Regulation of Non-Coding RNA in the Growth and Development of Skeletal Muscle in Domestic Chickens
Abstract
:1. Introduction
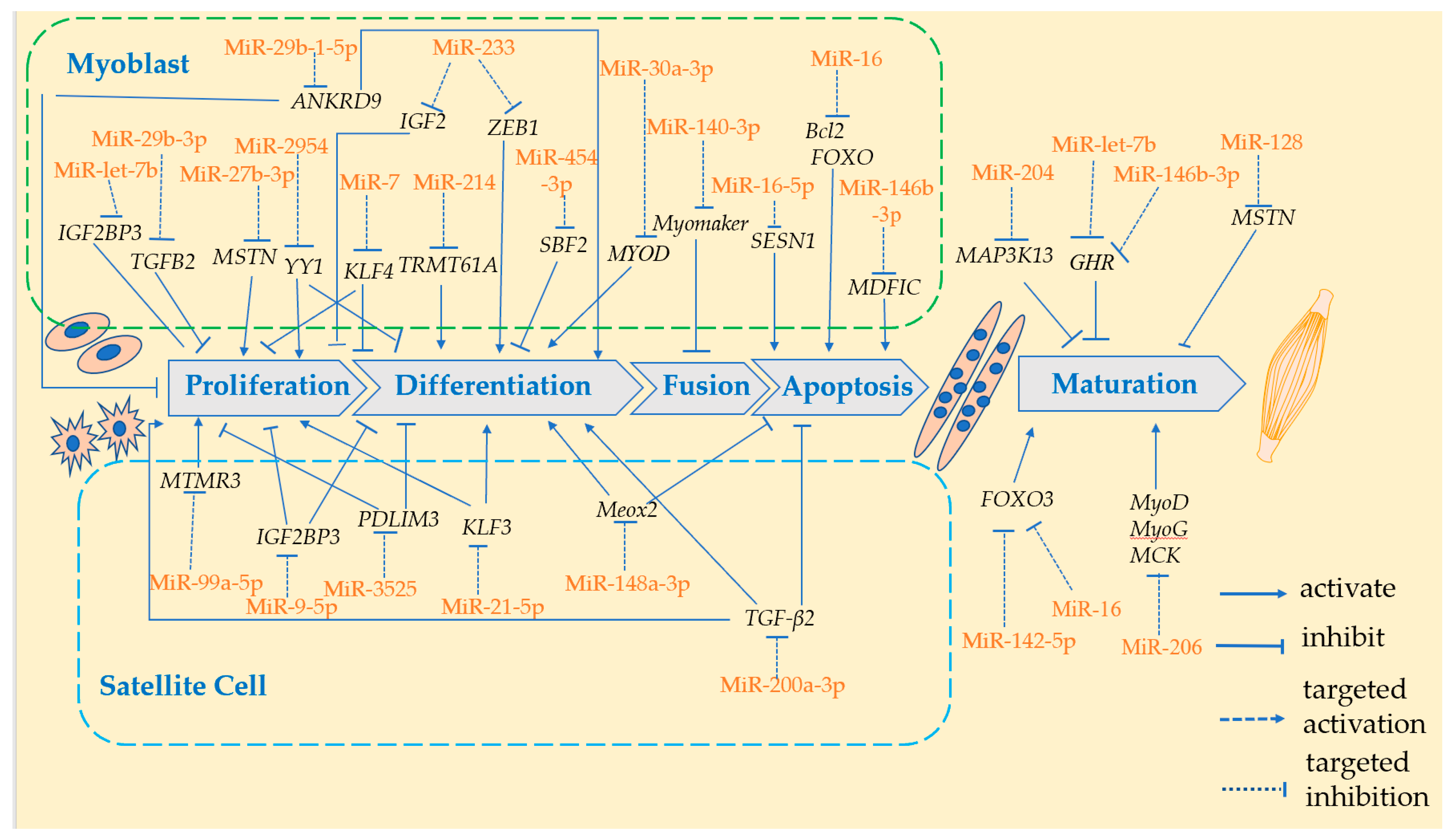
2. MiRNA Modulates Skeletal Muscle in Domestic Chicken
2.1. MiRNA Modulates Skeletal Muscle through Regulating Hormone Levels
2.2. MiRNA Modulates Skeletal Muscle through Regulating Myoblasts
2.2.1. Proliferation and Differentiation of Myoblasts
2.2.2. Fusion of Myoblasts
2.2.3. Apoptosis of Myoblasts
2.3. MiRNA Modulates Skeletal Muscle through Regulating Satellite Cells
2.3.1. Proliferation and Differentiation of Satellite Cells
2.3.2. Apoptosis of Satellite Cells
2.4. MiRNA Modulates Skeletal Muscle through Specificating and Maintaining Muscle Fiber Type
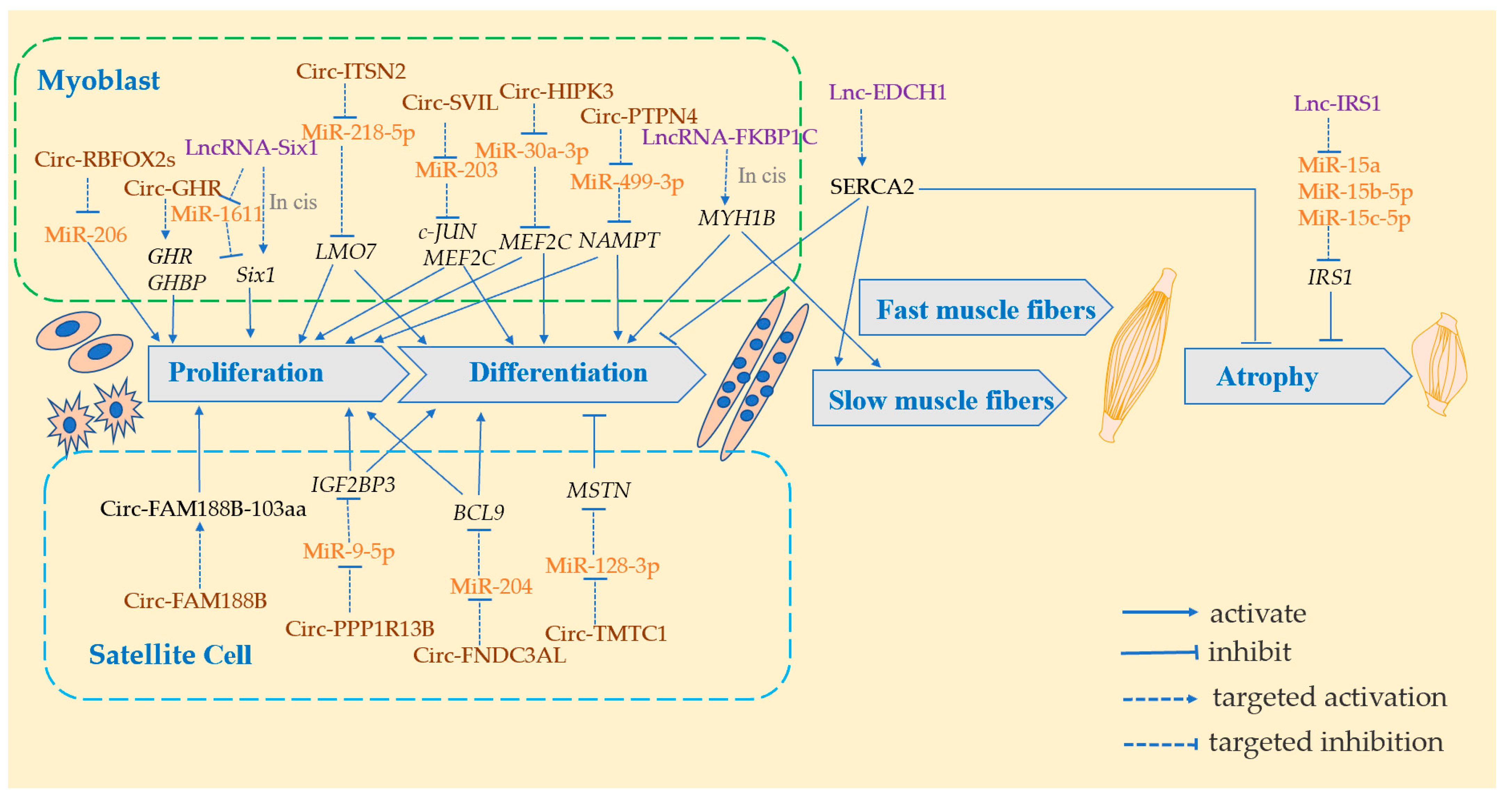
3. LncRNA Modulates Skeletal Muscle in Domestic Chicken
3.1. LncRNA Modulates Skeletal Muscle through Sponging miRNAs on Myoblasts
3.2. LncRNA Modulates Skeletal Muscle through Regulating Gene Expression in Cis or Trans
4. CircRNA Modulates Skeletal Muscle in Domestic Chickens
4.1. CircRNA Modulates Skeletal Muscle through Regulating Parental Genes
4.2. CircRNA Modulates Skeletal Muscle through Sponging miRNAs on Myoblasts
4.3. CircRNA Modulates Skeletal Muscle through Sponging miRNAs on Satellite Cells
4.4. CircRNA Modulates Skeletal Muscle through Translating Directly into Protein
5. Prospect
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, Y.; Zhang, M.; Shan, Y.; Ji, G.; Ju, X.; Tu, Y.; Sheng, Z.; Xie, J.; Zou, J.; Shu, J. miRNA-mRNA network regulation in the skeletal muscle fiber phenotype of chickens revealed by integrated analysis of miRNAome and transcriptome. Sci. Rep. 2020, 10, 10619. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Zhang, G.; Wu, P.; Duan, L.; Li, G.; Liu, Q.; Wang, J. Dissection of Myogenic Differentiation Signatures in Chickens by RNA-Seq Analysis. Genes (Basel) 2018, 9, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braun, T.; Gautel, M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat. Rev. Mol. Cell Biol. 2011, 12, 349–361. [Google Scholar] [CrossRef]
- Buckingham, M. Gene regulatory networks and cell lineages that underlie the formation of skeletal muscle. Proc. Natl. Acad. Sci. USA 2017, 114, 5830–5837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ran, J.; Li, J.; Yin, L.; Zhang, D.; Yu, C.; Du, H.; Jiang, X.; Yang, C.; Liu, Y. Comparative Analysis of Skeletal Muscle DNA Methylation and Transcriptome of the Chicken Embryo at Different Developmental Stages. Front Physiol. 2021, 12, 697121. [Google Scholar] [CrossRef]
- Scaal, M.; Marcelle, C. Chick muscle development. Int. J. Dev. Biol. 2018, 62, 127–136. [Google Scholar] [CrossRef] [Green Version]
- Relaix, F.; Zammit, P.S. Satellite cells are essential for skeletal muscle regeneration: The cell on the edge returns centre stage. Development 2012, 139, 2845–2856. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Li, F.; Hu, X.; Cao, D.; Liu, W.; Han, H.; Zhou, Y.; Lei, Q. Deciphering the miRNA transcriptome of breast muscle from the embryonic to post-hatching periods in chickens. BMC Genom. 2021, 22, 64. [Google Scholar] [CrossRef]
- Panni, S.; Lovering, R.C.; Porras, P.; Orchard, S. Non-coding RNA regulatory networks. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194417. [Google Scholar] [CrossRef]
- Chen, R.; Lei, S.; Jiang, T.; Zeng, J.; Zhou, S.; She, Y. Roles of lncRNAs and circRNAs in regulating skeletal muscle development. Acta Physiol. (Oxf.) 2020, 228, e13356. [Google Scholar] [CrossRef]
- Li, X.; Lian, L.; Zhang, D.; Qu, L.; Yang, N. gga-miR-26a targets NEK6 and suppresses Marek’s disease lymphoma cell proliferation. Poult. Sci. 2014, 93, 1097–1105. [Google Scholar] [CrossRef] [PubMed]
- Horak, M.; Novak, J.; Bienertova-Vasku, J. Muscle-specific microRNAs in skeletal muscle development. Dev. Biol. 2016, 410, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Vasudevan, S.; Tong, Y.; Steitz, J.A. Switching from repression to activation: MicroRNAs can up-regulate translation. Science 2007, 318, 1931–1934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orom, U.A.; Nielsen, F.C.; Lund, A.H. MicroRNA-10a binds the 5’UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 2008, 30, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Krek, A.; Grun, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; Macmenamin, P.; Da, P.I.; Gunsalus, K.C.; Stof-fel, M.; et al. Combinatorial microRNA target predictions. Nat. Genet. 2005, 37, 495–500. [Google Scholar] [CrossRef]
- Oclon, E.; Hrabia, A. miRNA expression profile in chicken ovarian follicles throughout development and miRNA-mediated MMP expression. Theriogenology 2021, 160, 116–127. [Google Scholar] [CrossRef]
- Xu, L.; Guo, Q.; Chang, G.; Qiu, L.; Liu, X.; Bi, Y.; Zhang, Y.; Wang, H.; Lu, W.; Ren, L.; et al. Discovery of mi-croRNAs during early spermatogenesis in chicken. PLoS ONE 2017, 12, e177098. [Google Scholar] [CrossRef]
