Screening of Differentially Expressed Genes and miRNAs in Hypothalamus and Pituitary Gland of Sheep under Different Photoperiods
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. RNA Extraction, Library Construction, Sequencing and Raw Data Processing
2.3. Differential Expression Analysis of mRNAs
2.4. Differential Expression Analysis of miRNAs and Target Gene Prediction
2.5. Functional Annotation and Enrichment Analysis of Target Genes of DE miRNAs and DE mRNAs
2.6. Construction of Integral miRNA-mRNA Interaction Networks
2.7. Quantitative PCR Validation
3. Results
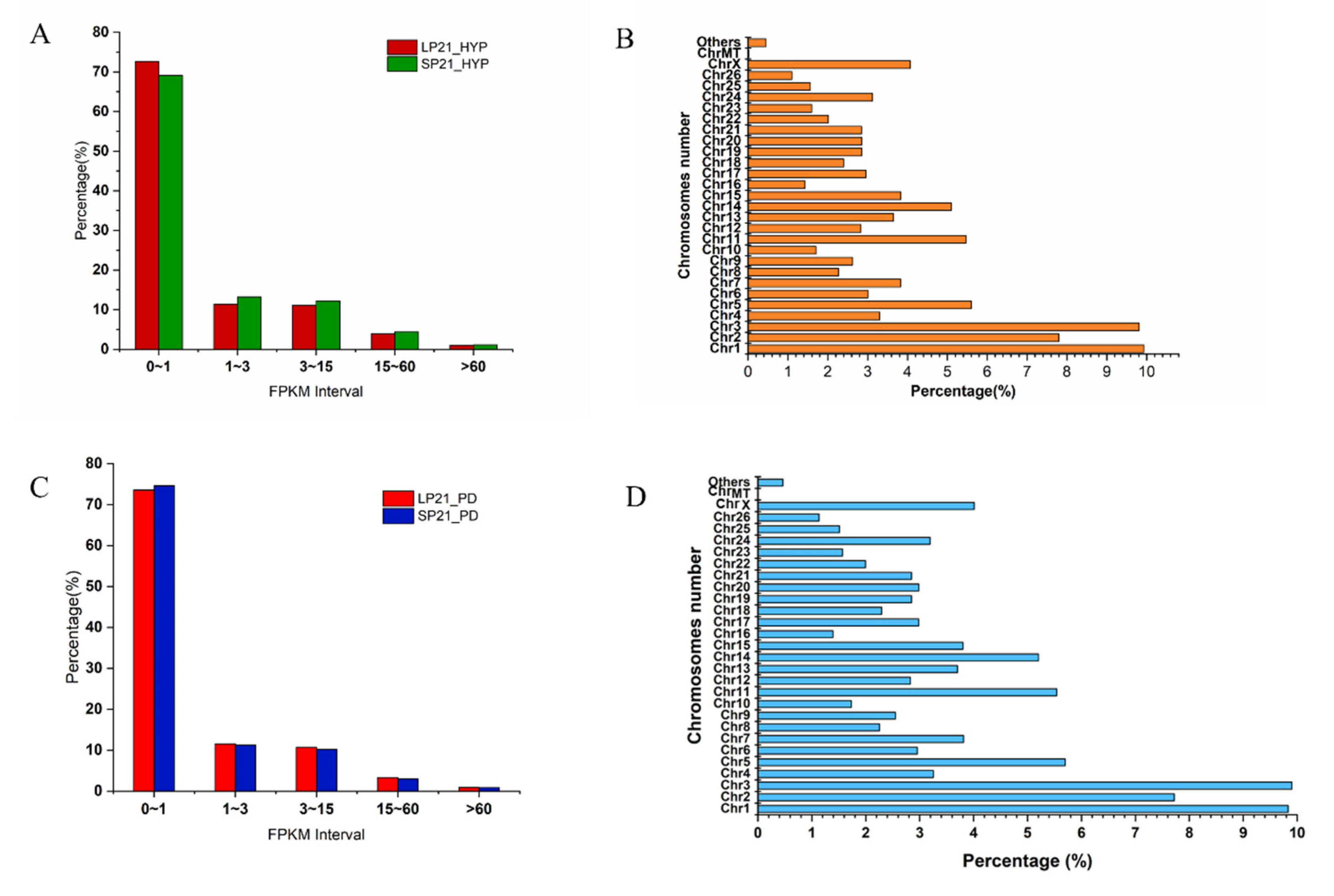
3.1. Summary of Sequencing Data for mRNA and miRNA
3.2. Identification of Differential Expressed Genes and miRNAs
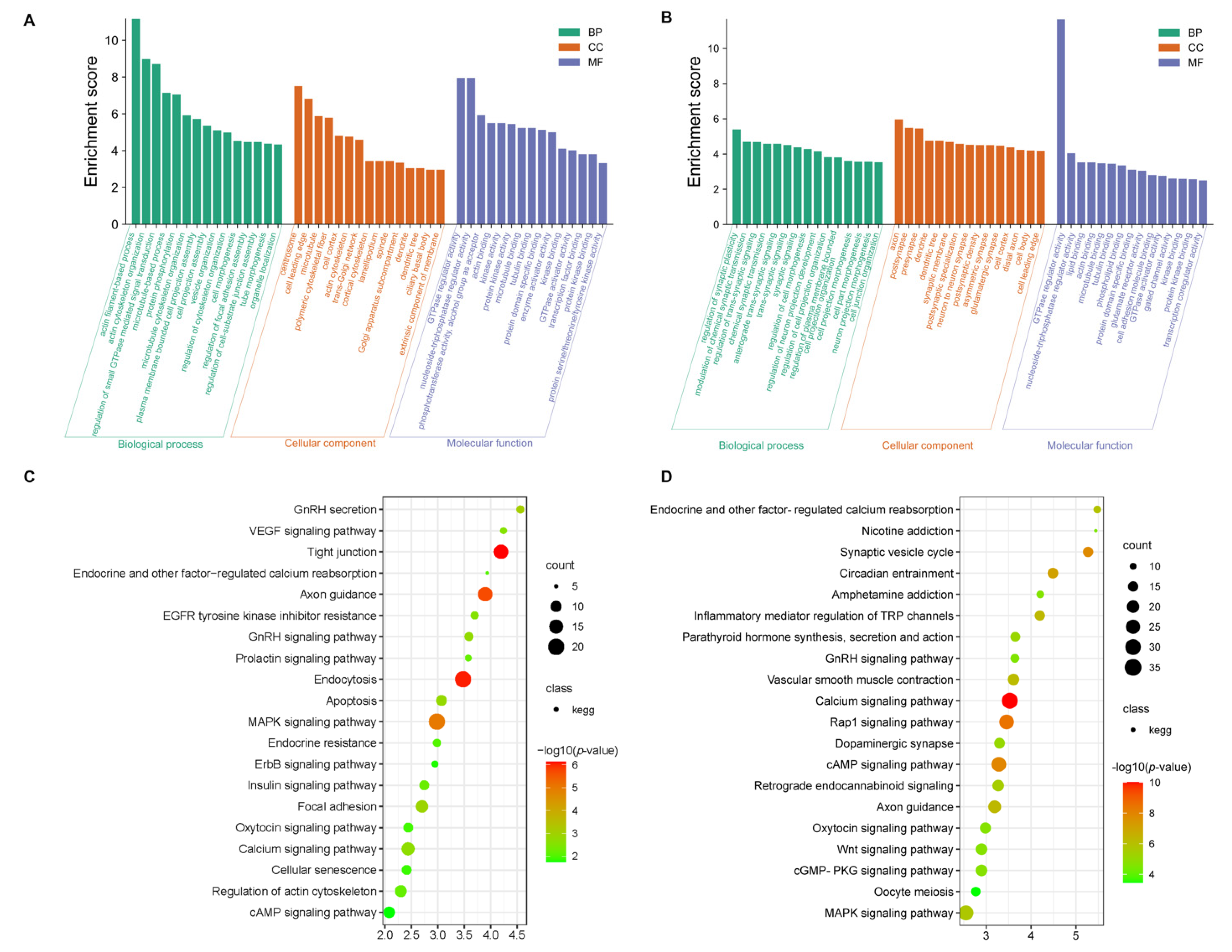
3.3. Functional Enrichment Analysis of the DEGs
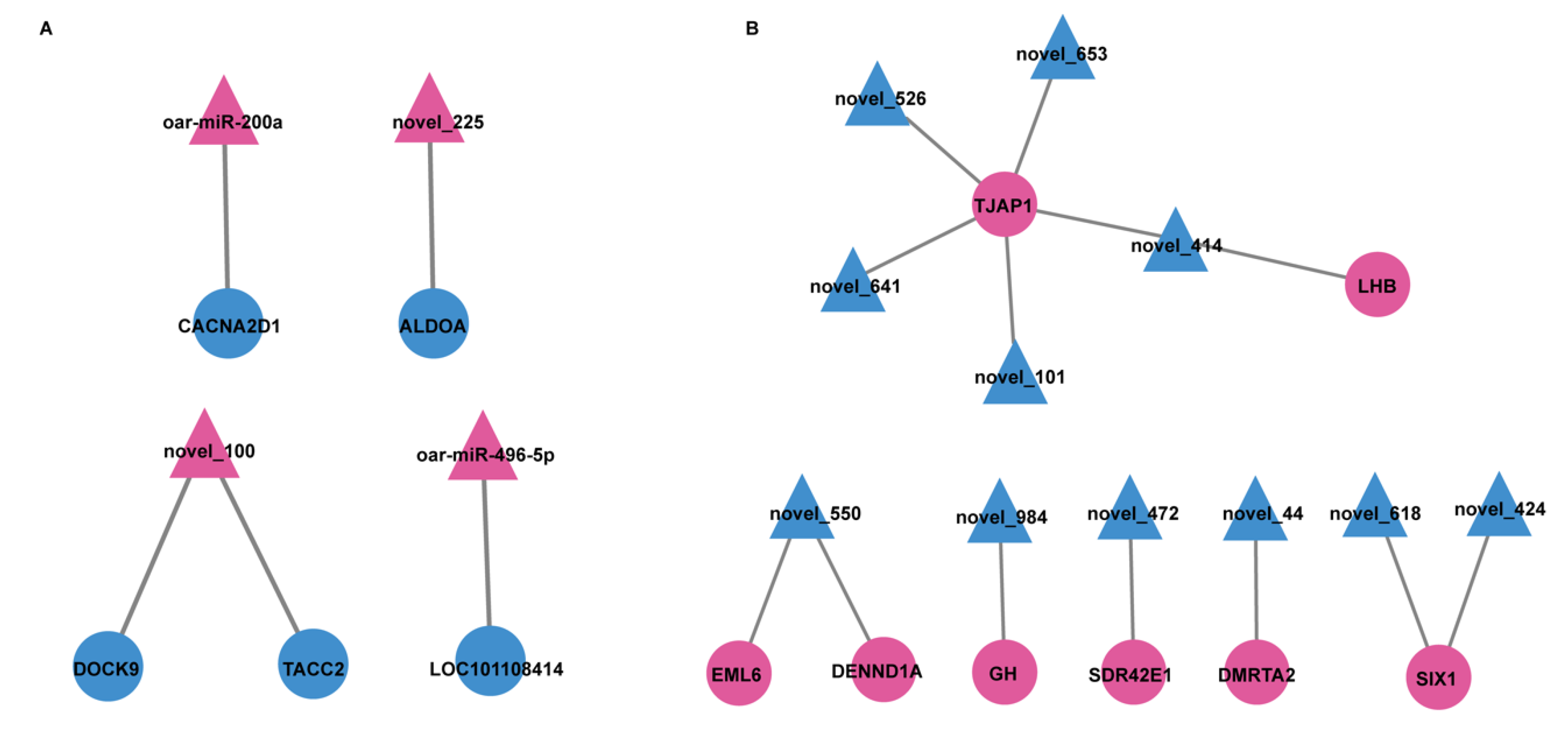
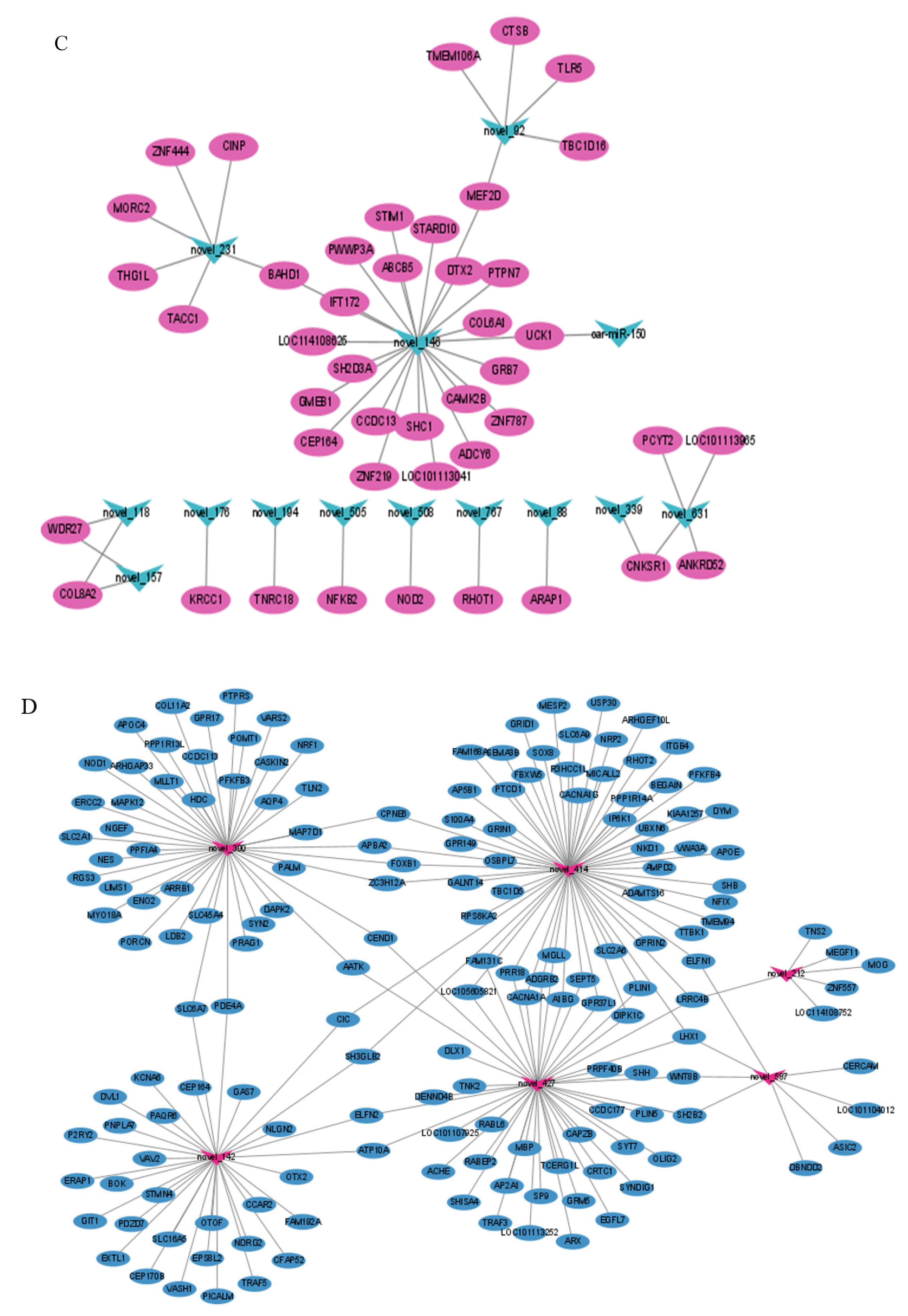
3.4. miRNA-mRNA Interaction Network
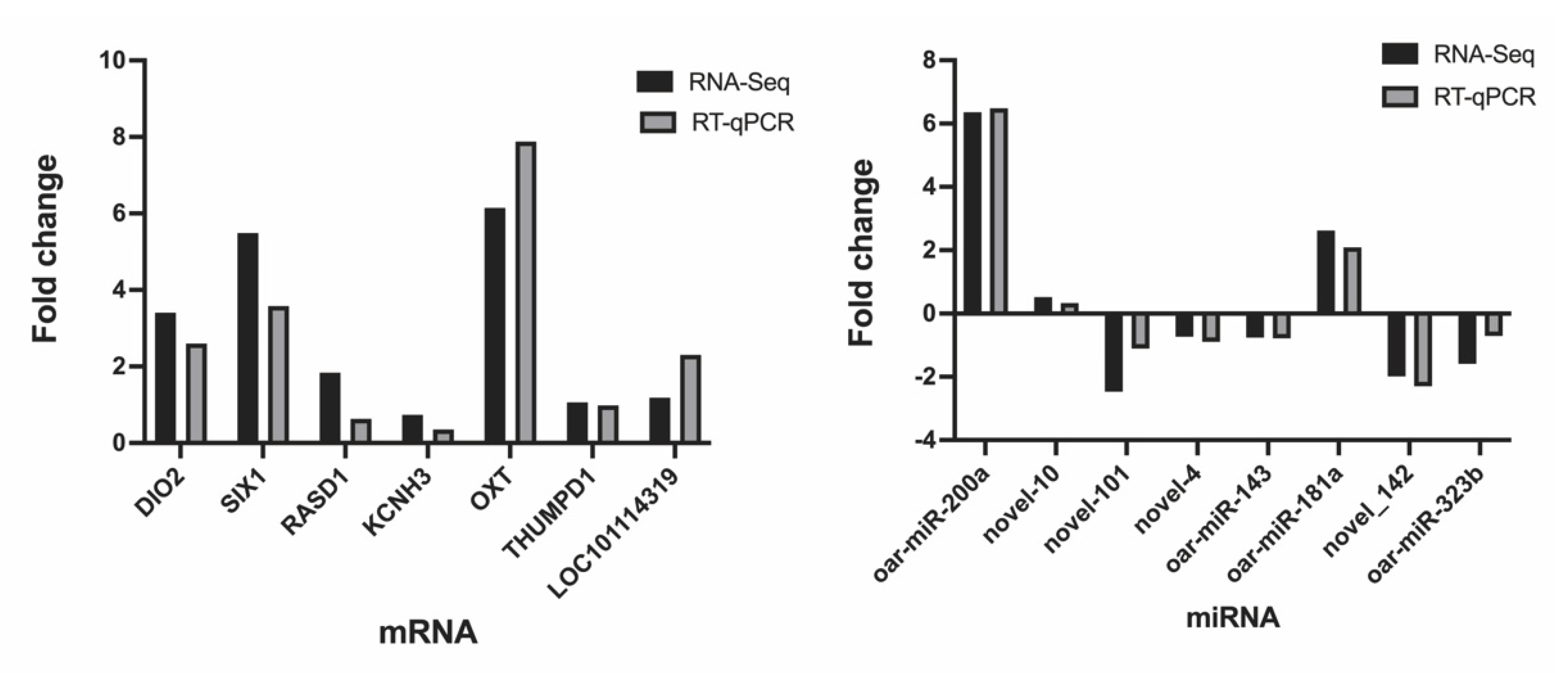
3.5. Data Validation
4. Discussion
4.1. Identification and Analysis of mRNAs and miRNAs Data in SP21− HYP vs. LP21− HYP
4.2. Analysis of miRNA-mRNA Interaction Network in SP21−HPY vs. LP21−HPY
4.3. Identification and Analysis of mRNAs and miRNAs Data in SP21−PD vs. LP21−PD
4.4. Analysis of miRNA-mRNA Interaction Network in SP−PD vs. LP−PD
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Matthews, C.D.; Guerin, M.V.; Deed, J.R. Melatonin and photoperiodic time measurement: Seasonal breeding in the sheep. J. Pineal Res. 1993, 14, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Brenna, A.; Ripperger, J.A.; Saro, G.; Glauser, D.A.; Yang, Z.; Albrecht, U. PER2 mediates CREB-dependent light induction of the clock gene Per1. Sci. Rep. 2021, 11, 21766. [Google Scholar] [CrossRef] [PubMed]
- Pittendrigh, C.S. Circadian surfaces and the diversity of possible roles of circadian organization in photoperiodic induction. Proc. Natl. Acad. Sci. USA 1972, 69, 2734–2737. [Google Scholar] [CrossRef] [PubMed]
- Dulcis, D.; Jamshidi, P.; Leutgeb, S.; Spitzer, N.C. Neurotransmitter switching in the adult brain regulates behavior. Science 2013, 340, 449–453. [Google Scholar] [CrossRef]
- Landgraf, D.; Long, J.E.; Proulx, C.D.; Barandas, R.; Malinow, R.; Welsh, D.K. Genetic Disruption of Circadian Rhythms in the Suprachiasmatic Nucleus Causes Helplessness, Behavioral Despair, and Anxiety-like Behavior in Mice. Biol Psychiatry 2016, 80, 827–835. [Google Scholar] [CrossRef]
- Tian, N.; Copenhagen, D.R. Visual deprivation alters development of synaptic function in inner retina after eye opening. Neuron 2001, 32, 439–449. [Google Scholar] [CrossRef]
- Wang, R.M.; Wu, Y.N.; Liang, H.J.; Lin, L.; Zheng, W.H.; Liu, J.S. Effects of temperature and photoperiod on body mass, energy budget and digestive tract morphology in Pycnonotus sinensis. J. Appl. Ecol. 2016, 27, 1959–1967. [Google Scholar]
