressaceae includes four woody plants (dark blue), Juglandaceae includes four woody plants (light green), Oleaceae includes four woody plants (orange), Fabaceae includes three woody plants (pink), Lauraceae includes three woody plants (light blue), Malvaceae includes two woody plants (dark green), Rutaceae includes two woody plants (grey), and Berberidaceae, Calycanthaceae, Cycadaceae, Euphorbiaceae, Fagaceae, Poaceae, Podocarpaceae, Salicaceae, and Sapindaceae each include one woody plant, respectively.
As shown in Figure 10, there were ten (AG3T3)3 signal pattern types in total: Type I, chromosome only includes signal at both ends; Type II, chromosome not only includes signal at both ends but also includes a non-telomeric signal location; Type III, chromosome includes single end signal, and the other telomeric signals outside of the chromosome; Type IV, chromosome not only includes signal at both ends, but also includes telomeric signal deviating from chromosome; Type V, chromosome not only includes signal at both ends, but also includes a large primary constriction; Type VI: chromosome not only includes signal at both ends signal, but also includes a large primary constriction, as well as a non-telomeric signal location; Type VII, chromosome only includes telomeric signal outside of the chromosome; Type VIII, chromosome only includes single end signal; Type IX, chromosome only includes non-telomeric signal location; and Type X, chromosome includes no signals. These types of signal pattern indicate that there is an abundant diversity in (AG3T3)3 signal arrangement.
All 42 plants possessed the 12 signal pattern types or type combinations shown in Figure 10. Ten woody plants only possessed signal pattern type I; Z. mays and 11 woody plants possessed the combination of type I + type II; eight woody plants possessed the combination of type I + type III; P. macrophyllus possessed the combination of type I + type II + type III; C. campanulatus possessed the combination of type I + type II + type IV; J. sigillata ‘Muzhilinhe’ possessed the combination of type I + type II + type III + type IV; B. diaphana possessed the combination of type I + type V; T. media and T. yunnanensis possessed the combination of type I + type II + type V; T. chinensis, T. cuspidata, and T. wallichiana possessed the combination of type I + type II + type VI; K. paniculate possessed the combination of type I + type III + type VII + type VIII; C. revolute possessed the combination of type VIII + type IX; and C. funebris possessed the combination of type VIII + type IX + type X.
There were diverse signal patterns of (AG3T3)3 among 37 species, indicating a complex genome architecture. For example, considering (i) Elaeagnaceae, five plants of H. rhamnoides possessed type I + type II, but E. lanceolata possessed type I; (ii) in Taxaceae, T. media and T. yunnanensis possessed the combination type I + type II + type V, but T. chinensis, T. cuspidata, and T. wallichiana possessed the combination type I + type II + type VI; (iii) in Cupressaceae, C. fortune, C. japonica, and P. orientalis possessed type I, but C. funebris possessed the combination type VIII + type IX + type X; (iv) in Juglandaceae, J. regia ‘Chuanzao1’, J. regia ‘Yanyuanzao’, and J. sigillata ‘Maerkang’ possessed type I, but J. sigillata ‘Muzhilinhe’ possessed the combination type I + type II + type III + type IV; (v) in Oleaceae, L. lucidum possessed the combination type I + type II, L. × vicaryi and S. oblata possessed the combination type I + type III, and F. pennsylvanica possessed the combination type I + type II + type III; (vi) in Fabaceae, C. chinensis possessed type I, R. pseudoacacia possessed the combination type I + type II, and E. crista-galli possessed the combination type I + type III; and (vii) in Malvaceae, H. mutabilis possessed the combination type I + type II, but F. simplex possessed the combination type I + type III.
The number shown after the species name in Figure 10 represents the ratio of longest to shortest chromosome length, indicating karyotype asymmetry. Type I included ten plants with ratio ranging from 1.12–4.13, with variance of 0.94. Type I + Type II included 12 plants with ratio ranging from 1.39–3.65, with variance of 0.38. Type I + Type II + Type III included eight plants with ratio ranging from 1.34–3.46, with variance of 0.48. These results indicate that chromosomes with conserved telomeric signal (Type I), in fact, have a wider range of karyotype asymmetry (VAR 0.94), while chromosomes with non-telomeric signals (Type II, III) have relatively concentrated karyotype asymmetry (VAR 0.38 and 0.48, respectively). In addition, no correlation between non-telomeric signals and karyotype asymmetry was observed.
3.3. Proposed Origin of (AG3T3)3 Signal Diversity
Based on Figure 7, Figure 8, Figure 9 and Figure 10, the proposed origin of (AG3T3)3 signal diversity is illustrated in Figure 11. There are three major groups. (i) Signal number: Increase signal number is likely caused by chromosome duplication, inversion, translocation, and/or sequence changes; a constant signal number indicates chromosome conservation and a decreased signal number is likely caused by chromosome deletion and/or sequence changes. (ii) Signal location: End signals on chromosomes indicate chromosome conservation; non-end signals are likely caused by chromosome deletion, duplication, inversion, translocation, and/or sequence changes; end signals deviating from the chromosome are probably caused by chromosome satellites, while end signal loss is probably caused by chromosome end deletion and/or sequence changes. (iii) Primary constriction: Normal primary constriction indicates chromosome conservation, while primary constriction likely becomes large due to chromosome breakage and the formation of new chromosomes (e.g., in Figure 8, T. media chromosome 9 and B. diaphana chromosome 9, T. cuspidata chromosome 9 and T. wallichiana chromosome 9 possibly indicate the formation of new chromosomes, while T. cuspidata chromosome 12 and T. wallichiana chromosome 12 were possibly formed in a reverse manner).