- Sun, G.; Li, F.; Ma, X.; Sun, J.; Jiang, R.; Tian, Y.; Han, R.; Li, G.; Wang, Y.; Li, Z.; et al. gga-miRNA-18b-3p Inhibits Intramuscular Adipocytes Differentiation in Chicken by Targeting the ACOT13 Gene. Cells 2019, 8, 556. [Google Scholar] [CrossRef] [Green Version]
- Dong, X.; Cheng, Y.; Qiao, L.; Wang, X.; Zeng, C.; Feng, Y. Male-Biased gga-miR-2954 Regulates Myoblast Prolifera-tion and Differentiation of Chicken Embryos by Targeting YY1. Genes (Basel) 2021, 12, 1325. [Google Scholar] [CrossRef]
- Hong, Y.; Truong, A.D.; Lee, J.; Vu, T.H.; Lee, S.; Song, K.D.; Lillehoj, H.S.; Hong, Y.H. Exosomal miRNA profiling from H5N1 avian influenza virus-infected chickens. Vet. Res. 2021, 52, 36. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Yao, X.; Lin, Y.; Zhang, D.; Cui, R.; Zhang, X. Interactive functions of microRNAs in the miR-23a-27a-24-2 cluster and the potential for targeted therapy in cancer. J. Cell. Physiol. 2020, 235, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010, 466, 835–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; Luo, W.; Abdalla, B.A.; Ouyang, H.; Yu, J.; Hu, F.; Nie, Q.; Zhang, X. miRNA-223 upregulated by MYOD inhibits myoblast proliferation by repressing IGF2 and facilitates myoblast differentiation by inhibiting ZEB1. Cell Death Dis. 2017, 8, e3094. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Qiu, M.; Du, H.; Li, Q.; Gan, W.; Xiong, X.; Yu, C.; Peng, H.; Xia, B.; Song, X.; et al. Small RNA sequencing of pectoral muscle tissue reveals microRNA-mediated gene modulation in chicken muscle growth. J. Anim. Physiol. Anim. Nutr. (Berl.) 2020, 104, 867–875. [Google Scholar] [CrossRef]
- Li, Z.; Abdalla, B.A.; Zheng, M.; He, X.; Cai, B.; Han, P.; Ouyang, H.; Chen, B.; Nie, Q.; Zhang, X. Systematic transcriptome-wide analysis of mRNA-miRNA interactions reveals the involvement of miR-142-5p and its target (FOXO3) in skeletal muscle growth in chickens. Mol. Genet. Genom. 2018, 293, 69–80. [Google Scholar] [CrossRef]
- Yin, H.; He, H.; Shen, X.; Zhao, J.; Cao, X.; Han, S.; Cui, C.; Chen, Y.; Wei, Y.; Xia, L.; et al. miR-9-5p Inhibits Skeletal Muscle Satellite Cell Proliferation and Differentiation by Targeting IGF2BP3 through the IGF2-PI3K/Akt Signaling Pathway. Int. J. Mol. Sci. 2020, 21, 1655. [Google Scholar] [CrossRef] [Green Version]
- Yin, H.; He, H.; Shen, X.; Tang, S.; Zhao, J.; Cao, X.; Han, S.; Cui, C.; Chen, Y.; Wei, Y.; et al. MicroRNA Profiling Reveals an Abundant miR-200a-3p Promotes Skeletal Muscle Satellite Cell Development by Targeting TGF-beta2 and Regulating the TGFbeta2/SMAD Signaling Pathway. Int. J. Mol. Sci. 2020, 21, 3274. [Google Scholar] [CrossRef]
- Hicks, J.A.; Tembhurne, P.; Liu, H.C. MicroRNA expression in chicken embryos. Poult. Sci. 2008, 87, 2335–2343. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Y.; Jin, W.; Fu, S.; Li, D.; Zhang, Y.; Sun, G.; Jiang, R.; Han, R.; Li, Z.; et al. Analyses of MicroRNA and mRNA Expression Profiles Reveal the Crucial Interaction Networks and Pathways for Regulation of Chicken Breast Muscle Development. Front Genet. 2019, 10, 197. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.G.; Yu, J.F.; Zhang, Y.; Gong, D.Q.; Gu, Z.L. Identification and characterization of microRNA from chicken adipose tissue and skeletal muscle. Poult. Sci. 2012, 91, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Li, H.; Mu, H.; Luo, W.; Li, Y.; Jia, X.; Wang, S.; Jia, X.; Nie, Q.; Li, Y.; et al. Let-7b regulates the expression of the growth hormone receptor gene in deletion-type dwarf chickens. BMC Genom. 2012, 13, 306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouyang, H.; He, X.; Li, G.; Xu, H.; Jia, X.; Nie, Q.; Zhang, X. Deep Sequencing Analysis of miRNA Expression in Breast Muscle of Fast-Growing and Slow-Growing Broilers. Int. J. Mol. Sci. 2015, 16, 16242–16262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, W.; Lin, S.; Li, G.; Nie, Q.; Zhang, X. Integrative Analyses of miRNA-mRNA Interactions Reveal let-7b, miR-128 and MAPK Pathway Involvement in Muscle Mass Loss in Sex-Linked Dwarf Chickens. Int. J. Mol. Sci. 2016, 17, 276. [Google Scholar] [CrossRef] [Green Version]
- Jia, X.; Lin, H.; Abdalla, B.A.; Nie, Q. Characterization of miR-206 Promoter and Its Association with Birthweight in Chicken. Int. J. Mol. Sci. 2016, 17, 559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, G.; Chen, F.; Wu, P.; Li, T.; He, M.; Yin, X.; Shi, H.; Duan, Y.; Zhang, T.; Wang, J.; et al. MicroRNA-7 Targets the KLF4 Gene to Regulate the Proliferation and Differentiation of Chicken Primary Myoblasts. Front Genet. 2020, 11, 842. [Google Scholar] [CrossRef]
- Duan, Y.; Wu, Y.; Yin, X.; Li, T.; Chen, F.; Wu, P.; Zhang, S.; Wang, J.; Zhang, G. MicroRNA-214 Inhibits Chicken Myoblasts Proliferation, Promotes Their Differentiation, and Targets the TRMT61A Gene. Genes (Basel) 2020, 11, 1400. [Google Scholar] [CrossRef]