- Cerasale, D.J.; Zajac, D.M.; Guglielmo, C.G. Behavioral and physiological effects of photoperiod-induced migratory state and leptin on a migratory bird, Zonotrichia albicollis: I. Anorectic effects of leptin administration. Gen. Comp. Endocrinol. 2011, 174, 276–286. [Google Scholar] [CrossRef]
- Nisembaum, L.G.; Martin, P.; Lecomte, F.; Falcón, J. Melatonin and osmoregulation in fish: A focus on Atlantic salmon Salmo salar smoltification. J. Neuroendocrinol. 2021, 33, e12955. [Google Scholar] [CrossRef]
- Webster, J.R.; Corson, I.D.; Littlejohn, R.P.; Stuart, S.K.; Suttie, J.M. Effects of photoperiod on the cessation of growth during autumn in male red deer and growth hormone and insulin-like growth factor-I secretion. Gen. Comp. Endocrinol. 1999, 113, 464–477. [Google Scholar] [CrossRef]
- Ikegami, K.; Yoshimura, T. Circadian clocks and the measurement of daylength in seasonal reproduction. Mol. Cell. Endocrinol. 2012, 349, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Abrieux, A.; Xue, Y.; Cai, Y.; Lewald, K.M.; Nguyen, H.N.; Zhang, Y.; Chiu, J.C. EYES ABSENT and TIMELESS integrate photoperiodic and temperature cues to regulate seasonal physiology in Drosophila. Proc. Natl. Acad. Sci. USA 2020, 117, 15293–15304. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.T. The role of kisspeptin and gonadotropin inhibitory hormone in the seasonal regulation of reproduction in sheep. Domest. Anim. Endocrinol. 2012, 43, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Harter, C.J.L.; Kavanagh, G.S.; Smith, J.T. The role of kisspeptin neurons in reproduction and metabolism. J. Endocrinol. 2018, 238, R173–R183. [Google Scholar] [CrossRef]
- He, X.; Liu, Q.; Li, X.; Guo, X.; Wang, X.; Hu, W.; Di, R.; Chu, M. Molecular cloning and epigenetic change detection of Kiss1 during seasonal reproduction in Chinese indigenous sheep. Reprod. Fertil. Dev. 2018, 30, 734–743. [Google Scholar] [CrossRef]
- Gao, X.; Ye, J.; Yang, C.; Luo, L.; Liu, Y.; Ding, J.; Zhang, Y.; Ling, Y.; Huang, W.; Zhang, X.; et al. RNA-seq analysis of lncRNA-controlled developmental gene expression during puberty in goat & rat. BMC Genet. 2018, 19, 19. [Google Scholar]
- Hill, J.W.; Elias, C.F. Neuroanatomical Framework of the Metabolic Control of Reproduction. Physiol. Rev. 2018, 98, 2349–2380. [Google Scholar] [CrossRef]
- Yamamura, T.; Hirunagi, K.; Ebihara, S.; Yoshimura, T. Seasonal morphological changes in the neuro-glial interaction between gonadotropin-releasing hormone nerve terminals and glial endfeet in Japanese quail. Endocrinology 2004, 145, 4264–4267. [Google Scholar] [CrossRef]
- Dardente, H.; Hazlerigg, D.G.; Ebling, F.J. Thyroid hormone and seasonal rhythmicity. Front. Endocrinol. 2014, 5, 19. [Google Scholar] [CrossRef]
