Effects of SNPs in AANAT and ASMT Genes on Milk and Peripheral Blood Melatonin Concentrations in Holstein Cows (Bos taurus)
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Genomic DNA Extraction
2.2. Primer Design
2.3. SNP Screening and SNP Genotyping
3. Statistical Analysis
4. Results
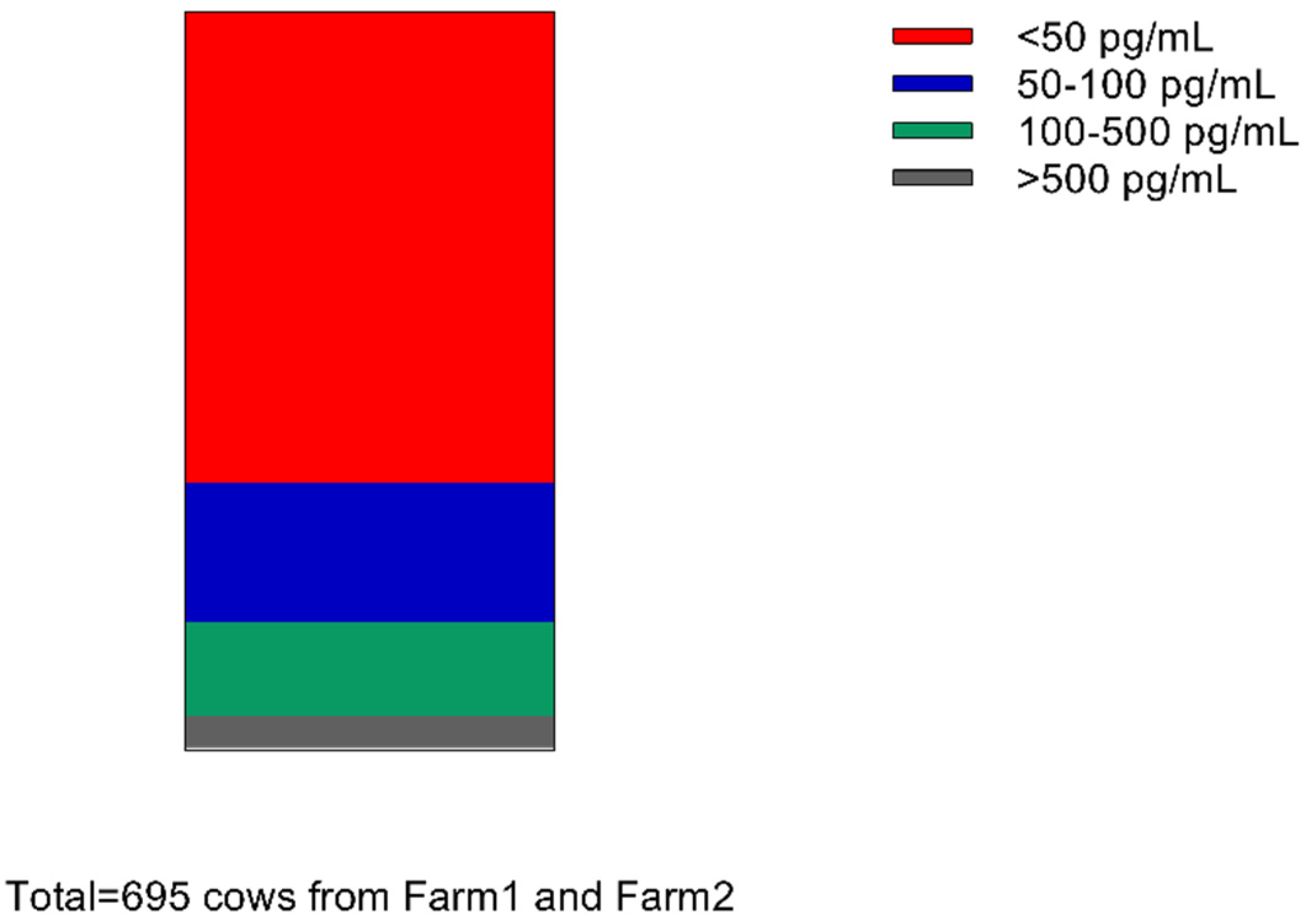
4.1. Distribution of MT Levels in Milk of 695 Cows
4.2. Correlation Analysis between Milk MT Levels and the Index of DHI
4.3. Association between Milk and Serum MT Levels
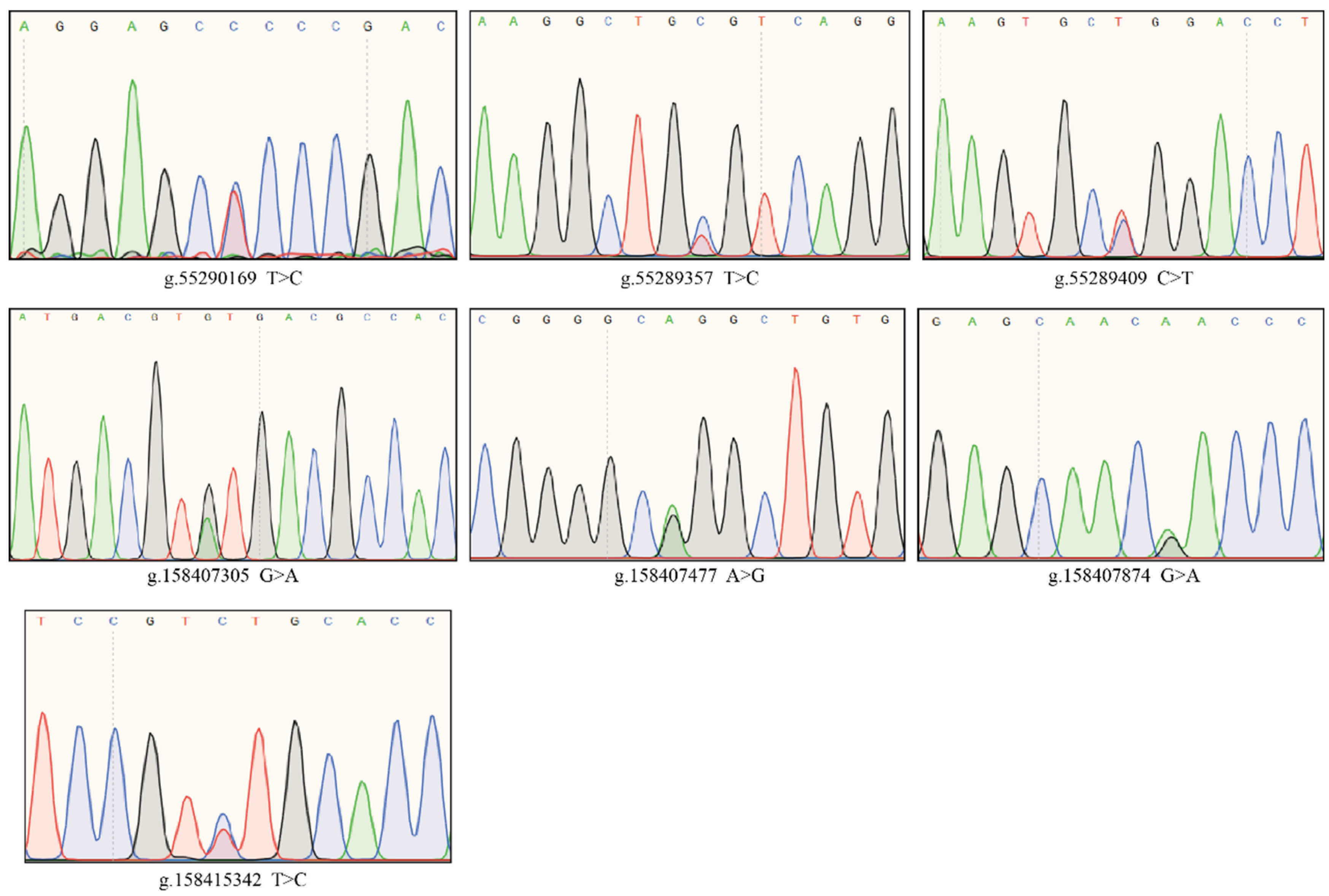
4.4. Identification of Three Significantly Associated SNPs within AANAT and Four SNPs within ASMT in Cows
4.5. Genotypic and Allelic Frequencies and Genetic Parameter
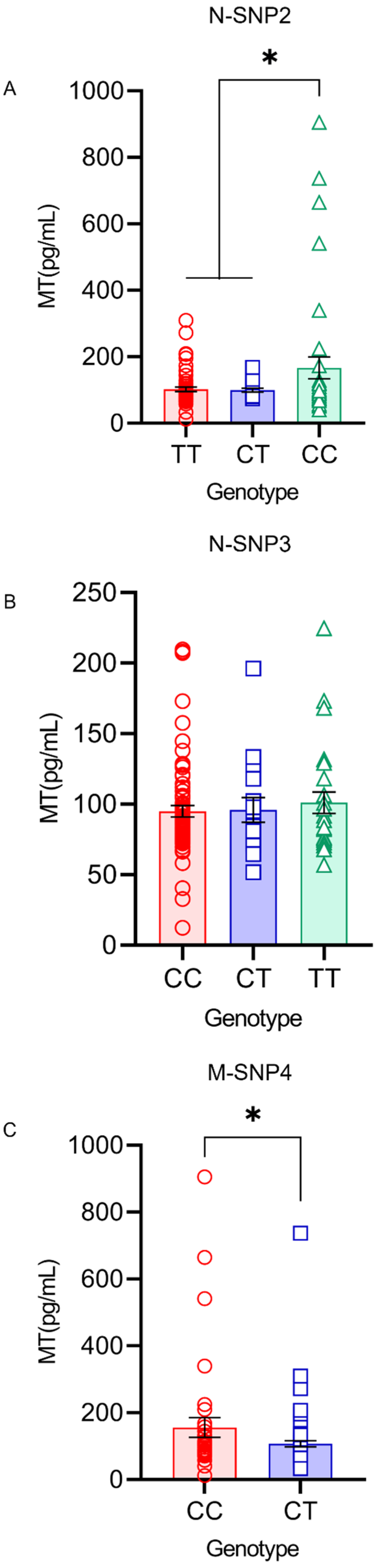
4.6. MT Levels in the Serum and Milk under Different Genotypes of SNP
4.7. Linkage Disequilibrium Analysis of SNP Loci within Cow AANAT and ASMT Genes
5. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Konturek, S.J.; Konturek, P.C.; Brzozowska, I.; Pawlik, M.; Sliwowski, Z.; Cześnikiewicz-Guzik, M.; Kwiecień, S.; Brzozowski, T.; Bubenik, G.A.; Pawlik, W.W. Localization and biological activities of melatonin in intact and diseased Gastrointestinal Tract (GIT). J. Physiol. Pharmacol. 2007, 58, 381–405. [Google Scholar] [PubMed]
- Cipolla-Neto, J.; Amaral, F.G. do Melatonin as a Hormone: New Physiological and Clinical Insights. Endocr. Rev. 2018, 39, 990–1028. [Google Scholar] [CrossRef] [PubMed]
- Kvetnoy, I.M. Extrapineal melatonin: Location and role within diffuse neuroendocrine system. Histochem. J. 1999, 31, 1–12. [Google Scholar] [CrossRef]
- Kvetnoy, I.; Ivanov, D.; Mironova, E.; Evsyukova, I.; Nasyrov, R.; Kvetnaia, T.; Polyakova, V. Melatonin as the Cornerstone of Neuroimmunoendocrinology. Int. J. Mol. Sci. 2022, 22, 1835. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Chen, B.; Li, Z.; Lin, R.; Ruan, S.; Wang, F.; Liang, H.; Tao, J. Electroacupuncture Ameliorates Cognitive Impairment Through the Inhibition of NLRP3 Inflammasome Activation by Regulating Melatonin-Mediated Mitophagy in Stroke Rats. Neurochem. Res. 2022, 47, 1917–1930. [Google Scholar] [CrossRef] [PubMed]