- Li, Y.; Zhai, B.; Yuan, P.; Fan, S.; Jin, W.; Li, W.; Sun, G.; Tian, Y.; Liu, X.; Kang, X.; et al. MiR-29b-1-5p regulates the proliferation and differentiation of chicken primary myoblasts and analysis of its effective targets. Poult. Sci. 2022, 101, 101557. [Google Scholar] [CrossRef]
- Li, Y.; Yuan, P.; Fan, S.; Zhai, B.; Li, S.; Li, H.; Zhang, Y.; Li, W.; Sun, G.; Han, R.; et al. miR-30a-3p can inhibit the proliferation and promote the differentiation of chicken primary myoblasts. Br. Poult. Sci. 2022, 28, 1–9. [Google Scholar] [CrossRef]
- Zhang, G.; He, M.; Wu, P.; Zhang, X.; Zhou, K.; Li, T.; Zhang, T.; Xie, K.; Dai, G.; Wang, J. MicroRNA-27b-3p Targets the Myostatin Gene to Regulate Myoblast Proliferation and Is Involved in Myoblast Differentiation. Cells 2021, 10. [Google Scholar] [CrossRef]
- Chen, M.; Zhang, S.; Xu, Z.; Gao, J.; Mishra, S.K.; Zhu, Q.; Zhao, X.; Wang, Y.; Yin, H.; Fan, X.; et al. MiRNA Profil-ing in Pectoral Muscle Throughout Pre- to Post-Natal Stages of Chicken Development. Front Genet. 2020, 11, 570. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Li, E.; Nie, Q.; Zhang, X. Myomaker, Regulated by MYOD, MYOG and miR-140-3p, Promotes Chicken My-oblast Fusion. Int. J. Mol. Sci. 2015, 16, 26186–26201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, B.; Ma, M.; Chen, B.; Li, Z.; Abdalla, B.A.; Nie, Q.; Zhang, X. MiR-16-5p targets SESN1 to regulate the p53 signaling pathway, affecting myoblast proliferation and apoptosis, and is involved in myoblast differentiation. Cell Death Dis. 2018, 9, 367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, X.; Ouyang, H.; Abdalla, B.A.; Xu, H.; Nie, Q.; Zhang, X. miR-16 controls myoblast proliferation and apoptosis through directly suppressing Bcl2 and FOXO1 activities. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 674–684. [Google Scholar] [CrossRef]
- Huang, W.; Guo, L.; Zhao, M.; Zhang, D.; Xu, H.; Nie, Q. The Inhibition on MDFIC and PI3K/AKT Pathway Caused by miR-146b-3p Triggers Suppression of Myoblast Proliferation and Differentiation and Promotion of Apoptosis. Cells 2019, 8, 656. [Google Scholar] [CrossRef] [Green Version]
- Yin, H.; Zhao, J.; He, H.; Chen, Y.; Wang, Y.; Li, D.; Zhu, Q. Gga-miR-3525 Targets PDLIM3 through the MAPK Signaling Pathway to Regulate the Proliferation and Differentiation of Skeletal Muscle Satellite Cells. Int. J. Mol. Sci. 2020, 21, 5573. [Google Scholar] [CrossRef]
- Cao, X.; Tang, S.; Du, F.; Li, H.; Shen, X.; Li, D.; Wang, Y.; Zhang, Z.; Xia, L.; Zhu, Q.; et al. miR-99a-5p Regulates the Proliferation and Differentiation of Skeletal Muscle Satellite Cells by Targeting MTMR3 in Chicken. Genes (Basel) 2020, 11, 369. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Ran, J.; Li, J.; Yu, C.; Cui, Z.; Amevor, F.K.; Wang, Y.; Jiang, X.; Qiu, M.; Du, H.; et al. miR-21-5p Regulates the Proliferation and Differentiation of Skeletal Muscle Satellite Cells by Targeting KLF3 in Chicken. Genes (Basel) 2021, 12, 814. [Google Scholar] [CrossRef]
- Yin, H.; He, H.; Cao, X.; Shen, X.; Han, S.; Cui, C.; Zhao, J.; Wei, Y.; Chen, Y.; Xia, L.; et al. MiR-148a-3p Regulates Skeletal Muscle Satellite Cell Differentiation and Apoptosis via the PI3K/AKT Signaling Pathway by Targeting Me-ox2. Front Genet. 2020, 11, 512. [Google Scholar] [CrossRef]
- Martin, A.I.; Priego, T.; Lopez-Calderon, A. Hormones and Muscle Atrophy. Adv. Exp. Med. Biol. 2018, 1088, 207–233. [Google Scholar] [CrossRef]
- Xu, H.D.; Li, T.; Wang, Z.; Adu-Asiamah, P.; Leng, Q.Y.; Zheng, J.H.; Zhao, Z.H.; An, L.L.; Zhang, X.Q.; Zhang, L. Roles of chicken growth hormone receptor antisense transcript in chicken muscle development and myoblast differentiation. Poult. Sci. 2019, 98, 6980–6988. [Google Scholar] [CrossRef] [PubMed]
- Clemmons, D.R. Role of IGF-I in skeletal muscle mass maintenance. Trends Endocrinol. Metab. 2009, 20, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Chen, J.; Wu, J.; Ren, X.; Li, L.; Lu, S.; Cheng, T.; Tan, L.; Liu, M.; Luo, Q.; et al. Integrative Analyses of mRNA Expression Profile Reveal SOCS2 and CISH Play Important Roles in GHR Mutation-Induced Excessive Abdominal Fat Deposition in the Sex-Linked Dwarf Chicken. Front Genet. 2020, 11, 610605. [Google Scholar] [CrossRef] [PubMed]
- Berg, M.A.; Argente, J.; Chernausek, S.; Gracia, R.; Guevara-Aguirre, J.; Hopp, M.; Perez-Jurado, L.; Rosenbloom, A.; Toledo, S.P.; Francke, U. Diverse growth hormone receptor gene mutations in Laron syndrome. Am. J. Hum. Genet. 1993, 52, 998–1005. [Google Scholar] [PubMed]