- Toni, R.; Malaguti, A.; Benfenati, F.; Martini, L. The human hypothalamus: A morpho-functional perspective. J. Endocrinol. Investig. 2004, 27, 73–94. [Google Scholar]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Lu, J.; Xiao, S.Y.; Han, X.Y.; Song, X.T.; Qi, M.Y.; Liu, G.S.; Yang, C.X.; Yao, Y.C. miRNA expression analysis of the sheep follicle during the prerecruitment, dominant, and mature stages of development under FSH stimulation. Theriogenology 2022, 181, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Gu, B.; Liu, H.; Han, Y.; Chen, Y.; Jiang, H. Integrated analysis of miRNA and mRNA expression profiles in 2-, 6-, and 12-month-old Small Tail Han Sheep ovaries reveals that oar-miR-432 downregulates RPS6KA1 expression. Gene 2019, 710, 76–90. [Google Scholar] [CrossRef] [PubMed]
- Xing, K.; Gao, M.; Li, X.; Feng, Y.; Ge, Y.; Qi, X.; Wang, X.; Ni, H.; Guo, Y.; Sheng, X. An integrated analysis of testis miRNA and mRNA transcriptome reveals important functional miRNA-targets in reproduction traits of roosters. Reprod. Biol. 2020, 20, 433–440. [Google Scholar] [CrossRef]
- Reza, A.; Choi, Y.J.; Han, S.G.; Song, H.; Park, C.; Hong, K.; Kim, J.H. Roles of microRNAs in mammalian reproduction: From the commitment of germ cells to peri-implantation embryos. Biol. Rev. Camb. Philos. Soc. 2019, 94, 415–438. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, P.; Li, L.; Che, D.; Li, T.; Li, H.; Li, Q.; Jia, H.; Tao, S.; Hua, J.; et al. miRNA editing landscape reveals miR-34c regulated spermatogenesis through structure and target change in pig and mouse. Biochem. Biophys. Res. Commun. 2018, 502, 486–492. [Google Scholar] [CrossRef]
- He, X.; Tao, L.; Zhong, Y.; Di, R.; Xia, Q.; Wang, X.; Guo, X.; Gan, S.; Zhang, X.; Zhang, J.; et al. Photoperiod induced the pituitary differential regulation of lncRNAs and mRNAs related to reproduction in sheep. PeerJ. 2021, 9, e10953. [Google Scholar] [CrossRef]
- He, X.; Di, R.; Guo, X.; Cao, X.; Zhou, M.; Li, X.; Xia, Q.; Wang, X.; Zhang, J.; Zhang, X.; et al. Transcriptomic changes of photoperiodic response in the hypothalamus were identified in ovariectomized and estradiol-treated sheep. Front. Mol. Biosci. 2022, 9, 848144. [Google Scholar] [CrossRef]
- Liang, C.; Han, M.; Zhou, Z.; Liu, Y.; He, X.; Jiang, Y.; Ouyang, Y.; Hong, Q.; Chu, M. Hypothalamic transcriptome analysis reveals the crucial microRNAs and mRNAs affecting litter size in goats. Front. Vet. Sci. 2021, 8, 747100. [Google Scholar] [CrossRef]
- Greives, T.J.; Mason, A.O.; Scotti, M.A.; Levine, J.; Ketterson, E.D.; Kriegsfeld, L.J.; Demas, G.E. Environmental control of kisspeptin: Implications for seasonal reproduction. Endocrinology 2007, 148, 1158–1166. [Google Scholar] [CrossRef]
- Di, R.; He, J.; Song, S.; Tian, D.; Liu, Q.; Liang, X.; Ma, Q.; Sun, M.; Wang, J.; Zhao, W.; et al. Characterization and comparative profiling of ovarian microRNAs during ovine anestrus and the breeding season. BMC Genom. 2014, 15, 899. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Hu, M.; Zhou, H.; Li, C.; Li, X.; Liu, X.; Zhao, Y.; Zhao, S. Neuronal Signal Transduction-Involved Genes in Pig Hypothalamus Affect Feed Efficiency as Revealed by Transcriptome Analysis. BioMed Res. Int. 2018, 2018, 5862571. [Google Scholar] [CrossRef] [PubMed]