- Sakimura, K.; Maekawa, T.; Kume, S.-I.; Ohta, T. Spontaneously Diabetic Torii (SDT) Fatty Rat, a Novel Animal Model of Type 2 Diabetes Mellitus, Shows Blunted Circadian Rhythms and Melatonin Secretion. Int. J. Endocrinol. 2018, 2018, 9065690. [Google Scholar] [CrossRef]
- Al Twaijri, W.A.; Ba Armah, D.; Bin Sabbar, N.; Aldebasi, T.; Shaheen, N.; Al Bekairy, A.K.S.; Al-Jeraisy, M. Effect of melatonin in children with neurodevelopmental disabilities and sleep disorders. J. Fam. Med. Prim. Care 2022, 11, 299–304. [Google Scholar] [CrossRef]
- Li, H.; Liu, M.; Zhang, C. Women with polycystic ovary syndrome (PCOS) have reduced melatonin concentrations in their follicles and have mild sleep disturbances. BMC Women’s Health 2022, 22, 79. [Google Scholar] [CrossRef]
- Erdem, Y.; Altunay, İ.K.; Özkur, E.; Şekerlisoy, G.; Karabay, E.A.; Özdemir, F.T.; Çerman, A.A. The Association between Melatonin Levels and Sleep Quality in Patients with Pruritus: A Potential Biomarker on a Candidate Future Treatment. Indian J. Dermatol. 2021, 66, 609–615. [Google Scholar] [CrossRef]
- Ares, G.; Giménez, A.; Gámbaro, A. Consumer perceived healthiness and willingness to try functional milk desserts. Influence of ingredient, ingredient name and health claim. Food Qual. Prefer. 2009, 20, 50–56. [Google Scholar] [CrossRef]
- Asher, A.; Shabtay, A.; Brosh, A.; Eitam, H.; Agmon, R.; Cohen-Zinder, M.; Zubidat, A.E.; Haim, A. “Chrono-functional milk”: The difference between melatonin concentrations in night-milk versus day-milk under different night illumination conditions. Chronobiol. Int. 2015, 32, 1409–1416. [Google Scholar] [CrossRef] [PubMed]