- Ben-Yair, R.; Kalcheim, C. Lineage analysis of the avian dermomyotome sheet reveals the existence of single cells with both dermal and muscle progenitor fates. Development 2005, 132, 689–701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, H.; Price, F.; Rudnicki, M.A. Satellite cells and the muscle stem cell niche. Physiol. Rev. 2013, 93, 23–67. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Kim, S.W.; Han, J.S.; Shin, S.P.; Lee, S.I.; Park, T.S. Functional analyses of miRNA-146b-5p during myo-genic proliferation and differentiation in chicken myoblasts. BMC Mol. Cell Biol. 2020, 21, 40. [Google Scholar] [CrossRef]
- Lee, Y.; Lim, B.; Lee, S.W.; Lee, W.R.; Kim, Y.I.; Kim, M.; Ju, H.; Kim, M.Y.; Kang, S.J.; Song, J.J.; et al. ANKRD9 is associated with tumor suppression as a substrate receptor subunit of ubiquitin ligase. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 3145–3153. [Google Scholar] [CrossRef]
- Zhu, M.; Wang, M.; Shao, Y.; Nan, Y.; Blair, H.T.; Morris, S.T.; Zhao, Z.; Zhang, H. Characterization of muscle development and gene expression in early embryos of chicken, quail, and their hybrids. Gene 2021, 768, 145319. [Google Scholar] [CrossRef]
- Lehka, L.; Redowicz, M.J. Mechanisms regulating myoblast fusion: A multilevel interplay. Semin. Cell Dev. Biol. 2020, 104, 81–92. [Google Scholar] [CrossRef]
- Jiang, A.; Guo, H.; Wu, W.; Liu, H. The Crosstalk between Autophagy and Apoptosis Is Necessary for Myogenic Differentiation. J. Agric. Food Chem. 2021, 69, 3942–3951. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.X.; Dumont, N.A.; Rudnicki, M.A. Muscle stem cells at a glance. J. Cell Sci. 2014, 127, 4543–4548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scharner, J.; Zammit, P.S. The muscle satellite cell at 50: The formative years. Skelet. Muscle 2011, 1, 28. [Google Scholar] [CrossRef] [Green Version]
- Seale, P.; Sabourin, L.A.; Girgis-Gabardo, A.; Mansouri, A.; Gruss, P.; Rudnicki, M.A. Pax7 is required for the specification of myogenic satellite cells. Cell 2000, 102, 777–786. [Google Scholar] [CrossRef] [Green Version]
- Hosotani, M.; Kametani, K.; Ohno, N.; Hiramatsu, K.; Kawasaki, T.; Hasegawa, Y.; Iwasaki, T.; Watanabe, T. The unique physiological features of the broiler pectoralis major muscle as suggested by the three-dimensional ultra-structural study of mitochondria in type IIb muscle fibers. J. Vet. Med. Sci. 2021, 83, 1764–1771. [Google Scholar] [CrossRef]
- Ismail, I.; Joo, S.T. Poultry Meat Quality in Relation to Muscle Growth and Muscle Fiber Characteristics. Korean J. Food Sci. Anim. Resour. 2017, 37, 873–883. [Google Scholar] [CrossRef]
- Liu, J.; Zhou, Y.; Hu, X.; Yang, J.; Lei, Q.; Liu, W.; Han, H.; Li, F.; Cao, D. Transcriptome Analysis Reveals the Profile of Long Non-coding RNAs During Chicken Muscle Development. Front Physiol. 2021, 12, 660370. [Google Scholar] [CrossRef]
- Ma, M.; Cai, B.; Jiang, L.; Abdalla, B.A.; Li, Z.; Nie, Q.; Zhang, X. lncRNA-Six1 Is a Target of miR-1611 that Functions as a ceRNA to Regulate Six1 Protein Expression and Fiber Type Switching in Chicken Myogenesis. Cells 2018, 7, 243. [Google Scholar] [CrossRef] [Green Version]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011, 147, 358–369. [Google Scholar] [CrossRef] [Green Version]
- Cai, B.; Ma, M.; Zhang, J.; Wang, Z.; Kong, S.; Zhou, Z.; Lian, L.; Zhang, J.; Li, J.; Wang, Y.; et al. LncEDCH1 improves mitochondrial function to reduce muscle atrophy by interacting with SERCA2. Mol. Ther. Nucleic Acids 2022, 27, 319–334. [Google Scholar] [CrossRef]
- Jin, J.J.; Lv, W.; Xia, P.; Xu, Z.Y.; Zheng, A.D.; Wang, X.J.; Wang, S.S.; Zeng, R.; Luo, H.M.; Li, G.L.; et al. Long noncoding RNA SYISL regulates myogenesis by interacting with polycomb repressive complex 2. Proc. Natl. Acad. Sci. USA 2018, 115, E9802–E9811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Li, B.; Jiang, A.; Cao, Y.; Hou, L.; Zhang, Z.; Zhang, X.; Liu, H.; Kim, K.H.; Wu, W. Exploring the lncRNAs Related to Skeletal Muscle Fiber Types and Meat Quality Traits in Pigs. Genes (Basel) 2020, 11, 883. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Deng, M.; Fan, Y.; Yang, H.; Zhang, G.; Feng, X.; Li, F.; Wang, D.; Wang, F.; Zhang, Y. Genome-Wide Analysis Reveals Extensive Changes in LncRNAs during Skeletal Muscle Development in Hu Sheep. Genes (Basel) 2017, 8, 191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, J.; Lv, Q.; Khas, E.; Bai, C.; Ma, B.; Li, W.; Cao, Q.; Fan, Z.; Ao, C. Tissue-specific regulatory mechanism of LncRNAs and methylation in sheep adipose and muscle induced by Allium mongolicum Regel extracts. Sci. Rep. 2021, 11, 9186. [Google Scholar] [CrossRef]
- Ling, Y.; Zheng, Q.; Sui, M.; Zhu, L.; Xu, L.; Zhang, Y.; Liu, Y.; Fang, F.; Chu, M.; Ma, Y.; et al. Comprehensive Analysis of LncRNA Reveals the Temporal-Specific Module of Goat Skeletal Muscle Development. Int. J. Mol. Sci. 2019, 20, 3950. [Google Scholar] [CrossRef] [Green Version]
- Shi, T.; Hu, W.; Hou, H.; Zhao, Z.; Shang, M.; Zhang, L. Identification and Comparative Analysis of Long Non-Coding RNA in the Skeletal Muscle of Two Dezhou Donkey Strains. Genes (Basel) 2020, 11, 508. [Google Scholar] [CrossRef]