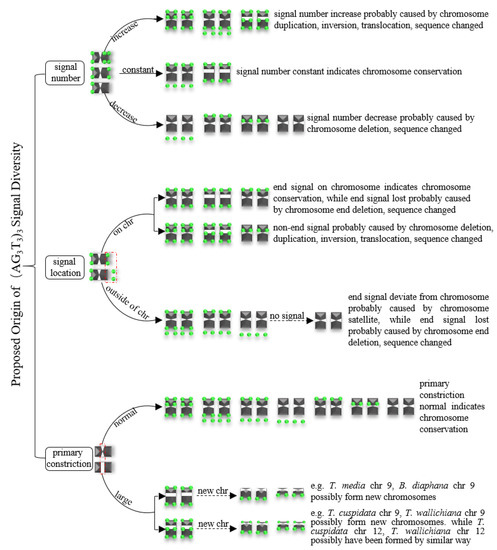
Figure 11.
Proposed origin of (AG3T3)3 signal diversity. The variations in signal number and signal location were likely caused by chromosome deletion, duplication, inversion, translocation, as sequence changes, as well as chromosome satellites, while primary constriction became large due to chromosome breakage and the formation of new chromosomes.
In brief, the variations in signal number and signal location were probably caused by chromosome deletion, duplication, inversion, translocation, and sequence changes, as well as chromosome satellites. It is likely that large primary constriction was due to chromosome breakage and the formation of new chromosomes.
4. Discussion
4.1. Karyotype Analysis of Z. mays and 41 Woody Plants
Chromosome number, size, centromere location, long/short arm ratio, and satellites are basic characteristics of karyotype. The presence of telomeric (AG3T3)3 in both chromosome ends may guarantee the accuracy of chromosome counting. The chromosome number in the 42 plants ranged from 14 (C. chinensis) to 136 (Z. bungeanum). There were six woody plants that presented chromosome numbers different to that reported in previous studies: J. regia ‘Chuanzao1’, J. regia ‘Yanyuanzao’, J. sigillata ‘Maerkang’, and J. sigillata ‘Muzhilinhe’ presented 2n = 34, a result supported by the work of Luo and Chen [48] but contradicted by Mu and Xi [54] and Mu et al. [55] (who reported 2n = 32). The differences were probably caused by hybridization, aneuploidization/among-population variation, or inaccurate chromosome number counts. P. macrophyllus presented 2n = 36, a result supported by the work of Hizume et al. [56] but contradicted by Zhu et al. [57] for eight small chromosomes, which were treated as satellite chromosomes in the latter. Z. armatum ‘Jinyang Qinghuajiao’ presented 2n = 98 in this study, which is contradicted by the work of Luo et al. [58], who reported 2n = ~128 for another variety, Z. armatum ‘Hanyuan Putao Qingjiao’. After excluding the experimental count error, we infer this big difference to have been caused by the confusion and complexity between Z. armatum varieties. We tested 16 Z. armatum varieties by FISH and SSR in another study in order to address this issue. Another reason why the chromosome numbers differed from previous studies is because of the limited available research on the chromosome numbers of woody plants. Compared to herbaceous plants, it is more difficult to obtain their chromosome preparation due to root lignification.
The chromosome lengths of the 42 plants in this study ranged from 0.56 μm (K. paniculate) to 14.48 μm (C. revoluta), while the ratio of longest chromosome to shortest chromosome ranged from 1.12 (K. paniculate) to 4.28 (C. revoluta), both of which indicate large differences among the considered species. Previous studies have reported the chromosome sizes of 12 species: R. pseudoacacia presented a size of 1.12–1.74 μm, which was close to that of He et al. [51] (0.94–1.67 μm); H. mutabilis presented 1.22–2.90 μm, close to that of Luo and He [47] (1.18–3.0 μm); B. diaphana presented 2.09–2.93 μm, close to that of Liu and Luo [50] (1.82–2.85 μm); F. pennsylvanica presented 0.98–2.06 μm, close to that of Luo and Liu [49] (1.12–2.06 μm); H. rhamnoide presented 1.15–2.72 μm, close to that of Xing et al. [59] (0.97–2.77 μm), but varying widely from that of Liu and Sheng [60] (1.67–4.44 μm); C. campanulatus presented 1.60–2.52 μm, differing from that of Luo and Chen [52] (1.07–2.41 μm); Z. armatum presented 0.98–1.99 μm, differing from that of Luo et al. [58] (1.23–2.34 μm); S. oblata presented 1.64–2.20 μm, differing from that of Luo and Liu [49] (1.25–2.32 μm); L. lucidum presented 0.98–1.36 μm, differing from that of Luo and Liu [49] (1.05–1.85 μm); L. × vicaryi presented 1.37–3.07 μm, differing from that of Luo and Liu [49] (1.25–2.83 μm); J. regia presented 0.63–2.60 μm, differing from that of Luo and Chen [48] (0.97–2.16 μm); finally, J. sigillata presented 0.66–2.79 μm, differing from that of Luo and Chen [48] (0.98–2.65 μm). Hence, chromosome length was more suitable for using in qualitative analysis than quantitative analysis.
