Identifying Genes Related to Acute Myocardial Infarction Based on Network Control Capability
Abstract
1. Introduction
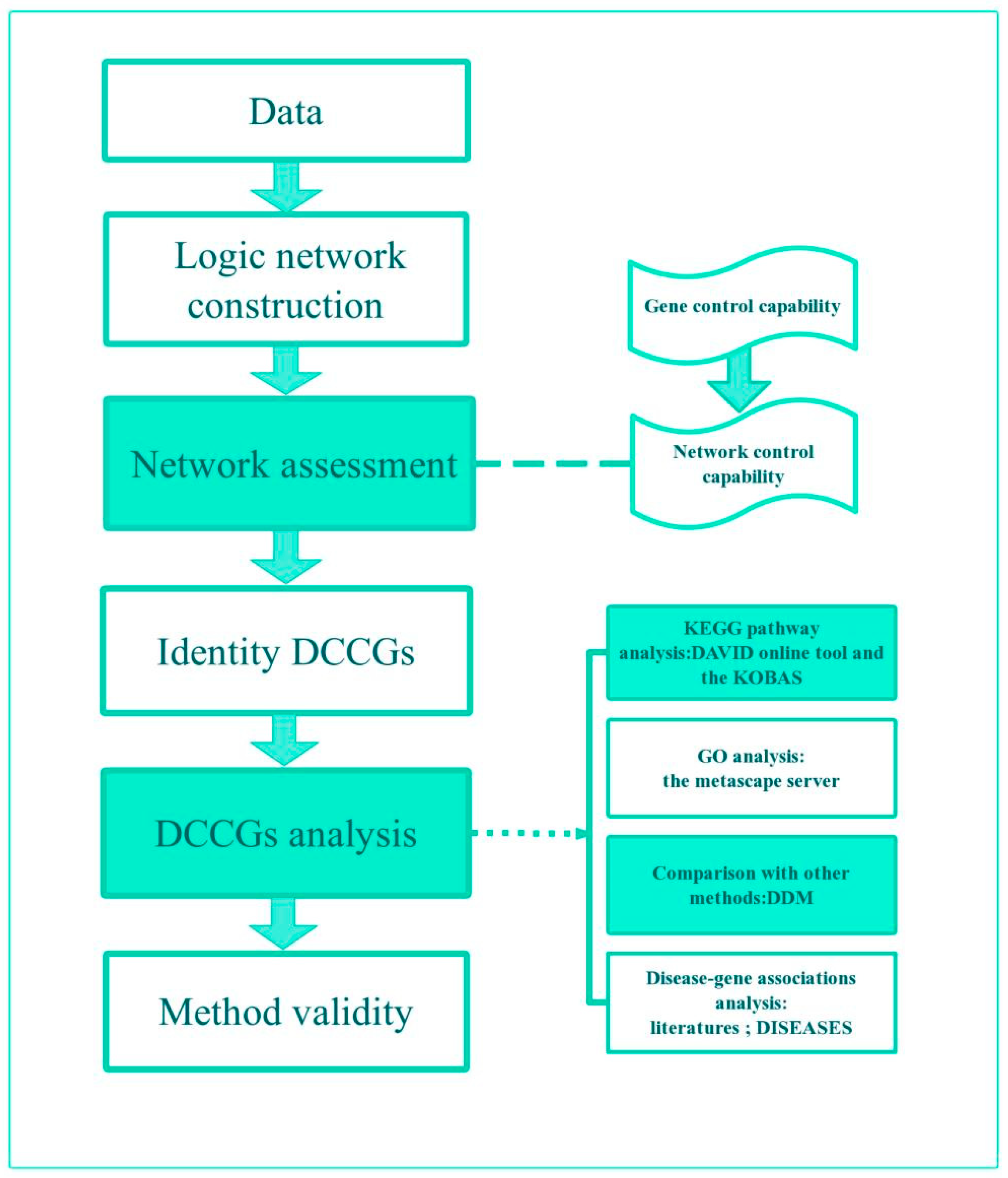
2. Materials and Methods
2.1. Study Design
2.2. Control Capabilities
2.3. Construction of Logic Networks
2.4. Identify DCCGs
3. Case and Results
3.1. Data
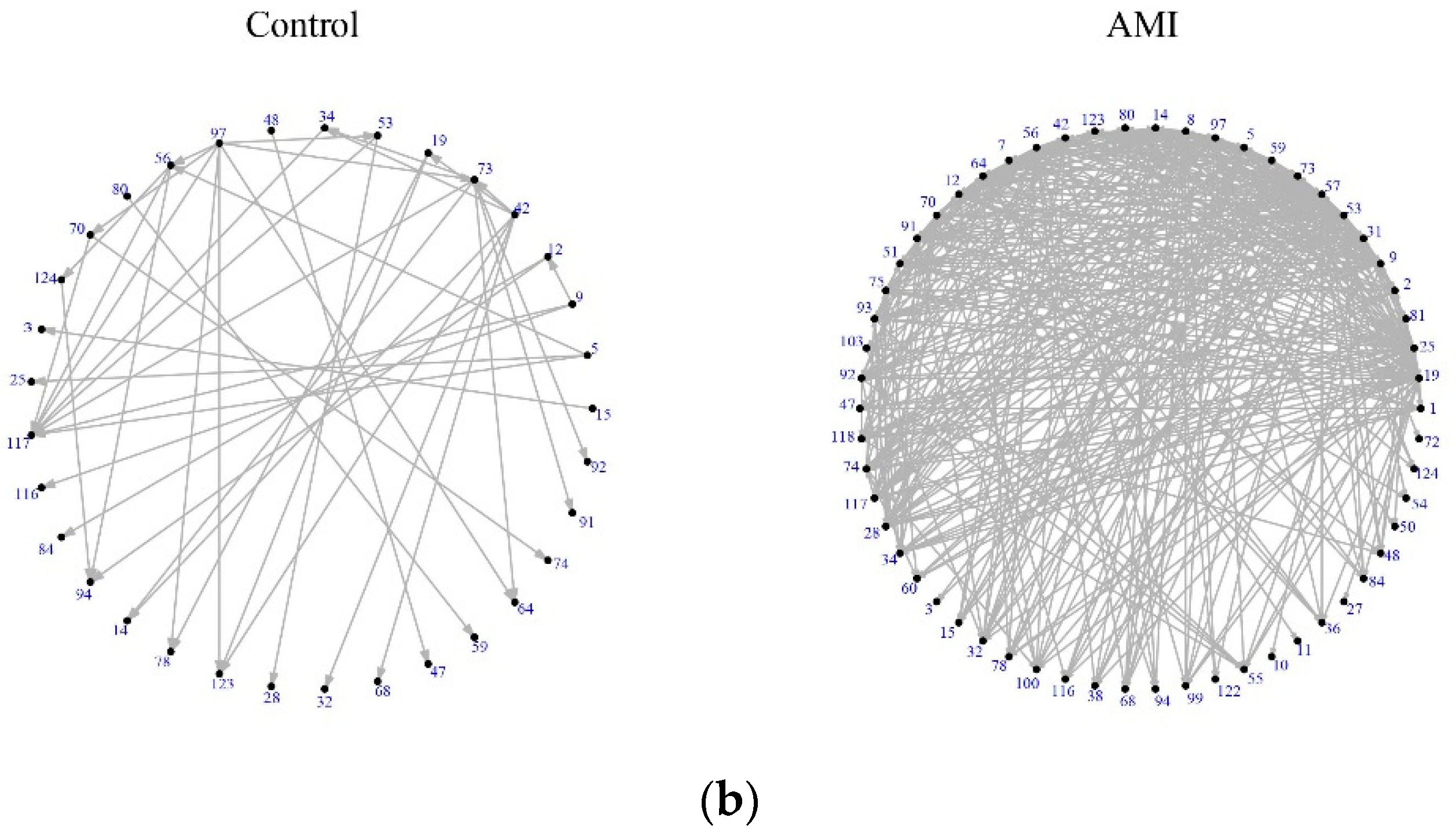
3.2. Assessment of DEGs’ Networks
3.3. DCCGs Identification
3.4. DCCGs Analysis
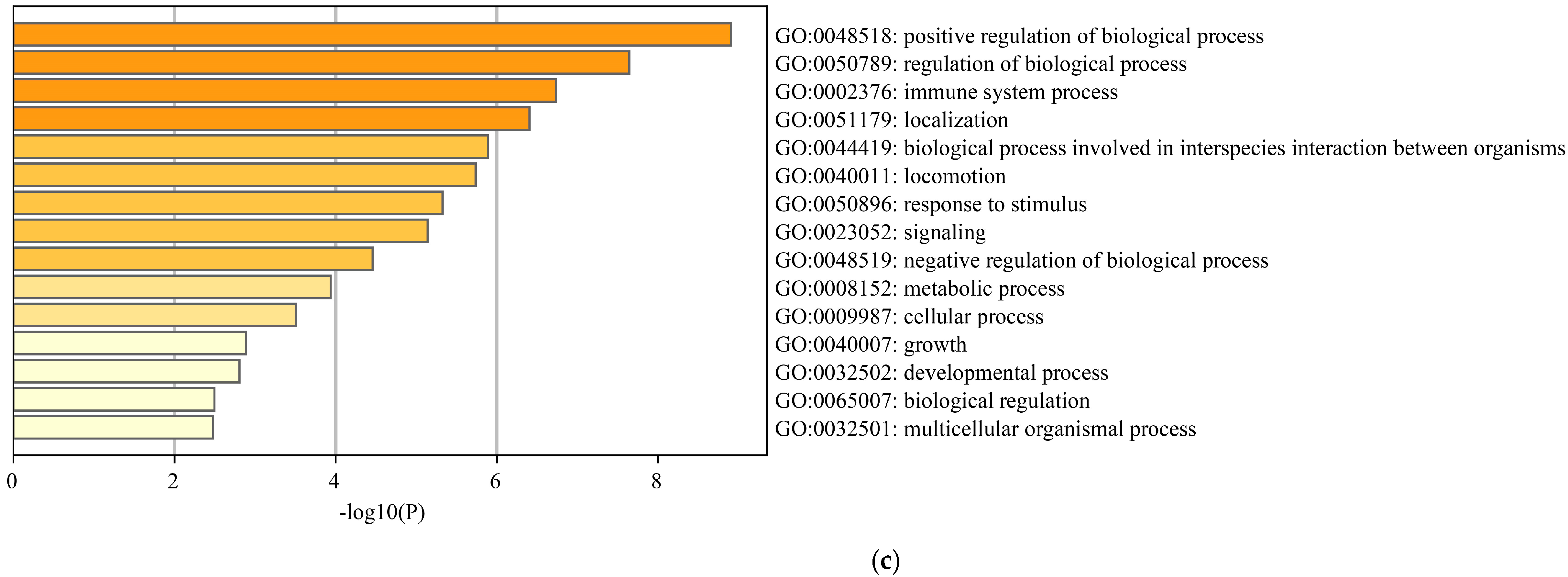
3.4.1. Enrichment Analysis
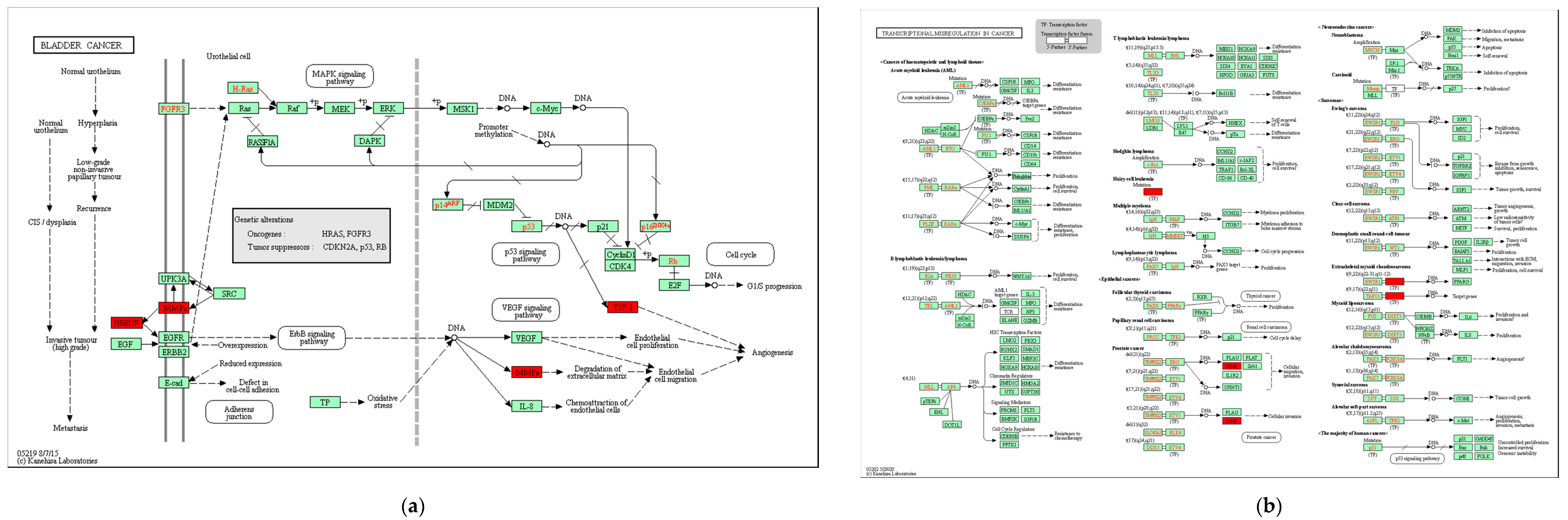
3.4.2. KEGG Pathway
3.4.3. Comparison with Other Methods
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mehta, L.S.; Beckie, T.M.; DeVon, H.A.; Grines, C.L.; Krumholz, H.M.; Johnson, M.N.; Lindley, K.J.; Vaccarino, V.; Wang, T.Y.; Watson, K.E.; et al. Acute Myocardial Infarction in Women. Circulation 2016, 133, 916–947. [Google Scholar] [CrossRef] [PubMed]
- Andreadou, I.; Cabrera-Fuentes, H.A.; Devaux, Y.; Frangogiannis, N.G.; Frantz, S.; Guzik, T.; Liehn, E.A.; Gomes, C.P.C.; Schulz, R.; Hausenloy, D.J. Immune cells as targets for cardioprotection: New players and novel therapeutic opportunities. Cardiovasc. Res. 2019, 115, 1117–1130. [Google Scholar] [CrossRef] [PubMed]
- Ong, S.B.; Hernández-Reséndiz, S.; Crespo-Avilan, G.E.; Mukhametshina, R.T.; Kwek, X.Y.; Cabrera-Fuentes, H.A.; Hausenloy, D.J. Inflammation following acute myocardial infarction: Multiple players, dynamic roles, and novel therapeutic opportunities. Pharmacol. Ther. 2018, 186, 73–87. [Google Scholar] [CrossRef] [PubMed]
- Westman, P.C.; Lipinski, M.J.; Luger, D.; Waksman, R.; Bonow, R.O.; Wu, E.; Epstein, S.E. Inflammation as a driver of adverse left ventricular remodeling after acute myocardial infarction. J. Am. Coll. Cardiol. 2016, 67, 2050–2060. [Google Scholar] [CrossRef] [PubMed]
- Keller, T.; Zeller, T.; Peetz, D.; Tzikas, S.; Roth, A.; Czyz, E.; Bickel, C.; Baldus, S.; Warnholtz, A.; Fröhlich, M.; et al. Sensitive troponin I assay in early diagnosis of acute myocardial infarction. N. Engl. J. Med. 2009, 361, 868–877. [Google Scholar] [CrossRef]
- Park, K.C.; Gaze, D.C.; Collinson, P.O.; Marber, M.S. Cardiac troponins: From myocardial infarction to chronic disease. Cardiovasc. Res. 2017, 113, 1708–1718. [Google Scholar] [CrossRef]