- Yan, X.M.; Zhang, Z.; Liu, J.B.; Li, N.; Yang, G.W.; Luo, D.; Zhang, Y.; Yuan, B.; Jiang, H.; Zhang, J.B. Genome-wide identification and analysis of long noncoding RNAs in longissimus muscle tissue from Kazakh cattle and Xinjiang brown cattle. Anim. Biosci. 2021, 34, 1739–1748. [Google Scholar] [CrossRef]
- Cai, B.; Li, Z.; Ma, M.; Wang, Z.; Han, P.; Abdalla, B.A.; Nie, Q.; Zhang, X. LncRNA-Six1 Encodes a Micropeptide to Activate Six1 in Cis and Is Involved in Cell Proliferation and Muscle Growth. Front. Physiol. 2017, 8, 230. [Google Scholar] [CrossRef]
- Zhou, R.; Wang, Y.X.; Long, K.R.; Jiang, A.A.; Jin, L. Regulatory mechanism for lncRNAs in skeletal muscle development and progress on its research in domestic animals. Yi Chuan 2018, 40, 292–304. [Google Scholar] [CrossRef]
- Li, T.; Wang, S.; Wu, R.; Zhou, X.; Zhu, D.; Zhang, Y. Identification of long non-protein coding RNAs in chicken skeletal muscle using next generation sequencing. Genomics 2012, 99, 292–298. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Ouyang, H.; Zheng, M.; Cai, B.; Han, P.; Abdalla, B.A.; Nie, Q.; Zhang, X. Integrated Analysis of Long Non-coding RNAs (LncRNAs) and mRNA Expression Profiles Reveals the Potential Role of LncRNAs in Skeletal Muscle Development of the Chicken. Front Physiol. 2016, 7, 687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Jin, W.; Zhai, B.; Chen, Y.; Li, G.; Zhang, Y.; Guo, Y.; Sun, G.; Han, R.; Li, Z.; et al. LncRNAs and their regulatory networks in breast muscle tissue of Chinese Gushi chickens during late postnatal development. BMC Genom. 2021, 22, 44. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Li, F.; Sun, J.W.; Li, D.H.; Li, W.T.; Jiang, R.R.; Li, Z.J.; Liu, X.J.; Han, R.L.; Li, G.X.; et al. LncRNA IM-FNCR Promotes Intramuscular Adipocyte Differentiation by Sponging miR-128-3p and miR-27b-3p. Front Genet. 2019, 10, 42. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Cai, B.; Abdalla, B.A.; Zhu, X.; Zheng, M.; Han, P.; Nie, Q.; Zhang, X. LncIRS1 controls muscle atrophy via sponging miR-15 family to activate IGF1-PI3K/AKT pathway. J. Cachexia Sarcopenia Muscle 2019, 10, 391–410. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.A.; Wang, Z.; Yang, X.; Ma, M.; Li, Z.; Nie, Q. LncRNA-FKBP1C regulates muscle fiber type switching by affecting the stability of MYH1B. Cell Death Discov. 2021, 7, 73. [Google Scholar] [CrossRef]
- Patop, I.L.; Wust, S.; Kadener, S. Past, present, and future of circRNAs. EMBO J. 2019, 38, e100836. [Google Scholar] [CrossRef]
- Rybak-Wolf, A.; Stottmeister, C.; Glazar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R.; et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Ex-pressed. Mol. Cell 2015, 58, 870–885. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Schuman, E. Circular RNAs in Brain and Other Tissues: A Functional Enigma. Trends Neurosci. 2016, 39, 597–604. [Google Scholar] [CrossRef]
- Bose, R.; Ain, R. Regulation of Transcription by Circular RNAs. Adv. Exp. Med. Biol. 2018, 1087, 81–94. [Google Scholar] [CrossRef]
- Pamudurti, N.R.; Bartok, O.; Jens, M.; Ashwal-Fluss, R.; Stottmeister, C.; Ruhe, L.; Hanan, M.; Wyler, E.; Perez-Hernandez, D.; Ramberger, E.; et al. Translation of CircRNAs. Mol. Cell 2017, 66, 9–21. [Google Scholar] [CrossRef] [Green Version]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, A.; Das, A.; Das, D.; Abdelmohsen, K.; Panda, A.C. Circular RNAs in myogenesis. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194372. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Li, H.; Yang, J.; Hao, D.; Dong, D.; Huang, Y.; Lan, X.; Plath, M.; Lei, C.; Lin, F.; et al. Circular RNA profiling reveals an abundant circLMO7 that regulates myoblasts differentiation and survival by sponging miR-378a-3p. Cell Death Dis. 2017, 8, e3153. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Li, X.; Yang, L.; Chen, L.L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cell 2018, 71, 428–442. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Chen, Y.; Nie, L.; Ding, X.; Zhang, X.; Zhao, W.; Xu, X.; Kyei, B.; Dai, D.; Zhan, S.; et al. MyoD-induced circular RNA CDR1as promotes myogenic differentiation of skeletal muscle satellite cells. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 807–821. [Google Scholar] [CrossRef]
- Gao, M.; Li, X.; Yang, Z.; Zhao, S.; Ling, X.; Li, J.; Xing, K.; Qi, X.; Wang, X.; Xiao, L.; et al. circHIPK3 regulates proliferation and differentiation of myoblast through the miR-7/TCF12 pathway. J. Cell. Physiol. 2021, 236, 6793–6805. [Google Scholar] [CrossRef]
- Liu, J.; Li, M.; Kong, L.; Cao, M.; Zhang, M.; Wang, Y.; Song, C.; Fang, X.; Chen, H.; Zhang, C. CircARID1A regulates mouse skeletal muscle regeneration by functioning as a sponge of miR-6368. FASEB J. 2021, 35, e21324. [Google Scholar] [CrossRef]