A total of 23 woody plants (nearly one third) had chromosome lengths less than 3 μm, thus belonging to the small chromosome group. Due to the indistinct location of the centromere and the small size of chromosomes in many of the woody plants, as well as the chromosome length fluctuating according to the chromosome division phase and measuring tool, further karyotype analysis (e.g., long/short arm length and karyotype formula) was not carried out. Chromosome length is also not discussed further.
4.2. Occurrence of (AG3T3)3 in Woody Plants
AG3T3 is a telomeric repetitive tandem that is conserved in higher plant chromosome telomeres. This oligos is included in a B chromosome-specific sequence, named M8-2D, in Z. mays [53]. M8-2D has shown low homology to sequences from the chromosome 4 centromere in Z. mays but has been detected in B chromosome centromeric and telomeric regions in Z. mays. Nonetheless, (AG3T3)3 had not been previously tested in Z. mays before this study.
Overall, (AG3T3)3 was found to occur in Z. mays and 41 woody plants, belonging to 37 species, 27 genera, and 18 families. Previous research has reported the occurrence of (AG3T3)3 in 14 woody species, including A. fruticose [51], B. soulieana [50], B. diaphana [50], C. campanulatus [52], F. pennsylvanica [49], H. mutabilis [47]), H. rhamnoides [11], J. regia [48], J. sigillata [48], L. lucidum [49], L. × vicaryi [49], R. pseudoacacia [51], S. oblata [49], and S. japonicum [51].
The Arabidopsis-type telomeric repeat sequences T3AG3 are most like AG3T3, which are still the most frequent even in woody plants—such as species of Abies [37], Aralia [41], Cycas [39], Dendropanax [41], Eleutherococcus [41], Kalopanax [41], Malus [23], Picea [23], Pinus [23,40,42,43,44], Populus [35], Podocarpus [46], and Zamia [23,61]—but has been shown to be absent in species of Cestrum, Sessea, and Vestia [14]. Nevertheless, these sequences cannot be entirely equated, due to the existence of at least ten variable telomere sequences (TxAyGz)n—including TAG3 [4], TA2G2 [5], T2AG2 [6], T2AG3 [7], TA2G3 [5], T3AG3 [8,9,10], T4AG3 [12], A2TG6 [13], T2AT2AG3 [7], and T6AG3 [14,15]—and at least seven variants, such as TCAG2 [16], T2AG2C [17], T2CAG2 [18], T3CAG2 [18], C3TA3 [19], T2N4AG3 [20], and CTCG2T2ATG3 [21].
4.3. The Chromosomally Diverse Distribution of (AG3T3)3 Reveals the Complex Genome Architecture of Woody Plants
The (AG3T3)3 distribution is mostly conserved in the chromosome termini of woody plants. (AG3T3)3 has only been detected at all chromosome termini in B. soulieana, B. diaphana [50], F. pennsylvanica [49], H. mutabilis [47], J. regia [48], J. sigillata [48], L. lucidum [49], L. × vicaryi [49], and S. oblata [49]. The location of (AG3T3)3 at chromosome termini may indicate the integrality of the chromosome, thus ensuring the accuracy of chromosome counting. Occasionally, (AG3T3)3 has been detected not only at chromosome termini, but also as intercalary bands in A. fruticose, R. pseudoacacia, S. japonicum [51], C. campanulatus [52], and H. rhamnoides [11]. In A. fruticose, R. pseudoacacia, and S. japonicum, only two weak non-telomeric signals have been described, while H. rhamnoides presented two very strong, two clear, and another two weak non-telomeric signals in six chromosomes. Furthermore, (AG3T3)3 has shown exceptionally high levels of diversity in C. campanulatus, with signal intensity from weak to very strong, signals located outside the chromosome (satellites), and signals located at chromosome termini, sub-telomeric regions, and proximal regions. All 22 chromosomes presented telomeric signals, while 16 chromosomes presented non-telomeric signals in C. campanulatus [52]. (AG3T3)3 in intercalary sites may be used as informative markers to distinguish woody plants. The telomeric and non-telomeric (AG3T3)3 reveal the complex genome architecture of woody plants.