- Legan, S.J.; Peng, X.; Yun, C.; Duncan, M.J. Effect of arousing stimuli on circulating corticosterone and the circadian rhythms of luteinizing hormone (LH) surges and locomotor activity in estradiol-treated ovariectomized (ovx + EB) Syrian hamsters. Horm. Behav. 2015, 72, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Scotti, M.A.; Place, N.J.; Demas, G.E. Short-day increases in aggression are independent of circulating gonadal steroids in female Siberian hamsters (Phodopus sungorus). Horm. Behav. 2007, 52, 183–190. [Google Scholar] [CrossRef]
- Jackson, L.M.; Mytinger, A.; Roberts, E.K.; Lee, T.M.; Foster, D.L.; Padmanabhan, V.; Jansen, H.T. Developmental programming: Postnatal steroids complete prenatal steroid actions to differentially organize the GnRH surge mechanism and reproductive behavior in female sheep. Endocrinology 2013, 154, 1612–1623. [Google Scholar] [CrossRef][Green Version]
- Zhai, M.; Xie, Y.; Liang, H.; Lei, X.; Zhao, Z. Comparative profiling of differentially expressed microRNAs in estrous ovaries of Kazakh sheep in different seasons. Gene 2018, 664, 181–191. [Google Scholar] [CrossRef]
- Zou, X.; Lu, T.; Zhao, Z.; Liu, G.; Lian, Z.; Guo, Y.; Sun, B.; Liu, D.; Li, Y. Comprehensive analysis of mRNAs and miRNAs in the ovarian follicles of uniparous and multiple goats at estrus phase. BMC Genom. 2020, 21, 267. [Google Scholar] [CrossRef]
- Yuan, L.; Feng, F.; Mao, Z.; Huang, J.Z.; Liu, Y.; Li, Y.L.; Jiang, R.X. Regulation mechanism of miR-494-3p on endometrial receptivity in mice via PI3K/AKT/mTOR pathway. Gen. Physiol. Biophys. 2021, 40, 351–363. [Google Scholar] [CrossRef]
- Zhang, X.; Dong, C.; Yang, J.; Li, Y.; Feng, J.; Wang, B.; Zhang, J.; Guo, X. The Roles of the miRNAome and Transcriptome in the Ovine Ovary Reveal Poor Efficiency in Juvenile Superovulation. Animals 2021, 11, 239. [Google Scholar] [CrossRef]
- Huang, D.W.; Wang, J.X.; Liu, Q.Y.; Chu, M.X.; Di, R.; He, J.N.; Cao, G.L.; Fang, L.; Feng, T.; Li, N. Analysis on DNA sequence of TSHB gene and its association with reproductive seasonality in goats. Mol. Biol. Rep. 2013, 40, 1893–1904. [Google Scholar] [CrossRef]
- Chen, S.; Guo, X.; He, X.; Di, R.; Zhang, X.; Zhang, J.; Wang, X.; Chu, M. Transcriptome analysis reveals differentially expressed genes and long non-coding RNAs associated with fecundity in sheep hypothalamus with different FecB genotypes. Front. Cell Dev. Biol. 2021, 9, 633747. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Gaur, U.; Zhu, Q.; Chen, B.; Xu, Z.; Zhao, X.; Yang, M.; Li, D. Expressed microRNA associated with high rate of egg production in chicken ovarian follicles. Anim. Genet. 2017, 48, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Tarnopolsky, M.A. Myopathies Related to Glycogen Metabolism Disorders. Neurotherapeutics 2018, 15, 915–927. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, Y.; Zhou, Z.; He, X.; Tao, L.; Jiang, Y.; Lan, R.; Hong, Q.; Chu, M. chi-miR-324-3p Regulates Goat Granulosa Cell Proliferation by Targeting DENND1A. Front. Vet. Sci. 2021, 8, 732440. [Google Scholar] [CrossRef]