- Valtonen, M.; Niskanen, L.; Kangas, A.-P.; Koskinen, T. Effect of melatonin-rich night-time milk on sleep and activity in elderly institutionalized subjects. Nord. J. Psychiatry 2005, 59, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Vignal, A.; Milan, D.; SanCristobal, M.; Eggen, A. A review on SNP and other types of molecular markers and their use in animal genetics. Genet. Sel. Evol. 2002, 34, 275–305. [Google Scholar] [CrossRef] [PubMed]
- Kruglyak, L. The use of a genetic map of biallelic markers in linkage studies. Nat. Genet. 1997, 17, 21–24. [Google Scholar] [CrossRef]
- Abeygunawardana, D.I.; Ranasinghe, R.M.S.B.K.; De Silva, S.N.T.; Deshapriya, R.M.C.; Gamika, P.A.; Rajapakse, J. Effect of LHCGR and FSHR gene polymorphisms on fertility traits and milk yield of cross-bred dairy cows in Sri Lanka. Anim. Biotechnol. 2022, 1–8. [Google Scholar] [CrossRef]
- Davis, S.R.; Ward, H.E.; Kelly, V.; Palmer, D.; Ankersmit-Udy, A.E.; Lopdell, T.J.; Berry, S.D.; Littlejohn, M.D.; Tiplady, K.; Adams, L.F.; et al. Screening for phenotypic outliers identifies an unusually low concentration of a β-lactoglobulin B protein isoform in bovine milk caused by a synonymous SNP. Genet. Sel. Evol. 2022, 54, 22. [Google Scholar] [CrossRef]
- Wang, C.; Yuan, Z.; Hu, R.; Li, F.; Yue, X. Association of SNPs within PTPN3 gene with wool production and growth traits in a dual-purpose sheep population. Anim. Biotechnol. 2022, 1–7. [Google Scholar] [CrossRef]
- Basson, A.; Strydom, P.E.; van Marle-Köster, E.; Webb, E.C.; Frylinck, L. Sustained Effects of Muscle Calpain System Genotypes on Tenderness Phenotypes of South African Beef Bulls during Ageing up to 20 Days. Animals 2022, 12, 686. [Google Scholar] [CrossRef]
- Molik, E.; Misztal, T.; Romanowicz, K.; Wierzchoś, E. Dependence of the lactation duration and efficiency on the season of lambing in relation to the prolactin and melatonin secretion in ewes. Livest. Sci. 2007, 107, 220–226. [Google Scholar] [CrossRef]