- Romaine, S.P.; Tomaszewski, M.; Condorelli, G.; Samani, N.J. MicroRNAs in cardiovascular disease: An introduction for clinicians. Heart 2015, 101, 921–928. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, W.; Liu, X.; Qi, J.; Deng, C. Biomarkers identification for acute myocardial infarction detection via weighted gene co-expression network analysis. Medicine 2017, 96, e8375. [Google Scholar] [CrossRef]
- Xie, H.; Zha, E.; Zhang, Y. Identification of featured metabolism-related genes in patients with acute myocardial infarction. Dis. Markers 2020, 2020, 8880004. [Google Scholar] [CrossRef]
- Stagljar, I.; Zilles, S.; Babu, M.; Houry, W.A. Systems analysis of the genetic interaction network of yeast molecular chaperones. Mol. Omics 2018, 14, 82–94. [Google Scholar]
- Ding, Q.; Shang, J.; Sun, Y.; Liu, G.; Li, F.; Yuan, X.; Liu, J.X. NIPMI: A network method based on interaction part mutual information to detect characteristic genes from integrated data on multi-cancers. IEEE Access 2019, 7, 135845–135854. [Google Scholar] [CrossRef]
- Shen, C. Controllability of Directed Complex Networks Based on Driver Nodes. Master’s Thesis, Qingdao University, Qingdao, China, 2020. (In Chinese). [Google Scholar]
- Liu, Y.Y.; Slotine, J.J.; Barabási, A.L. Controllability of complex networks. Nature 2011, 473, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Bowers, P.M.; Cokus, S.J.; Eisenberg, D.; Yeates, T.O. Use of logic relationships to decipher protein network organization. Science 2004, 306, 2246–2249. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Feng, Y.P.; Tang, L.X.; Yan, Y.L.; Bai, J.W. The protective role of NR4A3 in acute myocardial infarction by suppressing inflammatory responses via JAK2-STAT3/NF-κB pathway. Biochem. Biophys. Res. Commun. 2019, 517, 697–702. [Google Scholar] [CrossRef]
- Mouton, A.J.; Ma, Y.; Rivera Gonzalez, O.J.; Daseke, M.J., II; Flynn, E.R.; Freeman, T.C.; Garrett, M.R.; DeLeon-Pennell, K.Y.; Lindsey, M.L. Fibroblast polarization over the myocardial infarction time continuum shifts roles from inflammation to angiogenesis. Basic Res. Cardiol. 2019, 114, 6. [Google Scholar] [CrossRef] [PubMed]
- Qian, C.; Chang, D.; Li, H.; Wang, Y. Identification of potentially critical genes in the development of heart failure after ST-segment elevation myocardial infarction (STEMI). J. Cell. Biochem. 2019, 120, 7771–7777. [Google Scholar] [CrossRef] [PubMed]