- 4Shi, J.; Gao, Q.; Cao, Y.; Fu, J. Dennd1a, a susceptibility gene for polycystic ovary syndrome, is essential for mouse embryogenesis. Dev. Dyn. 2019, 248, 351–362. [Google Scholar]
- Zhu, Y.N.; Zhang, Y.T.; Liu, Q.; Shen, S.M.; Zou, X.; Cao, Y.X.; Wang, W.J.; Yi, L.; Gao, Q.; Yang, W.D.; et al. Association analysis between the tag single nucleotide polymorphisms of DENND1A and the risk of polycystic ovary syndrome in Chinese Han women. BMC Med. Genet. 2020, 21, 14. [Google Scholar] [CrossRef]
- Zheng, J.; Deng, T.; Jiang, E.; Li, J.; Wijayanti, D.; Wang, Y.; Ding, X.; Lan, X. Genetic variations of bovine PCOS-related DENND1A gene identified in GWAS significantly affect female reproductive traits. Gene 2021, 802, 145867. [Google Scholar] [CrossRef]
- Yin, H.; Hou, X.; Zhang, T.; Shi, L.; Su, Y.Q. Participation of EML6 in the regulation of oocyte meiotic progression in mice. J. Biomed. Res. 2019, 34, 44–53. [Google Scholar]
- Blumenfeld, Z.; Amit, T. The role of growth hormone (GH), GH-receptor and GH-binding protein in reproduction and ovulation induction. J. Pediatric Endocrinol. Metab. 1996, 9, 145–162. [Google Scholar]
- Hull, K.L.; Harvey, S. GH as a co-gonadotropin: The relevance of correlative changes in GH secretion and reproductive state. J. Endocrinol. 2002, 172, 1–19. [Google Scholar] [CrossRef]
- Hull, K.L.; Harvey, S. Growth hormone: Roles in female reproduction. J. Endocrinol. 2001, 168, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Valentinis, B.; Baserga, R. IGF-I receptor signalling in transformation and differentiation. Mol. Pathol. 2001, 54, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Gomez-León, V.E.; Ginther, O.J.; Guimarães, J.D.; Wiltbank, M.C. Hormonal mechanisms regulating follicular wave dynamics II: Progesterone decreases diameter at follicle selection regardless of whether circulating FSH or LH are decreased or elevated. Theriogenology 2020, 143, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Dupré, S.M.; Miedzinska, K.; Duval, C.V.; Yu, L.; Goodman, R.L.; Lincoln, G.A.; Davis, J.R.; McNeilly, A.S.; Burt, D.D.; Loudon, A.S. Identification of Eya3 and TAC1 as long-day signals in the sheep pituitary. Curr. Biol. 2010, 20, 829–835. [Google Scholar] [CrossRef] [PubMed]
- García, I.A.; Torres Demichelis, V.; Viale, D.L.; Di Giusto, P.; Ezhova, Y.; Polishchuk, R.S.; Sampieri, L.; Martinez, H.; Sztul, E.; Alvarez, C. CREB3L1-mediated functional and structural adaptation of the secretory pathway in hormone-stimulated thyroid cells. J. Cell Sci. 2017, 130, 4155–4167. [Google Scholar] [CrossRef] [PubMed]
- Neumann, I.D. Brain oxytocin: A key regulator of emotional and social behaviours in both females and males. J. Neuroendocrinol. 2008, 20, 858–865. [Google Scholar] [CrossRef]