- Murphy, B.A.; Herlihy, M.M.; Nolan, M.B.; O’Brien, C.; Furlong, J.G.; Butler, S.T. Identification of the blue light intensity administered to one eye required to suppress bovine plasma melatonin and investigation into effects on milk production in grazing dairy cows. J. Dairy Sci. 2021, 104, 12127–12138. [Google Scholar] [CrossRef]
- Singh, V.P.; Sachan, N.; Verma, A.K. Melatonin milk; a milk of intrinsic health benefit: A review. Int. J. Dairy Sci. 2011, 6, 246–252. [Google Scholar] [CrossRef][Green Version]
- Milagres, M.P.; Minim, V.P.R.; Simiqueli, A.A.; Espeschit, A.C.R.; Minim, L.A. Milk with increased concentration of melatonin packaging factors on consumer purchase intention. Rev. Inst. Laticínios Cândido Tostes 2014, 69, 25–36. [Google Scholar]
- Dustmann, H. Milk melatonin: Creation of higher value required by native ingredient. DMZ Leb. Milchwirtsch. 2010, 131, 20–21. [Google Scholar]
- Zhang, W.; Zhao, G.; Zhang, X.; Mo, F. Technology for production of cow’s milk rich in naturally expressed melatonin. Chin. J. Anim. Nutr. 2016, 28, 635–640. [Google Scholar]
- Chen, J.; Zhao, F.; Wei, K.; Tan, S.; Yang, K. Effects of tryptophan and rumen-protected tryptophan supplementations on lactation performance, plasma indexes and milk melatonin content of dairy cows. Chin. J. Anim. Nutr. 2017, 29, 3921–3931. [Google Scholar]
- Sangsopha, J.; Johns, N.P.; Johns, J.; Moongngarm, A. Dietary sources of melatonin and benefits from production of high melatonin pasteurized milk. J. Food Sci. Technol. 2020, 57, 2026–2037. [Google Scholar] [CrossRef]
- Hardeland, R. Aging, Melatonin, and the Pro- and Anti-Inflammatory Networks. Int. J. Mol. Sci. 2019, 20, 1223. [Google Scholar] [CrossRef]
- Tao, J.; Yang, M.; Wu, H.; Ma, T.; He, C.; Chai, M.; Zhang, X.; Zhang, J.; Ding, F.; Wang, S.; et al. Effects of AANAT overexpression on the inflammatory responses and autophagy activity in the cellular and transgenic animal levels. Autophagy 2018, 14, 1850–1869. [Google Scholar] [CrossRef]