- Farbehi, N.; Patrick, R.; Xaymardan, M.; Harvey, R.; Nordon, R. Large-scale, single cell RNA sequencing defines novel cellular subsets required for cardiac repair. Cytotherapy 2018, 20, S14. [Google Scholar] [CrossRef]
- Kadoglou, N.P.E.; Tahmatzidis, D.K.; Giannakoulas, C.; Kapelouzou, A.; Gkontopoulos, A.; Parissis, J.; Lampropoulos, S.; Kottas, G. Serum levels of novel adipokines, omentin-1 and chemerin, in patients with acute myocardial infarction: KOZANI STUDY. J. Cardiovasc. Med. 2015, 16, 341–346. [Google Scholar] [CrossRef]
- Yao, Y.; Zhao, J.; Zhou, X.; Hu, J.; Wang, Y. Potential role of a three-gene signature in predicting diagnosis in patients with myocardial infarction. Bioengineered 2021, 12, 2734–2749. [Google Scholar] [CrossRef]
- Dai, W.; Sun, Y.; Jiang, Z.; Du, K.; Xia, N.; Zhong, G. Key genes associated with non-alcoholic fatty liver disease and acute myocardial infarction. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2020, 26, e922492-1. [Google Scholar] [CrossRef]
- Feng, J.; Zhan, J.; Ma, S. LRG1 promotes hypoxia-induced cardiomyocyte apoptosis and autophagy by regulating hypoxia-inducible factor-1α. Bioengineered 2021, 12, 8897–8907. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.W.; Wang, B.C.; Hu, J.L.; Sun, J.J.; Wang, S.; Chen, X.J.; Meng, S.P.; Liu, L.; Cheng, Z.Y. IRAK3 gene silencing prevents cardiac rupture and ventricular remodeling through negative regulation of the NF-κB signaling pathway in a mouse model of acute myocardial infarction. J. Cell. Physiol. 2019, 234, 11722–11733. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Noh, J.H.; Eun, J.W.; Koh, Y.S.; Seo, S.M.; Park, W.S.; Lee, J.Y.; Chang, K.; Seung, K.B.; Kim, P.J.; et al. Assessment and diagnostic relevance of novel serum biomarkers for early decision of ST-elevation myocardial infarction. Oncotarget 2015, 6, 12970. [Google Scholar] [CrossRef] [PubMed]
- Ushikoshi, H.; Takahashi, T.; Chen, X.; Khai, N.C.; Esaki, M.; Goto, K.; Takemura, G.; Maruyama, R.; Minatoguchi, S.; Fujiwara, T.; et al. Local overexpression of HB-EGF exacerbates remodeling following myocardial infarction by activating noncardiomyocytes. Lab. Investig. 2005, 85, 862–873. [Google Scholar] [CrossRef][Green Version]