- Jurek, B.; Neumann, I.D. The Oxytocin Receptor: From Intracellular Signaling to Behavior. Physiol. Rev. 2018, 98, 1805–1908. [Google Scholar] [CrossRef]
- Neumann, I.D.; Slattery, D.A. Oxytocin in General Anxiety and Social Fear: A Translational Approach. Biol. Psychiatry 2016, 79, 213–221. [Google Scholar] [CrossRef]
- Olazábal, D.E. Role of oxytocin in parental behaviour. J. Neuroendocrinol. 2018, 30, e12594. [Google Scholar] [CrossRef]
- Yang, H.; Fu, L.; Luo, Q.; Li, L.; Zheng, F.; Liu, X.; Zhao, Z.; Wang, Z.; Xu, H. Comparative analysis and identification of differentially expressed microRNAs in the hypothalamus of Kazakh sheep exposed to different photoperiod conditions. Biochem. Biokhimiia 2021, 86, 1315–1325. [Google Scholar] [CrossRef]
- Korf, H.W.; Schomerus, C.; Maronde, E.; Stehle, J.H. Signal transduction molecules in the rat pineal organ: Ca2+, pCREB, and ICER. Naturwissenschaften 1996, 83, 535–543. [Google Scholar] [CrossRef] [PubMed]
- Kroeber, S.; Meissl, H.; Maronde, E.; Korf, H.W. Analyses of signal transduction cascades reveal an essential role of calcium ions for regulation of melatonin biosynthesis in the light-sensitive pineal organ of the rainbow trout (Oncorhynchus mykiss). J. Neurochem. 2000, 74, 2478–2489. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Tang, J.; Di, R.; Liu, Q.; Wang, X.; Gan, S.; Zhang, X.; Zhang, J.; Chu, M.; Hu, W. Integrated hypothalamic transcriptome profiling reveals the reproductive roles of mRNAs and miRNAs in sheep. Front. Genet. 2019, 10, 1296. [Google Scholar] [CrossRef] [PubMed]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Müller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [CrossRef]
- Hertel, J.; Bartschat, S.; Wintsche, A.; Otto, C.; Stadler, P.F. Evolution of the let-7 microRNA family. RNA Biol. 2012, 9, 231–241. [Google Scholar] [CrossRef]
- Zhou, R.; Miao, Y.; Li, Y.; Li, X.; Xi, J.; Zhang, Z. MicroRNA-150 promote apoptosis of ovine ovarian granulosa cells by targeting STAR gene. Theriogenology 2019, 127, 66–71. [Google Scholar] [CrossRef]
- Kool, M.J.; Proietti Onori, M.; Borgesius, N.Z.; van de Bree, J.E.; Elgersma-Hooisma, M.; Nio, E.; Bezstarosti, K.; Buitendijk, G.H.S.; Aghadavoud Jolfaei, M.; Demmers, J.A.A.; et al. CAMK2-Dependent Signaling in Neurons Is Essential for Survival. J. Neurosci. 2019, 39, 5424–5439. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.; Feng, C.; Zhao, Y.X.; Liu, X.Y. Camk2b protects neurons from homocysteine-induced apoptosis with the involvement of HIF-1α signal pathway. Int. J. Clin. Exp. Med. 2014, 7, 1659–1668. [Google Scholar]
- Kutsche, L.K.; Gysi, D.M.; Fallmann, J.; Lenk, K.; Petri, R.; Swiersy, A.; Klapper, S.D.; Pircs, K.; Khattak, S.; Stadler, P.F.; et al. Combined experimental and system-level analyses reveal the complex regulatory network of miR-124 during human neurogenesis. Cell Syst. 2018, 7, 438–452.e438. [Google Scholar] [CrossRef]