In this study of 42 plants, the first class (90.5% of plants) presented telomeric signals at each chromosome terminus; this result is supported by previous studies [11,47,48,49,50,51,52]. Only four woody plants presented telomeric signal absence at several chromosome termini (C. funebris, C. revoluta, K. paniculata, and T. yunnanensis). The probable reasons for the absence of signals were: (i) Telomeric signal occurrence which was lost during experiment; and (ii) changed end sequences, such that the ends presented no (AG3T3)3 signal. The second class (54.8% of plants) presented non-telomeric signal at several chromosome locations. C. campanulatus, H. rhamnoides, and R. pseudoacacia had non-telomeric signals, as supported by Luo and Chen [52], Luo et al. [11], and He et al. [51]. However, F. pennsylvanica, H. mutabilis, J. sigillata ‘Muzhilinhe‘, and L. lucidum presented non-telomeric signals in this study, but only showed telomeric signals in previous research [47,48,49]. The probable reasons for such discrepant signals were: (i) non-telomeric signal occurrence was lost during the previous experiments; and (ii) the variation in different batches of materials in the same species (i.e., intraspecific variation). Previous evidence suggested several species with a chromosome number of n = 21 or higher may present interstitial telomeric sequences [62]. In the present work, five species, F. pennsylvanica (2n = 46), L. × vicaryi (2n = 46), H. mutabilis (2n = 90), Z. armatum (2n = 98), and Z. bungeanum (2n = 136), presented interstitial telomeric signals, all having a chromosome number 2n > 42. It is possible that high chromosome number species have not been used as frequently in previous studies. The third class (26.2% of plants) presented telomeric signal deviating from the chromosome. C. campanulatus, F. pennsylvanica, L. × vicaryi, and S. oblata showed this type of signal, as supported by the results of Luo and Liu [49] and Luo and Chen [52]. J. sigillata ‘Muzhilinhe’ also presented this type of signal, in contrast to the results of Luo and Chen [48]. The probable reasons for this discrepancy are similar to those for the second class.
Unusual telomere sequences described by non-telomeric signals are, in many cases, connected with high C-values [63]; for example, in species of Cestrum and Allium [64]. In the present study, Z. mays, Z. armatum, and Z. bungeanum presented non-telomeric signals along with giant C-values (Zea, 3.8 pg; Zanthoxylum, 4.57 pg) [65] (https://cvalues.science.kew.org/, accessed on 17 May 2022). Nevertheless, the small C-values of Juglans (0.64 pg), Robinia (0.74 pg), and Chimonanthus (0.86 pg) were also accompanied by non-telomeric signals (C. campanulatus, J. sigillata ‘Muzhilinhe‘, R. pseudoacacia); especially for C. campanulatus, which presented highly diverse non-telomeric signals. Hence, to the best of our knowledge, there is no correlation between the presence of non-telomeric signals and the C-value in woody plants; this result agrees with the conclusion of Gorelick et al. [66].
4.4. Proposed Origin of (AG3T3)3 Diversity in Woody Plants
Telomeric sequences are not found exclusively at chromosome termini, but also in non-terminal sites of chromosomes in many species [23,24,25]. Interstitial telomeric sequences may represent a significant part of telomeric DNA. Bolzán [3] has revealed two major types of interstitial telomeric sequences: one is heterochromatic and is largely observed in centromeric or pericentromeric regions (e.g., in this study, C. campanulatus, five species of Taxus, and five plants of H. rhamnoides). The other type is short and distributed at various sites throughout chromosomes, such as in J. sigillata ‘Muzhilinhe’ and R. pseudoacacia.
Interstitial telomeric sequences could be considered the result of chromosomal rearrangements [23,24,67,68]. Messier et al. [69] explained interstitial telomeric sequences through the creation of a small number of repeats by random mutations followed by repeat expansion towards two flanks. In the present work, the non-terminal (AG3T3)3 signals were likely caused by chromosome deletion, duplication, inversion, translocation, and sequence changes, all of which are chromosomal reorganizations. Previous researchers have hypothesized that interstitial telomeric sequences in the heterochromatic region could be the trace of the chromosome end fusion, causing descendent hypoploidy (a decrease in the chromosome number) [28,29,30]. In the present work, we observed large primary constriction in chromosome 9 of B. diaphana and five species of Taxus. We may infer that these chromosomes with large primary constriction will possibly lead to ascent hyperploidyc. Thus, in the case of large primary constriction with non-end (AG3T3)3 signal, such as T. cuspidate chromosome 9 or T. wallichiana chromosome 9, ascent hyperploidy may occur. Similarly, we may also infer that the telocentric chromosome 12 in five species of Taxus was possibly formed in a reverse manner. However, in the case of large primary constriction with no signal, such as that observed in T. media chromosome 9 and B. diaphana chromosome 9, chromosome breakage is likely unrelated to interstitial (AG3T3)3.
5. Conclusions
In this paper, we examined Z. mays and 41 woody plants, established FISH physical mapping, described the diverse distribution of (AG3T3)3, and disclosed the complex genome architecture of woody plants. We inferred that the observed non-telomeric signals were probably caused by chromosome arrangements. We intend to continue our research by testing more woody plants, such as species of Calycanthaceae, to explore the abundant non-telomeric (AG3T3)3, as well as Rutaceae, to determine the chromosome number.