- Wang, C.; Bai, X.; Liu, S.; Wang, J.; Su, Z.; Zhang, W.; Bu, D.; Yan, Y.; Song, X. RNA-seq based transcriptome analysis of the protective effect of compound longmaining decoction on acute myocardial infarction. J. Pharm. Biomed. Anal. 2018, 158, 339–345. [Google Scholar] [CrossRef]
- Yadav, S.K.; Kambis, T.N.; Kar, S.; Park, S.Y.; Mishra, P.K. MMP9 mediates acute hyperglycemia-induced human cardiac stem cell death by upregulating apoptosis and pyroptosis in vitro. Cell Death Dis. 2020, 11, 1–15. [Google Scholar] [CrossRef]
- Mezzaroma, E.; Toldo, S.; Farkas, D.; Seropian, I.M.; Tassell, B.W.V.; Salloum, F.N.; Kannan, H.R.; Menna, M.C.; Voelkel, N.F.; Abbate, A. The inflammasome promotes adverse cardiac remodeling following acute myocardial infarction in the mouse. Proc. Natl. Acad. Sci. USA 2011, 108, 19725–19730. [Google Scholar] [CrossRef]
- Sandanger, Ø.; Ranheim, T.; Vinge, L.E.; Bliksøen, M.; Alfsnes, K.; Finsen, A.V.; Dahl, C.P.; Askevold, E.T.; Florholmen, G.; Christensen, G.; et al. The NLRP3 inflammasome is up-regulated in cardiac fibroblasts and mediates myocardial ischaemia–reperfusion injury. Cardiovasc. Res. 2013, 99, 164–174. [Google Scholar] [CrossRef]
- Zhang, K.Z.; Shen, X.Y.; Wang, M.; Wang, L.; Sun, H.X.; Li, X.Z.; Huang, J.J.; Li, X.Q.; Wu, C.; Zhao, C.; et al. Retinol-Binding Protein 4 Promotes Cardiac Injury After Myocardial Infarction Via Inducing Cardiomyocyte Pyroptosis through an Interaction With NLRP3. J. Am. Heart Assoc. 2021, 10, e022011. [Google Scholar] [CrossRef]
- Lee, E.J.; Hwang, I.; Kim, G.H.; Moon, D.; Kang, S.Y.; Hwang, I.C.; Lee, S.Y.; Marie, P.J.; Kim, H.S. Endothelin-1 augments therapeutic potency of human mesenchymal stem cells via CDH2 and VEGF signaling. Mol. Ther.-Methods Clin. Dev. 2019, 13, 503–511. [Google Scholar] [CrossRef]
- Marketou, M.; Kintsurashvili, E.; Papanicolaou, K.N.; Lucero, H.A.; Gavras, I.; Gavras, H. Cardioprotective effects of a selective B2 receptor agonist of bradykinin post-acute myocardial infarct. Am. J. Hypertens. 2010, 23, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Luo, M.; Li, Y.; Su, Z.; Wang, Y.; Chen, X.; Zhang, S.; Sun, W.; Kong, X. Bcl6 knockdown aggravates hypoxia injury in cardiomyocytes via the P38 pathway. Cell Biol. Int. 2019, 43, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Kalman, R.E. Mathematical description of linear dynamical systems. J. Soc. Ind. Appl. Math. Ser. A Control. 1963, 1, 152–192. [Google Scholar] [CrossRef]