- Jääskeläinen, M.; Nieminen, A.; Pökkylä, R.M.; Kauppinen, M.; Liakka, A.; Heikinheimo, M.; Vaskivuo, T.E.; Klefström, J.; Tapanainen, J.S. Regulation of cell death in human fetal and adult ovaries—Role of Bok and Bcl-X(L). Mol. Cell. Endocrinol. 2010, 330, 17–24. [Google Scholar] [CrossRef][Green Version]
- Govindaraj, V.; Rao, A.J. Comparative proteomic analysis of primordial follicles from ovaries of immature and aged rats. Syst. Biol. Reprod. Med. 2015, 61, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Bedont, J.L.; LeGates, T.A.; Buhr, E.; Bathini, A.; Ling, J.P.; Bell, B.; Wu, M.N.; Wong, P.C.; Van Gelder, R.N.; Mongrain, V.; et al. An LHX1-regulated transcriptional network controls sleep/wake coupling and thermal resistance of the central circadian clockworks. Curr. Biol. 2017, 27, 128–136. [Google Scholar] [CrossRef]
- Hanon, E.A.; Lincoln, G.A.; Fustin, J.M.; Dardente, H.; Masson-Pévet, M.; Morgan, P.J.; Hazlerigg, D.G. Ancestral TSH mechanism signals summer in a photoperiodic mammal. Curr. Biol. 2008, 18, 1147–1152. [Google Scholar] [CrossRef] [PubMed]
- Radwańska, P.; Kosior-Korzecka, U. Relationships between leptin, KiSS-1/GPR54 expression and TSH secretion from pituitary cells of pubertal ewes in vitro. Res. Vet. Sci. 2016, 105, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Caraty, A.; Lomet, D.; Sébert, M.E.; Guillaume, D.; Beltramo, M.; Evans, N.P. Gonadotrophin-releasing hormone release into the hypophyseal portal blood of the ewe mirrors both pulsatile and continuous intravenous infusion of kisspeptin: An insight into kisspeptin’s mechanism of action. J. Neuroendocrinol. 2013, 25, 537–546. [Google Scholar] [CrossRef]




| Gene | Primer Sequences (5′-3′) | Product Size (bp) | Tm (℃) |
|---|---|---|---|
| KCNH3 | F:CGCAGAACACCTTCCTGGACAC | 122 | 60 |
| R:GCAGAAGCCATCAGAGCAGTAGAC | |||
| SIX1 | F:CATCGTTCGGCTTCACACAGGAG | 142 | 60 |
| R:GCCTTGAGCACGCTTTCATTCTTG | |||
| DIO2 | F:CCAGAGCTGTTCCAAGGCAAGTC | 109 | 60 |
| R:CTCCAGTGCTGCTGTCCAAGATG | |||
| RASD1 | F: GGAGACGTGTTCATCCTGGTGTTC | 86 | 60 |
| R: GTGTCGAGAATCTGCCGCTTGAG | |||
| OXT | F:GCCTTCTCCCAGCACTGAGA | 81 | 60 |
| R: TCCTGGGGATGATTACAGAGGGA | |||
| THUMPD1 | F:TCAACGAATACGGCGACGACATG | 108 | 60 |
| R:TCTTCAAGGCAGCTTCCACATCATC | |||
| LOC101114319 | F:GTACACCTTGGTCTTGACAGATCCG | 121 | 60 |
| R: GAGAACCGTGCCACTGCTGATG | |||
| RPL19 | F:ATCGCCAATGCCAACTC | 154 | 60 |
| R: CCTTTCGCTTACCTATACC |
| The Name of the Primer | Primer Sequence | Tm (℃) |
|---|---|---|
| oar-miR-200a | F:GCTGCAACACTGTCTGGTAACGAT | 60 |
| novel_10 | F:CGCTTCACAGTGGCTAAGTTCTGC | 60 |
| novel_101 | F:AGCTCTGGGTCTGTGGGGA | 60 |
| novel_4 | F:CCGCGTCTTTGGTTATCTAGCTGTATG | 60 |
| oar-miR-143 | F:CGCTGAGATGAAGCACTGTAGCTC | 60 |
| novel_142 | F:CCACCTCCCCTGCAAACG | 60 |
| oar-miR-181a | F:TGCGAACATTCAACGCTGTCGGTGAG | 60 |
| oar-miR-323b | F:AGGCCTCCCAATACACGGTCGATCTC | 60 |
| U6 | F:CAAGGATGACACGCAAATTCG | 60 |
| Group | mRNA | log2FoldChange | q-Value | Up/Down |
|---|---|---|---|---|
| SP21−HYP vs. LP21−HYP | COMMD5 | −7.48 | 3.93 × 10−72 | down |