Author Contributions
J.L. and H.W. collected the materials and conducted the experiments. X.L. designed the oligo-probes, wrote the manuscript, provided funding, and supervised the study. Z.H. and X.G. performed chromosome image analysis. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Natural Science Foundation of China (grant number 31500993).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data and materials are included in the form of graphs in this article.
Acknowledgments
The authors thank Yonghong Zhou for laboratory equipment.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zakian, V.A. Telomeres: Beginning to understand the end. Science 1995, 270, 1601–1607. [Google Scholar] [CrossRef]
- Wellinger, R.J.; Sen, D. The DNA structures at the ends of eukaryotic chromosomes. Eur. J. Cancer 1997, 33, 735–749. [Google Scholar] [CrossRef]
- Bolzán, A.D. Interstitial telomeric sequences in vertebrate chromosomes: Origin, function, instability and evolution. Mutat. Res. Mutat. Res. 2017, 773, 51–65. [Google Scholar] [CrossRef]
- Traut, W.; Szczepanowski, M.; Vitkova, M.; Opitz, C.; Marec, F.; Zrzavy, J. The telomere repeat motif of basal Metazoa. Chromosome Res. 2007, 15, 371–382. [Google Scholar] [CrossRef]
- Uzlikova, M.; Fulneckova, J.; Weisz, F.; Sykorova, E.; Nohynkova, E.; Tumova, P. Characterization of telomeres and telomerase from the single-celled eukaryote Giardia intestinalis. Mol. Biochem. Parasitol. 2017, 211, 31–38. [Google Scholar] [CrossRef]
- Vitkova, M.; Kral, J.; Traut, W.; Zrzavy, J.; Marec, F. The evolutionary origin of insect telomeric repeats, (TTAGG)n. Chromosome Res. 2005, 13, 145–156. [Google Scholar] [CrossRef]
- Fulneckova, J.; Sevcikova, T.; Fajkus, J.; Lukesova, A.; Lukes, M.; Vlcek, C.; Lang, B.F.; Kim, E.; Eliás, M.; Sykorová, E. A broad phylogenetic survey unveils the diversity and evolution of telomeres in eukaryotes. Genome Biol. Evol. 2013, 5, 468–483. [Google Scholar] [CrossRef]
- Higashiyama, T.; Noutoshi, Y.; Akiba, M.; Yamada, T. Telomere and LINE-like elements at the termini of the Chlorella chromosome I. Nucleic. Acids Symp. Ser. 1995, 34, 71–72. [Google Scholar]
- Derelle, E.; Ferraz, C.; Rombauts, S.; Rouzé, P.; Worden, A.Z.; Robbens, S.; Partensky, F.; Degroeve, S.; Echeynié, S.; Cooke, R.; et al. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc. Natl. Acad. Sci. USA 2006, 103, 11647–11652. [Google Scholar] [CrossRef] [Green Version]
- Campomayor, N.B.; Waminal, N.E.; Kang, B.Y.; Nguyen, T.H.; Lee, S.S.; Huh, J.H.; Kim, H.H. Subgenome discrimination in Brassica and Raphanus allopolyploids using microsatellites. Cells 2021, 10, 2358. [Google Scholar] [CrossRef]
- Luo, X.; Liu, J.; He, Z. Oligo-FISH can identify chromosomes and distinguish Hippophaë rhamnoides L. taxa. Genes 2022, 13, 195. [Google Scholar] [CrossRef]
- Petracek, M.E.; Lefebvre, P.A.; Silflow, C.D.; Berman, J. Chlamydomonas telomere sequences are A+T-rich but contain three consecutive G-C base pairs. Proc. Natl. Acad. Sci. USA 1990, 87, 8222–8226. [Google Scholar] [CrossRef] [Green Version]
- Nozaki, H.; Takano, H.; Misumi, O.; Terasawa, K.; Matsuzaki, M.; Maruyama, S.; Nishida, K.; Yagisawa, F.; Yoshida, Y.; Fujiwara, T.; et al. A 100%-complete sequence reveals unusually simple genomic features in the hot-spring red alga Cyanidioschyzon merolae. BMC Biol. 2007, 5, 28. [Google Scholar] [CrossRef] [Green Version]
- Sykorova, E.; Lim, K.Y.; Kunicka, Z.; Chase, M.W.; Knapp, S.; Leitch, I.J.; Leitch, A.R.; Fajkus, J. The absence of Arabidopsis-type telomeres in Cestrum and closely related genera Vestia and Sessea (Solanaceae): First evidence from eudicots. Plant J. 2003, 34, 283–291. [Google Scholar] [CrossRef] [Green Version]
- Sykorova, E.; Lim, K.Y.; Fajkus, J.; Leitch, A.R. The signature of the Cestrum genome suggests an evolutionary response to the loss of (TTTAGGG)n telomeres. Chromosoma 2003, 112, 164–172. [Google Scholar] [CrossRef]
- Mravinac, B.; Mestrovic, N.; Cavrak, V.V.; Plohl, M. TCAGG, an alternative telomeric sequence in insects. Chromosoma 2011, 120, 367–376. [Google Scholar] [CrossRef]
- Muller, F.; Wicky, C.; Spicher, A.; Tobler, H. New telomere formation after developmentally regulated chromosomal breakage during the process of chromatin diminution in Ascaris lumbricoides. Cell 1991, 67, 815–822. [Google Scholar] [CrossRef]