- Czubak, K.; Taylor, K.; Piasecka, A.; Sobczak, K.; Kozlowska, K.; Philips, A.; Sedehizadeh, S.; Brook, J.D.; Wojciechowska, M.; Kozlowski, P. Global Increase in Circular RNA Levels in Myotonic Dystrophy. Front Genet 2019, 10, 649. [Google Scholar] [CrossRef] [Green Version]
- Shen, M.; Li, T.; Zhang, G.; Wu, P.; Chen, F.; Lou, Q.; Chen, L.; Yin, X.; Zhang, T.; Wang, J. Dynamic expression and functional analysis of circRNA in granulosa cells during follicular development in chicken. BMC Genom. 2019, 20, 96. [Google Scholar] [CrossRef]
- Shen, M.; Wu, P.; Li, T.; Wu, P.; Chen, F.; Chen, L.; Xie, K.; Wang, J.; Zhang, G. Transcriptome Analysis of circRNA and mRNA in Theca Cells during Follicular Development in Chickens. Genes (Basel) 2020, 11, 489. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.D.; Song, J.; Liu, X.; Shan, H. Research Note: Circular RNA expressing in different developmental stages of the chicken bursa of Fabricius. Poult. Sci. 2020, 99, 3846–3852. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Zhang, B.; Zhong, H.; Nie, R.; Ling, Y.; Zhang, H.; Wu, C. Dynamic Expression and Regulatory Network of Circular RNA for Abdominal Preadipocytes Differentiation in Chicken (Gallus gallus). Front Cell Dev. Biol. 2021, 9, 761638. [Google Scholar] [CrossRef] [PubMed]
- Ju, X.; Liu, Y.; Shan, Y.; Ji, G.; Zhang, M.; Tu, Y.; Zou, J.; Chen, X.; Geng, Z.; Shu, J. Analysis of potential regulatory LncRNAs and CircRNAs in the oxidative myofiber and glycolytic myofiber of chickens. Sci. Rep. 2021, 11, 20861. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Chang, G.; Bi, Y.; Liu, X.; Chen, G. Circular RNA and mRNA profiling reveal competing endogenous RNA networks during avian leukosis virus, subgroup J-induced tumorigenesis in chickens. PLoS ONE 2018, 13, e204931. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.U.; Agarwal, V.; Guo, H.; Bartel, D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014, 15, 409. [Google Scholar] [CrossRef]
- Li, K.; Huang, W.; Wang, Z.; Chen, Y.; Cai, D.; Nie, Q. circTAF8 Regulates Myoblast Development and Associated Carcass Traits in Chicken. Front Genet. 2021, 12, 743757. [Google Scholar] [CrossRef]
- Xu, H.; Leng, Q.; Zheng, J.; Adu-Asiamah, P.; Lin, S.; Li, T.; Wang, Z.; An, L.; Zhao, Z.; Zhang, L. Effects of Circular RNA of Chicken Growth Hormone Receptor Gene on Cell Proliferation. Front Genet. 2021, 12, 598575. [Google Scholar] [CrossRef]
- Ouyang, H.; Chen, X.; Li, W.; Li, Z.; Nie, Q.; Zhang, X. Circular RNA circSVIL Promotes Myoblast Proliferation and Differentiation by Sponging miR-203 in Chicken. Front Genet. 2018, 9, 172. [Google Scholar] [CrossRef] [Green Version]
- Cai, B.; Ma, M.; Zhou, Z.; Kong, S.; Zhang, J.; Zhang, X.; Nie, Q. circPTPN4 regulates myogenesis via the miR-499-3p/NAMPT axis. J. Anim. Sci. Biotechnol. 2022, 13, 2. [Google Scholar] [CrossRef]
- Ouyang, H.; Chen, X.; Wang, Z.; Yu, J.; Jia, X.; Li, Z.; Luo, W.; Abdalla, B.A.; Jebessa, E.; Nie, Q.; et al. Circular RNAs are abundant and dynamically expressed during embryonic muscle development in chickens. DNA Res. 2018, 25, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Yu, J.; Guo, L.; Byers, M.S.; Wang, Z.; Chen, X.; Xu, H.; Nie, Q. Circular RNA circHIPK3 Promotes the Proliferation and Differentiation of Chicken Myoblast Cells by Sponging miR-30a-3p. Cells 2019, 8, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, X.; Wei, Y.; Liu, W.; You, G.; Tang, S.; Su, Z.; Du, M.; He, J.; Zhao, J.; Tian, Y.; et al. A Novel Circular RNA cir-cITSN2 Targets the miR-218-5p/LMO7 Axis to Promote Chicken Embryonic Myoblast Proliferation and Differentiation. Front Cell Dev. Biol. 2021, 9, 748844. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Liu, Z.; Cao, X.; He, H.; Han, S.; Chen, Y.; Cui, C.; Zhao, J.; Li, D.; Wang, Y.; et al. Circular RNA profiling identified an abundant circular RNA circTMTC1 that inhibits chicken skeletal muscle satellite cell differentiation by sponging miR-128-3p. Int. J. Biol. Sci. 2019, 15, 2265–2281. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Wei, Y.; You, G.; Liu, W.; Amevor, F.K.; Zhang, Y.; He, H.; Ma, M.; Zhang, Y.; Li, D.; et al. Circular PPP1R13B RNA Promotes Chicken Skeletal Muscle Satellite Cell Proliferation and Differentiation via Targeting miR-9-5p. Animals (Basel) 2021, 11, 2396. [Google Scholar] [CrossRef]
- Wei, Y.; Tian, Y.; Li, X.; Amevor, F.K.; Shen, X.; Zhao, J.; Zhao, X.; Zhang, X.; Huang, W.; Hu, J.; et al. Circular RNA circFNDC3AL Upregulates BCL9 Expression to Promote Chicken Skeletal Muscle Satellite Cells Proliferation and Differentiation by Binding to miR-204. Front Cell Dev. Biol. 2021, 9, 736749. [Google Scholar] [CrossRef]
- Yin, H.; Shen, X.; Zhao, J.; Cao, X.; He, H.; Han, S.; Chen, Y.; Cui, C.; Wei, Y.; Wang, Y.; et al. Circular RNA CircFAM188B Encodes a Protein That Regulates Proliferation and Differentiation of Chicken Skeletal Muscle Satellite Cells. Front Cell Dev. Biol. 2020, 8, 522588. [Google Scholar] [CrossRef]