| EHBP1 | −6.30 | 1.54 × 10−51 | down | |
| LOC114108752 | 3.92 | 2.39 × 10−48 | up | |
| LOC114116052 | 5.90 | 8.05 × 10−45 | up | |
| MAPK6 | −14.07 | 5.16 × 10−24 | down | |
| MAX | 1.93 | 2.30 × 10−23 | up | |
| ASPSCR1 | 6.50 | 2.49 × 10−23 | up | |
| STK38 | 5.08- | 2.49 × 10−23 | up | |
| SV2A | −9.16 | 3.40 × 10−23 | down | |
| RBM42 | 13.67 | 3.76 × 10−21 | up | |
| SP21−PD vs. LP21−PD | LOC114116052 | 5.43 | 4.39 × 10−80 | up |
| LOC114108752 | 3.84 | 1.15 × 10−43 | up | |
| PRUNE2 | 9.87 | 6.13 × 10−38 | up | |
| AKAP9 | 15.95 | 9.79 × 10−36 | up | |
| GGNBP2 | 6.59 | 1.94 × 10−26 | up | |
| BROX | −13.02 | 2.59 × 10−21 | down | |
| RAPGEF6 | 13.83 | 2.03 × 10−18 | up | |
| PTEN | −4.09- | 6.83 × 10−17 | down | |
| PLEC | 13.59 | 6.83 × 10−17 | up | |
| CEP112 | −8.20 | 1.14 × 10−16 | down |
| Group | miRNA | log2FoldChange | p-Value | Up/Down |
|---|---|---|---|---|
| SP21−HYP vs. LP21−HYP | novel_154 | 7.34 | 1.17 × 10−5 | up |
| oar-miR-3956-5p | −2.32 | 1.33 × 10−5 | down | |
| oar-miR-544-5p | 3.86 | 3.61 × 10−5 | up | |
| novel_156 | 4.64 | 6.50 × 10−5 | up | |
| oar-miR-3956-3p | −3.68 | 1.61 × 10−4 | down | |
| oar-miR-376e-3p | 4.85 | 1.63 × 10−4 | up | |
| oar-miR-376b-3p | 5.55 | 1.70 × 10−4 | up | |
| oar-miR-374a | 3.85 | 1.77 × 10−4 | up | |
| novel_234 | 3.86 | 3.66 × 10−4 | up | |
| novel_199 | 5.67 | 6.00 × 10−4 | up | |
| SP21−PD vs. LP21−PD | novel_5 | 8.83 | 1.02 × 10−9 | up |
| novel_200 | 5.59 | 3.06 × 10−9 | up | |
| novel_62 | 9.67 | 1.85 × 10−6 | up | |
| novel_652 | 5.47 | 5.10 × 10−6 | up | |
| novel_172 | 6.38 | 7.94 × 10−6 | up | |
| novel_123 | 4.85 | 2.04 × 10−5 | up | |
| novel_203 | 5.69 | 4.00 × 10−5 | up | |
| novel_505 | 4.12 | 1.18 × 10−4 | up | |
| novel_357 | 6.41 | 1.78 × 10−4 | up | |
| novel_83 | 3.28 | 5.61 × 10−4 | up |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Q.; Di, R.; Ren, C.; He, X.; Wang, X.; Xia, Q.; Chu, M.; Zhang, Z. Screening of Differentially Expressed Genes and miRNAs in Hypothalamus and Pituitary Gland of Sheep under Different Photoperiods. Genes 2022, 13, 1091. https://doi.org/10.3390/genes13061091
Liu Q, Di R, Ren C, He X, Wang X, Xia Q, Chu M, Zhang Z. Screening of Differentially Expressed Genes and miRNAs in Hypothalamus and Pituitary Gland of Sheep under Different Photoperiods. Genes. 2022; 13(6):1091. https://doi.org/10.3390/genes13061091
Chicago/Turabian StyleLiu, Qingqing, Ran Di, Chunhuan Ren, Xiaoyun He, Xiangyu Wang, Qing Xia, Mingxing Chu, and Zijun Zhang. 2022. "Screening of Differentially Expressed Genes and miRNAs in Hypothalamus and Pituitary Gland of Sheep under Different Photoperiods" Genes 13, no. 6: 1091. https://doi.org/10.3390/genes13061091
APA StyleLiu, Q., Di, R., Ren, C., He, X., Wang, X., Xia, Q., Chu, M., & Zhang, Z. (2022). Screening of Differentially Expressed Genes and miRNAs in Hypothalamus and Pituitary Gland of Sheep under Different Photoperiods. Genes, 13(6), 1091. https://doi.org/10.3390/genes13061091