- Tran, T.D.; Cao, H.X.; Jovtchev, G.; Neumann, P.; Novák, P.; Fojtová, M.; Vu, G.T.; Macas, J.; Fajkus, J.; Schubert, I.; et al. Centromere and telomere sequence alterations reflect the rapid genome evolution within the carnivorous plant genus Genlisea. Plant J. 2015, 84, 1087–1099. [Google Scholar] [CrossRef] [Green Version]
- Castilho, A.; Vershinin, A.; Heslop-Harrison, J.S. Repetitive DNA and the chromosomes in the genome of oil Palm (Elaeis guineensis). Ann. Bot. 2000, 85, 837–844. [Google Scholar] [CrossRef] [Green Version]
- Červenák, F.; Juríková, K.; Devillers, H.; Kaffe, B.; Khatib, A.; Bonnell, E.; Sopkovičová, M.; Wellinger, R.J.; Nosek, J.; Tzfati, Y.; et al. Identification of telomerase RNAs in species of the Yarrowia clade provides insights into the co-evolution of telomerase, telomeric repeats and telomere-binding proteins. Sci. Rep. 2019, 9, 13365. [Google Scholar] [CrossRef] [Green Version]
- Fajkus, P.; Peška, V.; Sitová, Z.; Fulnečková, J.; Dvořáčková, M.; Gogela, R.; Sýkorová, E.; Hapala, J.; Fajkus, J. Allium telomeres unmasked: The unusual telomeric sequence (CTCGGTTATGGG)n is synthesized by telomerase. Plant J. 2016, 85, 337–347. [Google Scholar] [CrossRef] [Green Version]
- Gomes, N.M.V.; Shay, J.W.; Wright, W.E. Telomere biology in Metazoa. FEBS Lett. 2010, 584, 3741–3751. [Google Scholar] [CrossRef] [Green Version]
- Fuchs, J.; Brandes, A.; Schubert, I. Telomere sequence localization and karyotype evolution in higher plants. Plant Syst. Evol. 1995, 196, 227–241. [Google Scholar] [CrossRef]
- Uchida, W.; Matsunaga, S.; Sugiyama, R.; Kawano, S. Interstitial telomere-like repeats in the Arabidopsis thaliana genome. Genes Genet. Syst. 2002, 77, 63–67. [Google Scholar] [CrossRef] [Green Version]
- He, L.; Liu, J.; Torres, G.A.; Zhang, H.; Jiang, J.; Xie, C. Interstitial telomeric repeats are enriched in the centromeres of chromosomes in Solanum species. Chromosome Res. 2013, 21, 5–13. [Google Scholar] [CrossRef]
- Mandáková, T.; Gloss, A.D.; Whiteman, N.K.; Lysak, M.A. How diploidization turned a tetraploid into a pseudotriploid. Am. J. Bot. 2016, 103, 1187–1196. [Google Scholar] [CrossRef] [Green Version]
- Lysak, M.A.; Mandáková, T.; Schranz, M.E. Comparative paleo genomics of crucifers: Ancestral genomic blocks revisited. Curr. Opin. Plant Biol. 2016, 30, 108–115. [Google Scholar] [CrossRef]
- Guerra, M. Chromosome numbers in plant cytotaxonomy: Concepts and implications. Cytogenet. Genome Res. 2008, 120, 339–350. [Google Scholar] [CrossRef]
- Sousa, A.; Cusimano, N.; Renner, S.S. Combining FISH and model-based predictions to understand chromosome evolution in Typhonium (Araceae). Ann. Bot. 2014, 113, 669–680. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Jin, D.; Wang, Z.; Guo, H.; Zhang, L.; Wang, L.; Li, J.; Paterson, A.H. Telomere-centric genome repatterning determines recurring chromosome number reductions during the evolution of eukaryotes. New Phytol. 2015, 205, 378–389. [Google Scholar] [CrossRef]
- Fonsêca, A.; Pedrosa-Harand, A. Karyotype stability in the genus Phaseolus evidenced by the comparative mapping of the wild species Phaseolus microcarpus. Genome 2013, 56, 335–343. [Google Scholar] [CrossRef]
- Murat, F.; Xu, J.H.; Tannier, E.; Abrouk, M.; Guilhot, N.; Pont, C.; Messing, J.; Salse, J. Ancestral grass karyotype reconstruction unravels new mechanisms of genome shuffling as a source of plant. Genome Res. 2010, 20, 1545–1557. [Google Scholar] [CrossRef] [Green Version]
- Schweizer, D.; Loidl, J. A model for heterochromatin dispersion and the evolution of C-band patterns. Chromosome Today 1987, 9, 61–74. [Google Scholar] [CrossRef]
- Sousa, G.; Vanzela, A.L.; Crosa, O.; Guerra, M. Interstitial telomeric sites and Robertsonian translocations in species of Ipheion and Nothoscordum (Amaryllidaceae). Genetica 2016, 144, 157–166. [Google Scholar] [CrossRef]
- Islam-Faridi, M.N.; Nelson, C.D.; DiFazio, S.P.; Gunter, L.E.; Tuskan, G.A. Cytogenetic analysis of Populus trichocarpa--ribosomal DNA, telomere repeat sequence, and marker-selected BACs. Cytogenet. Genome Res. 2009, 125, 74–80. [Google Scholar] [CrossRef]