- Wu, H.; Cui, X.; Guan, S.; Li, G.; Yao, Y.; Wu, H.; Zhang, J.; Zhang, X.; Yu, T.; Li, Y.; et al. The Improved Milk Quality and Enhanced Anti-Inflammatory Effect in Acetylserotonin-O-methyltransferase (ASMT) Overexpressed Goats: An Association with the Elevated Endogenous Melatonin Production. Molecules 2022, 27, 572. [Google Scholar] [CrossRef]
- Ma, T.; Tao, J.; Yang, M.; He, C.; Tian, X.; Zhang, X.; Zhang, J.; Deng, S.; Feng, J.; Zhang, Z.; et al. An AANAT/ASMT transgenic animal model constructed with CRISPR/Cas9 system serving as the mammary gland bioreactor to produce melatonin-enriched milk in sheep. J. Pineal Res. 2017, 63, e12406. [Google Scholar] [CrossRef]
- Wang, X.; Yang, Q.; Zhang, S.; Zhang, X.; Pan, C.; Chen, H.; Zhu, H.; Lan, X. Genetic Effects of Single Nucleotide Polymorphisms in the Goat GDF9 Gene on Prolificacy: True or False Positive? Animals 2019, 9, 886. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, X.; Jiang, E.; Yan, H.; Zhu, H.; Chen, H.; Liu, J.; Qu, L.; Pan, C.; Lan, X. InDels within caprine IGF2BP1 intron 2 and the 3’-untranslated regions are associated with goat growth traits. Anim. Genet. 2020, 51, 117–121. [Google Scholar] [CrossRef] [PubMed]

| DHI Index | Sample Size | Pearson r | p Values |
|---|---|---|---|
| DIM | 593 | −0.013 | 0.754 |
| L# | 593 | −0.081 | 0.050 |
| HTM | 593 | −0.055 | 0.185 |
| HTACM | 593 | −0.032 | 0.438 |
| Prev.M | 593 | −0.070 | 0.105 |
| FP | 540 | −0.007 | 0.873 |
| PP | 593 | −0.017 | 0.680 |
| F/P | 593 | −0.011 | 0.781 |
| LP | 593 | 0.008 | 0.855 |
| SP | 593 | −0.009 | 0.824 |
| SCC | 593 | 0.017 | 0.679 |
| MLos | 593 | −0.025 | 0.539 |
| LSCC | 593 | −0.037 | 0.374 |
| PreSCC | 593 | −0.035 | 0.418 |
| 1stSCC | 539 | −0.031 | 0.455 |
| LTDM | 593 | −0.037 | 0.364 |
| 305M | 593 | −0.092 | 0.041 * |
| PeakM | 492 | −0.104 | 0.011 * |
| PeakD | 593 | 0.002 | 0.961 |
| Persist | 593 | 0.015 | 0.728 |
| LacLen | 540 | −0.013 | 0.754 |
| SNP Loci | Genotypic Frequencies | Allele Frequencies | Genetic Parameter | |||||
|---|---|---|---|---|---|---|---|---|