- Luo, W.; Wu, H.; Ye, Y.; Li, Z.; Hao, S.; Kong, L.; Zheng, X.; Lin, S.; Nie, Q.; Zhang, X. The transient expression of miR-203 and its inhibiting effects on skeletal muscle cell proliferation and differentiation. Cell Death Dis. 2014, 5, e1347. [Google Scholar] [CrossRef] [Green Version]
- Kong, S.; Tao, M.; Shen, X.; Ju, S. Translatable circRNAs and lncRNAs: Driving mechanisms and functions of their translation products. Cancer Lett. 2020, 483, 59–65. [Google Scholar] [CrossRef]
- Meganck, R.M.; Liu, J.; Hale, A.E.; Simon, K.E.; Fanous, M.M.; Vincent, H.A.; Wilusz, J.E.; Moorman, N.J.; Marzluff, W.F.; Asokan, A. Engineering highly efficient backsplicing and translation of synthetic circRNAs. Mol. Ther. Nucleic Acids 2021, 23, 821–834. [Google Scholar] [CrossRef]
- Shi, Y.; Jia, X.; Xu, J. The new function of circRNA: Translation. Clin. Transl. Oncol. 2020, 22, 2162–2169. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N (6)-methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, X.; Chen, L.; Zhou, Y.; Wang, Q.; Zheng, Z.; Xu, B.; Wu, C.; Zhou, Q.; Hu, W.; Wu, C.; et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol. Cancer 2019, 18, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paraboschi, E.M.; Cardamone, G.; Solda, G.; Duga, S.; Asselta, R. Interpreting Non-coding Genetic Variation in Mul-tiple Sclerosis Genome-Wide Associated Regions. Front Genet. 2018, 9, 647. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.L.; Wu, W.P.; Cheng, J.; Liang, L.L.; Cen, J.M.; Chen, C.; Liu, X.; Xiong, X.D. CircFOXO3 rs12196996, a polymorphism at the gene flanking intron, is associated with circFOXO3 levels and the risk of coronary artery disease. Aging (Albany N. Y.) 2020, 12, 13076–13089. [Google Scholar] [CrossRef]
- Shaker, F.; Nikravesh, A.; Arezumand, R.; Aghaee-Bakhtiari, S.H. Web-based tools for miRNA studies analysis. Comput. Biol. Med. 2020, 127, 104060. [Google Scholar] [CrossRef]
- Weikard, R.; Demasius, W.; Kuehn, C. Mining long noncoding RNA in livestock. Anim. Genet. 2017, 48, 3–18. [Google Scholar] [CrossRef]
- Zong, X.; Huang, L.; Tripathi, V.; Peralta, R.; Prasanth, K.V. Knockdown of Nuclear-Retained Long Noncoding RNAs Using Modified DNA Antisense Oligonucleotides. Methods Mol. Biol. (Clifton N. J.) 2015, 1262, 321–331. [Google Scholar]
- Zibitt, M.S.; Hartford, C.C.R.; Lal, A. Interrogating lncRNA functions via CRISPR/Cas systems. RNA Biol. 2021, 18, 2097–2106. [Google Scholar] [CrossRef]
- Luiza, C.P.; Dorota, S. CRISPR/Cas9 gene editing in a chicken model: Current approaches and applications. J. Appl. Genet. 2020, 61, 221–229. [Google Scholar] [CrossRef] [Green Version]
- Zuo, Q.; Jing, J.; Zhou, J.; Zhang, Y.; Wei, W.; Chen, G.; Li, B. Dual regulatory actions of LncBMP4 on BMP4 promote chicken primordial germ cell formation. EMBO Rep. 2022, 23, e52491. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Q.; Jin, J.; Jin, K.; Zhou, J.; Sun, C.; Song, J.; Chen, G.; Zhang, Y.; Li, B. P53 and H3K4me2 activate N6-methylated LncPGCAT-1 to regulate primordial germ cell formation via MAPK signaling. J. Cell. Physiol. 2020, 235, 9895–9909. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, Y.; Zhang, C.; Sun, X. Biological functions of circRNAs and their progress in livestock and poultry. Reprod. Domest. Anim. 2020, 55, 1667–1677. [Google Scholar] [CrossRef] [PubMed]
- Shu, L.; Zhou, C.; Yuan, X.; Zhang, J.; Deng, L. MSCFS: Inferring circRNA functional similarity based on multiple data sources. BMC Bioinform. 2021, 22, 371. [Google Scholar] [CrossRef]
- Hu, D.; Zhang, P.; Chen, M. Database Resources for Functional Circular RNAs. Methods Mol. Biol. 2021, 2284, 457–466. [Google Scholar] [CrossRef] [PubMed]
| Items | miRNA | lncRNA | circRNA |
|---|---|---|---|
| Chicken | 674 | 12850 | 494 |
| Reference Database | miRBase (https://www.mirbase.org/, accessed on 14 May 2022) | NONCODE (http://www.noncode.org/, accessed on 14 May 2022) | CircFunBase (http://bis.zju.edu.cn/CircFunBase/index.php, accessed on 14 May 2022) |
| Regulating Method | miRNA | Target Genes | Function | Chicken Breeds | Organs | Days of Age | References |
|---|---|---|---|---|---|---|---|
| Hormone-related genes | MiR-let7b | GHR | Inhibit the growth of skeletal muscle | Recessive White Rock and Xinghua | Leg muscles | E14, 7 w | [32] |
| MiR-146b-3p | Pectoral muscles | 7 w | [33] | ||||
| Myogenic factors | MiR-204 | MAP3K13 | Inhibit skeletal muscle growth | Sichuan mountainous black-bone and Dahen | Pectoral muscles | 10 w | [25] |
| MiR-142-5p | FOXO3 | Promotes skeletal muscle growth | Recessive White Rock and Xinghua | Pectoral and leg muscles | 7 w | [26] | |
| MiR-128 | MSTN | Muscle mass loss | Sex-linked dwarf | Leg muscles | E14, 7 w | [34] | |