- Muse, E.D.; Kramer, E.R.; Wang, H.; Barrett, P.; Parviz, F.; Novotny, M.A.; Lasken, R.S.; Jatkoe, T.A.; Oliveira, G.; Peng, H.; et al. A whole blood molecular signature for acute myocardial infarction. Sci. Rep. 2017, 7, 12268. [Google Scholar] [CrossRef]
- Wang, Y.; Li, C.; Stephenson, J.M.; Marrelli, S.P.; Kou, Y.M.; Meng, D.Z.; Wu, T. NR4A3 and CCL20 clusters dominate the genetic networks in CD146+ blood cells during acute myocardial infarction in humans. Eur. J. Med. Res. 2021, 26, 113. [Google Scholar] [CrossRef]
- Wang, Y.; Kou, Y.; Meng, D. Network structure analysis identifying key genes of autism and its mechanism. Comput. Math. Methods Med. 2020, 2020, 3753080. [Google Scholar] [CrossRef]
- Cardoso, A.C.; Lam, N.T.; Savla, J.J.; Nakada, Y.; Pereira, A.H.M.; Elnwasany, A.; Menendez-Montes, I.; Ensley, E.L.; Petric, U.B.; Sharma, G.; et al. Mitochondrial substrate utilization regulates cardiomyocyte cell-cycle progression. Nat. Metab. 2020, 2, 167–178. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Shoorei, H.; Mohaqiq, M.; Taheri, M. Non-coding RNAs regulate angiogenic processes. Vasc. Pharmacol. 2020, 133, 106778. [Google Scholar] [CrossRef]
- Yan, Y.; Zhang, B.; Liu, N.; Qi, C.; Xiao, Y.; Tian, X.; Li, T.; Liu, B. Circulating long noncoding RNA UCA1 as a novel biomarker of acute myocardial infarction. BioMed Res. Int. 2016, 2016, 8079372. [Google Scholar] [CrossRef]
- Lian, H.; Ma, Y.; Feng, J.; Dong, W.; Yang, Q.; Lu, D.; Zhang, L. Heparin-binding EGF-like growth factor induces heart interstitial fibrosis via an Akt/mTor/p70s6k pathway. PLoS ONE 2012, 7, e44946. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, N.; Masamura, K.; Yoshida, M.; Kato, M.; Kawai, Y.; Miyamori, I. A role of heparin-binding epidermal growth factor-like growth factor in cardiac remodeling after myocardial infarction. Biochem. Biophys. Res. Commun. 2002, 297, 375–381. [Google Scholar] [CrossRef]
- Rodríguez-Pérez, J.M.; Vargas-Alarcón, G.; Posadas-Sánchez, R.; Zagal-Jiménez, T.X.; Ortíz-Alarcón, R.; Valente-Acosta, B.; Tovilla-Zárate, C.; Nostroza-Hernández, C.; Pérez-Méndez, O.; Pérez-Hernández, N. rs3918242 MMP9 gene polymorphism is associated with myocardial infarction in Mexican patients. Genet. Mol. Res. 2016, 15, 15017776. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, L.; Chen, C.; Chi, Y.L.; Yang, X.Q.; Xu, Y.; Li, X.T.; Guo, S.L.; Xiong, S.H.; Shen, M.R.; et al. Interferon regulatory factor-1 together with reactive oxygen species promotes the acceleration of cell cycle progression by up-regulating the cyclin E and CDK2 genes during high glucose-induced proliferation of vascular smooth muscle cells. Cardiovasc. Diabetol. 2013, 12, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P. The string database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Degl’Innocenti, D.; Marzocchini, R.; Malentacchi, F.; Ramazzotti, M.; Raugei, G.; Ramponi, G. ACYP1 gene possesses two alternative splicing forms that induce apoptosis. IUBMB Life 2004, 56, 29–33. [Google Scholar]
- Zhou, W.; Thiery, J.P. Loss of Git2 induces epithelial–mesenchymal transition by miR146a-Cnot6L-controlled expression of Zeb1. J. Cell Sci. 2013, 126, 2740–2746. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Alimujiang, M.; Chen, Q.; Shi, H.; Luo, X. Exosomes derived from miR-146a-modified adipose-derived stem cells attenuate acute myocardial infarction− induced myocardial damage via downregulation of early growth response factor 1. J. Cell. Biochem. 2019, 120, 4433–4443. [Google Scholar] [CrossRef] [PubMed]







| Genes | Gene Names | Control Capability Fold-Change | Gene Expression Fold-Change | Functions in AMI/MI | AMI Z-Score | MI Z-Score |
|---|---|---|---|---|---|---|
| CAhighGeExphigh | ||||||
| HBEGF | Heparin-binding EGF-like growth factor | 11.5 | 0.215 | Upregulated HBEGF plays a pathophysiological role in injured hearts after MI [25]. | 2.1 | 3.4 |
| THBS1 | thrombospondin 1 | 8.714 | 0.395 | The development of heart failure after acute STEMI [17]; MI fibroblast secretome repressed angiogenesis through THBS1 signaling [16]. | 3.1 | 4.4 |
| NR4A3 | nuclear receptor subfamily 4 group A member 3 | 6.684 | 0.422 | Inhibiting post-AMI inflammation responses via JAK2-STAT3/NF-kappa B signaling may well be a therapeutic target for cardiac remodeling after AMI [15]. | 1.2 | 2 |
| BCL6 | BCL6 transcription repressor | 4.808 | 0.239 | —— | 1.2 | 1.9 |
| NLRP3 | NLR family pyrin domain-containing 3 | 3.216 | 0.411 | RBP4 as a novel modulator promoting cardiomyocyte pyroptosis via interaction with NLRP3 in AMI [30]; NLRP3 deletion reduces infarct size during AMI [28]; NLRP3 inflammasome is upregulated in myocardial fibroblasts post-MI [29]. | 3.7 | 4.7 |