| D | H | R | Reference | Mutation | Ho | He | HWE (p Value) | |
| N-SNP1 | 0.706 (72) | 0.225 (23) | 0.069 (7) | 0.819 | 0.181 | 0.704 | 0.296 | 0.0484 * |
| N-SNP2 | 0.508 (62) | 0.18 (22) | 0.312 (38) | 0.598 | 0.402 | 0.519 | 0.481 | <0.00001 ***** |
| N-SNP3 | 0.623 (76) | 0.148 (18) | 0.229 (28) | 0.697 | 0.303 | 0.578 | 0.422 | <0.00001 ***** |
| M-SNP1 | 0.51 (52) | 0.343 (35) | 0.147 (15) | 0.681 | 0.319 | 0.566 | 0.434 | 0.1079 |
| M-SNP2 | 0.51 (52) | 0.333 (34) | 0.157 (16) | 0.676 | 0.324 | 0.562 | 0.438 | 0.0547 |
| M-SNP3 | 0.569 (58) | 0.353 (36) | 0.078 (8) | 0.745 | 0.255 | 0.62 | 0.38 | 0.7658 |
| M-SNP4 | 0.303 (37) | 0.697 (85) | NA | 0.65 | 0.35 | 0.545 | 0.455 | 0.000051 **** |
| Gene | AANAT | ASMT | |||||||
|---|---|---|---|---|---|---|---|---|---|
| LD | N-SNP1 N-SNP2 | N-SNP1 N-SNP3 | N-SNP2 N-SNP3 | M-SNP1 M-SNP2 | M-SNP1 M-SNP3 | M-SNP1 M-SNP4 | M-SNP2 M-SNP3 | M-SNP2 M-SNP4 | M-SNP3 M-SNP4 |
| D′ | 0.004 | 0.009 | 0.27 | 0.98 | 0.05 | 0.0002 | 0.05 | 0.0005 | 0.0007 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, S.; Liu, Y.; Liu, X.; Liu, G. Effects of SNPs in AANAT and ASMT Genes on Milk and Peripheral Blood Melatonin Concentrations in Holstein Cows (Bos taurus). Genes 2022, 13, 1196. https://doi.org/10.3390/genes13071196
Yao S, Liu Y, Liu X, Liu G. Effects of SNPs in AANAT and ASMT Genes on Milk and Peripheral Blood Melatonin Concentrations in Holstein Cows (Bos taurus). Genes. 2022; 13(7):1196. https://doi.org/10.3390/genes13071196
Chicago/Turabian StyleYao, Songyang, Yunjie Liu, Xuening Liu, and Guoshi Liu. 2022. "Effects of SNPs in AANAT and ASMT Genes on Milk and Peripheral Blood Melatonin Concentrations in Holstein Cows (Bos taurus)" Genes 13, no. 7: 1196. https://doi.org/10.3390/genes13071196
APA StyleYao, S., Liu, Y., Liu, X., & Liu, G. (2022). Effects of SNPs in AANAT and ASMT Genes on Milk and Peripheral Blood Melatonin Concentrations in Holstein Cows (Bos taurus). Genes, 13(7), 1196. https://doi.org/10.3390/genes13071196