- Lubaretz, O.; Fuchs, J.; Ahne, R.; Meister, A.; Schubert, I. Karyotyping of three Pinaceae species via fluorescent in situ hybridization and computer-aided chromosome analysis. Theor. Appl. Genet. 1996, 92, 411–416. [Google Scholar] [CrossRef]
- Puizina, J.; Sviben, T.; Krajacić-Sokol, I.; Zoldos-Pećnik, V.; Siljak-Yakovlev, S.; Papes, D.; Besendorfer, V. Cytogenetic and molecular characterization of the Abies alba genome and its relationship with other members of the Pinaceae. Plant Biol. 2008, 10, 256–267. [Google Scholar] [CrossRef]
- Rastogi, S.; Ohri, D. Karyotype evolution in cycads. Nucleus 2020, 63, 131–141. [Google Scholar] [CrossRef]
- Shibata, F.; Hizume, M. Survey of Arabidopsis and human-type telomeric repeats using fluorescence in situ hybridization. Cytologia 2011, 76, 353–360. [Google Scholar] [CrossRef] [Green Version]
- Shibata, F.; Matsusaki, Y.; Hizume, M. A comparative analysis of multi-probe fluorescence in situ hybridization (FISH) karyotypes in 26 Pinus species (Pinaceae). Cytologia 2016, 81, 409–421. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.C.; Pellerin, R.C.; Waminal, N.E.; Yang, T.J.; Kim, H.H. Pre-labelled oligo probe-FISH karyotype analyses of four Araliaceae species using rDNA and telomeric repeat. Genes Genom. 2019, 41, 839–847. [Google Scholar] [CrossRef]
- Hizume, M.; Shibata, F.; Matsusaki, Y.; Garajova, Z. Chromosome identification and comparative karyotypic analyses of four Pinus species. Theor. Appl. Genet. 2002, 105, 491–497. [Google Scholar] [CrossRef]
- Islam-Faridi, M.N.; Nelson, C.D.; Kubisiak, T.L. Reference karyotype and cytomolecular map for loblolly pine (Pinus taeda L.). Genome 2007, 50, 241–251. [Google Scholar] [CrossRef]
- Shibata, F.; Matsusaki, Y.; Hizume, M. AT-rich sequences containing Arabidopsis -type telomere sequence and their chromosomal distribution in Pinus densiflora. Theor. Appl. Genet. 2005, 110, 1253–1258. [Google Scholar] [CrossRef]
- Nkongolo, K.K.; Mehes-Smith, M. Karyotype evolution in the Pinaceae: Implication with molecular phylogeny. Genome 2012, 55, 735–753. [Google Scholar] [CrossRef]
- Murray, B.G.; Friesen, N.; Heslop-Harrison, J.S. Molecular cytogenetic analysis of Podocarpus and comparison with other gymnosperm species. Ann. Bot. 2002, 89, 483–489. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.; He, Z.J. Distribution of FISH oligo-5S rDNA and oligo-(AGGGTTT)3 in Hibiscus mutabilis L. Genome 2021, 64, 655–664. [Google Scholar] [CrossRef]
- Luo, X.; Chen, J. Distinguishing Sichuan walnut cultivars and examining their relationships with Juglans regia and J. sigillata by FISH, early-fruiting gene analysis, and SSR analysis. Front. Plant Sci. 2020, 11, 27. [Google Scholar] [CrossRef]
- Luo, X.; Liu, J. Fluorescence in situ hybridization (FISH) analysis of the locations of the oligonucleotides 5S rDNA, (AGGGTTT)3, and (TTG)6 in three genera of Oleaceae and their phylogenetic framework. Genes 2019, 10, 375. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Luo, X. First report of bicolour FISH of Berberis diaphana and B. soulieana reveals interspecific differences and co-localization of (AGGGTTT)3 and rDNA 5S in B. diaphana. Hereditas 2019, 156, 13. [Google Scholar] [CrossRef]
- He, Z.; Zhang, W.; Luo, X.; Huan, J. Five Fabaceae karyotype and phylogenetic relationship analysis based on oligo-FISH for 5SrDNA and (AG3T3)3. Genes 2022, 13, 768. [Google Scholar] [CrossRef]
- Luo, X.; Chen, J. Physical map of FISH 5S rDNA and (AG3T3)3 signals displays Chimonanthus campanulatus R.H. Chang & C.S. Ding chromosomes, reproduces its metaphase dynamics and distinguishes its chromosomes. Genes 2019, 10, 904. [Google Scholar] [CrossRef] [Green Version]
- Qi, Z.X.; Zeng, H.; Li, X.L.; Chen, C.B.; Song, W.Q.; Chen, R.Y. The molecular characterization of maize B chromosome specific AFLPs. Cell Res. 2002, 12, 63–68. [Google Scholar] [CrossRef] [Green Version]
- Mu, Y.L.; Xi, R.T. Microsporogenesis studying and karyotype analysis of Juglans regia L. and J. hopeiensis. J. Agric. Univ. Hebei 1988, 11, 48–55. Available online: https://kns.cnki.net/kcms/detail/detail.aspx?dbcode=CJFD&dbname=CJFD8589&filename=CULT198804007&uniplatform=NZKPT&v=ACIdK6WTIUPilcSLGVU01ZAZrHj971fMa9A1vqYsgTQD-NovAXbNVrrEKJsUtcW3 (accessed on 17 May 2022).