| ITLN1 | intelectin 1 | 2.735 | 0.694 | The suppression of inflammation in the 6-month post-AMI period might have mediated the significant upregulation of omentin-1, implicating a novel target of treatment [19]. | 2.4 | 3.1 |
| PDK4 | pyruvate dehydrogenase kinase 4 | 2.375 | 0.348 | Following myocardial infarction, inducible deletion of PDK4 improved left ventricular function and decreased remodeling [38]. | 1.9 | 3.4 |
| CAhighGeExplow | ||||||
| ACYP1 | acylphosphatase 1 | 4.778 | −0.071 | —— | ||
| CNOT6L | CCR4-NOT transcription complex subunit 6 like | 3.905 | 0.095 | —— | ||
| CAlowGeExphigh | ||||||
| VNN3 | vanin 3, pseudogene | 1.833 | 0.332 | Diagnostic biomarkers for STEMI [24]. | —— | 1.3 |
| CXCL3 | C-X-C motif chemokine ligand 3 | 1.781 | 0.412 | Associated with reparative phases (post MI) [18]. | 1.1 | 2.6 |
| CLEC4D | C-type lectin domain family 4 member D | 1.691 | 0.545 | Playing an important role in the occurrence and progression AMI [21]. | ||
| LRG1 | Leucine-rich α-2-glycoprotein 1 | 1.630 | 0.344 | LRG1/HIF-1 α promoted H9c2 cell apoptosis and autophagy in hypoxia, potentially providing new ideas for the determination and treatment of AMI [22]. | 1.3 | 2.2 |
| IRAK3 | interleukin 1 receptor-associated kinase 3 | 1.607 | 0.456 | Silencing of IRAK3 inactivates the NF-B signaling pathway and prevents AMI progression [23]. | 2.2 | 2.8 |
| MMP9 | matrix metallopeptidase 9 | 1.575 | 0.377 | Inhibiting the chemokine signaling pathway and leukocyte transendothelial migration play a protective effect on AMI [26]. MMP9 is upregulated in the diabetic heart, and ablation of MMP9 decreases the infarct size in the non-diabetic myocardial infarction heart [27]. | 5 | 5.9 |
| EDN1 | endothelin 1 | 1.474 | 0.296 | EDN1 induces CDH2 and VEGF expression in hUCB-MSCs, leading to improved therapeutic efficacy in rat MI [31]. | 5.1 | 6.0 |
| AC079305.10 | unnamed | 1.389 | 0.417 | —— |
| Term | Description | LogP | Genes |
|---|---|---|---|
| GO:0048661 | positive regulation of smooth muscle cell proliferation | −8.93 | HBEGF, EDN1, MMP9, THBS1, NR4A3, BCL6, ITLN1, IRAK3, CXCL3 |
| GO:0002718 | regulation of cytokine production involved in immune response | −6.74 | BCL6, NR4A3, IRAK3, NLRP3, THBS1, CLEC4D, CXCL3, EDN1 |
| WP2865 | IL1 and megakaryocytes in obesity | −6.56 | HBEGF, MMP9, NLRP3, BCL6, THBS1, IRAK3, NR4A3, CLEC4D, LRG1 |
| GO:0009617 | response to bacterium | −5.91 | EDN1, CXCL3, IRAK3, NLRP3, LRG1, CLEC4D, THBS1, PDK4 |
| GO:1904707 | positive regulation of vascular associated smooth muscle cell proliferation | −5.74 | EDN1, CXCL3, IRAK3, NLRP3, LRG1, CLEC4D, THBS1, PDK4 |
| hsa04668 | TNF signaling pathway | −4.57 | EDN1, CXCL3, MMP9, NLRP3, LRG1 |
| M5885 | NABA MATRISOME ASSOCIATED | −4.49 | HBEGF, CXCL3, MMP9, ITLN1, CLEC4D |
| Genes | Gene Names | AMI Z-Score | MI Z-Score |
|---|---|---|---|
| IRAK3 | interleukin 1 receptor-associated kinase 3 | 2.2 | 2.8 |
| ITLN1 | intelectin 1 | 2.4 | 3.1 |
| BCL6 | BCL6 transcription repressor | 1.2 | 1.9 |
| CXCL3 | C-X-C motif chemokine ligand 3 | 1.1 | 2.6 |
| NR4A3 | nuclear receptor subfamily 4 group A member 3 | 1.2 | 2 |
| CLEC4D | C-type lectin domain family 4 member D | —— | —— |
| CDC25B | cell division cycle 25B | —— | —— |
| AC079305.10 | unnamed | —— | —— |
| MMP9 | matrix metallopeptidase 9 | 5 | 5.9 |
| GLUL | glutamate-ammonia ligase | —— | 1.9 |
| FITM2 | fat storage-inducing transmembrane protein 2 | —— | —— |
| ITPRIP | inositol 1,4,5-trisphosphate receptor-interacting protein | —— | —— |
| METRNL | meteorin-like, glial cell differentiation regulator | —— | —— |
| GABARAPL1 | GABA type A receptor-associated protein-like 1 | 1.5 | 2.9 |
| RNF144B | ring finger protein 144B | —— | —— |
| NLRP3 | NLR family pyrin domain-containing 3 | 3.7 | 3.7 |
| ANXA3 | annexin A3 | 2.1 | 2.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Xian, H. Identifying Genes Related to Acute Myocardial Infarction Based on Network Control Capability. Genes 2022, 13, 1238. https://doi.org/10.3390/genes13071238
Wang Y, Xian H. Identifying Genes Related to Acute Myocardial Infarction Based on Network Control Capability. Genes. 2022; 13(7):1238. https://doi.org/10.3390/genes13071238
Chicago/Turabian StyleWang, Yanhui, and Huimin Xian. 2022. "Identifying Genes Related to Acute Myocardial Infarction Based on Network Control Capability" Genes 13, no. 7: 1238. https://doi.org/10.3390/genes13071238
APA StyleWang, Y., & Xian, H. (2022). Identifying Genes Related to Acute Myocardial Infarction Based on Network Control Capability. Genes, 13(7), 1238. https://doi.org/10.3390/genes13071238