- Mu, Y.L.; Xi, R.T.; Lv, Z. Microsporogenesis observation and karyotype analysis of some species in genus Juglans L. J. Wuhan Bot. Res. 1990, 8, 301–310. Available online: https://en.cnki.com.cn/Article_en/CJFDTOTAL-WZXY199004000.htm (accessed on 17 May 2022).
- Hizume, M. Karyomorphological studies in the family Pinaceae. Mem. Fac. Educ. Ehime Univ. Ser. III Nat. Sci. 1988, 8, 1–108. [Google Scholar]
- Zhu, J.J.; Jiang, H.; Wang, Y.H.; Song, Y.C. Karyotype analyze Podocarpus macrophuus. Sichuan Forestry Technol. 1987, 8, 18–20. [Google Scholar] [CrossRef]
- Luo, X.; Liu, J.; Wang, J.; Gong, W.; Chen, L.; Wan, W. FISH analysis of Zanthoxylum armatum based on oligonucleotides for 5S rDNA and (GAA)6. Genome 2018, 61, 699–702. [Google Scholar] [CrossRef]
- Xing, M.S.; Xue, C.J.; Li, R.P. Karyotype analysis of sea buckthorn. J. Shanxi Univ. (Nat. Sci. Ed.) 1989, 12, 323–330. [Google Scholar] [CrossRef]
- Liu, H.Z.; Sheng, L.X. Studies on karyotype of three subspecies from Hippophae rhamnoides. J. Jilin Agr. Univ. 2011, 33, 628–631, 636. Available online: http://qikan.cqvip.com/Qikan/Article/Detail?id=39921155 (accessed on 17 May 2022).
- Kondo, K.; Tagashira, N. Regions in situ-hybridized by the Arabidopsis-type telomere sequence repeats in Zamia chromosomes. Chromosome Sci. 1998, 2, 87–99. Available online: https://xueshu.baidu.com/usercenter/paper/show?paperid=23e51cca2da947c9a694157d685eff7a&site=xueshu_se&hitarticle=1 (accessed on 17 May 2022).
- Maravilla, A.J.; Rosato, M.; Rosselló, J.A. Interstitial telomeric-like repeats (ITR) in seed plants as assessed by molecular cytogenetic techniques: A Review. Plants 2021, 10, 2541. [Google Scholar] [CrossRef] [PubMed]
- Peska, V.; Garcia, S. Origin, diversity, and evolution of telomere sequences in plants. Front. Plant Sci. 2020, 11, 117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sykorova, E.; Fajkus, J.; Meznikova, M.; Lim, K.Y.; Neplechova, K.; Blattner, F.R.; Chase, M.W.; Leitch, A.R. Minisatellite telomeres occur in the family Allianeae but are lost in Allium. A. J. Bot. 2006, 93, 814–823. [Google Scholar] [CrossRef] [PubMed]
- Pellicer, J.; Leitch, I.J. The Plant DNA C-values database (release 7.1): An updated online repository of plant genome size data for comparative studies. New Phytol. 2020, 226, 301–305. [Google Scholar] [CrossRef] [Green Version]
- Gorelick, R.; Fraser, D.; Zonneveld, B.J.M.; Little, D.P. Cycad (Cycadales) chromosome numbers are not correlated with genome size. Int. J. Plant Sci. 2014, 175, 986–997. [Google Scholar] [CrossRef]
- Pellerin, R.J.; Waminal, N.E.; Kim, H.H. FISH mapping of rDNA and telomeric repeats in 10 Senna species. Hortic. Environ. Biotechnol. 2019, 60, 253–260. [Google Scholar] [CrossRef]
- Waminal, N.E.; Pellerin, R.J.; Kang, S.H.; Kim, H.H. Chromosomal mapping of tandem repeats revealed massive chromosomal rearrangements and insights Into Senna tora dysploidy. Front. Plant Sci. 2021, 12, 629898. [Google Scholar] [CrossRef]
- Messier, W.; Li, S.H.; Steward, C.B. The birth of microsatellites. Nature 1996, 381, 483. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).