| MiR-206 | MyoD MyoG MCK | Increase muscle mass | F2(Recessive White Rock and Xinghua) | Pectoral and leg muscles | 30 w | [35] | |
| Proliferation and differentiation of myoblasts | MiR- 29b-3p | TGFB2 | Inhibits proliferation | Shouguang | Pectoral muscles | E12, E17, D1, D14, D56, D98 | [8] |
| MiR-2954 | YY1 | Promotes proliferation and inhibits differentiation | Jinghai | Leg muscles | E7, E10, E13, E15, E18, D1 | [20] | |
| MiR-7 | KLF4 | Inhibition proliferation and differentiation | Jinghai | Pectoral and leg muscles | E12, E14, E16, E18, E20, D1 | [36] | |
| MiR-214 | TRMT61A | Promotes differentiation | Haiyang | Pectoral and leg muscles | E12, E14, E16, E18, D1 | [37] | |
| MiR-29b-1-5p | ANKRD9 | Inhibits proliferation and promotes differentiatio | Gushi | Pectoral muscles | E10, E12, E14, E16, E18 | [38] | |
| MiR-30a-3p | MYOD | Promotes differentiation | Gushi | Pectoral muscles | E10, E12, E14, E16, E18 | [39] | |
| MiR-233 | IGF2 ZEB1 | Inhibits proliferation and promotes differentiation | Recessive White Rock and Xinghua | Leg muscles | [35] | ||
| MiR- 27b-3p | MSTN | Promote proliferation | Hangyang | Pectoral and leg muscles | E12, E16, E18, E20, D1, 4W, 8W, 16W | [40] | |
| MiR-454-3p | SBF2 | Inhibits differentiation | Tibetan | Pectoral muscles | D300 | [41] | |
| Fusion of myoblasts | MiR-140-3p | Myomaker | Inhibit fusion | Chick | Leg muscles | E10 | [42] |
| Apoptosis of myoblasts | MiR- 16-5p | SESN1 | Inhibits proliferation and differentiation, promotes apoptosis | Xinghua | Pectoral and leg muscles | E10- E20, D1 | [43] |
| MiR-16 | Bcl2 FOXO1 | Inhibits proliferation and promotes apoptosis | Commercial and heritage chickens | hypertrophic and normal pectoral muscle | 7 w | [44] | |
| MiR-146b-3p | MDFIC | Inhibits proliferation and differentiation, promotes apoptosis | Xinghua | Leg muscles | E11, E16 | [45] | |
| Proliferation and differentiation of satellite cells | MiR-9-5p | IGF2BP3 | Inhibits proliferation and differentiation | Recessive White Rock and Xinghua | Pectoral muscles | 7 w | [27] |
| MiR-3525 | PDLIM3 | Inhibits proliferation and differentiation | Ross 308 | Pectoral muscles | D4 | [46] | |
| MiR-99a-5p | MTMR3 | Promote proliferation | Ross 308 | Pectoral muscles | D4 | [47] | |
| MiR-21-5p | KLF3 | Promotes proliferation and differentiation | Dahen | Leg muscles | D3 | [48] | |
| Apoptosis of satellite cells | MiR-499-5p/MiR-196-5p | SOX6/CALM1 | Regulation of slow muscle fiber formation | Qinguan | PMM and SART | D140 | [1] |
| MiR-200a-3p | TGF-β2 | Promotes proliferation and differentiation, inhibits apoptosis | Ross 308 and White Longhorn | Pectoral muscles | E10, E13, E16, E19, | [28] | |
| MiR-148a-3p | Meox2 | Pomotes differentiation and inhibits apoptosis | Ross 308 | Pectoral muscles | D4 | [49] |
| Regulating Method | Lnc RNA | Ce RNA | Target Genes | Functions | Chicken Breeds | Organs | Days of Age | References |
|---|---|---|---|---|---|---|---|---|
| Sponges miRNAs | Lnc-Six1 | MiR-1611 | Six1 | Promotes myoblast proliferation and division | Xinghua | Pectoral and leg muscles | 7 w | [68] |
| Lnc-IMFNCR | MiR- 128-3p/ MiR- 27b-3p | PPARG | Promotes intramuscular adipocytes differentiation | Gushi | Pectoral and leg muscles | 3 w | [83] | |
| Lnc-IRS1 | MiR-15a/miR-15b-5p/MiR-15c-5p | IRS1 | Inhibits muscle atrophy | Hypertrophic broilers and leaner broilers | Pectoral muscles | 6 w | [84] | |
| Regulation of gene expression in cis or trans | Lnc-FKBP1C | MYH1B | Inhibits myoblast proliferation and promotes differentiation, upregulates slow muscle genes | Recessive White Rock and Xinghua | Pectoral and leg muscles | 7 w | [5] | |
| Lnc-EDCH1 | SERCA2 | Promotes myoblast proliferation, Inhibits differentiation, activates slow muscle phenotype, reduce muscle atrophy | Recessive White Rock and Xinghua | Leg muscles | D1, D4, D8 | [70] | ||
| Lnc-Six1 | Six1 | Promotes myoblast proliferation and division | Recessive White Rock and Xinghua | Pectoral and leg muscles | 7 w | [78] |
| Regulating Method | CircRNA | Target miRNAs | Target Genes | Functions | Chicken Breeds | Organs | Days of Age | References |
|---|---|---|---|---|---|---|---|---|
| Regulating parental genes | Circ-GHR | GHR GHBP | Promotes myoblast proliferation | Xinghua | Pectoral and leg muscles | E13, E16, E19, D1, 1 W, 2 W, 3 W, 4 W, 5 W, 6 W, 7 W | [108] | |
| Sponging miRNAs act on myoblasts | Circ-SVIL | MiR-203 | c-JUN MEF2C | Promotes myoblast proliferation and differentiation | Xinghua | Leg muscles | E10-D1 | [109] |
| Circ-PTPN4 | MiR-499-3p | NAMPT | Promotes myoblast proliferation and differentiation | Xinghua | Pectoralis major and soleus | 7 w | [110] | |
| Circ-RBFOX2s | MiR-206 | Promotes myoblast proliferation | Xinghua | Leg muscles | E10-D1 | [111] | ||
| Circ-HIPK3 | MiR-30a-3p | MEF2C | Promotes myoblast proliferation and differentiation | Yuhe | Leg muscles | E10-D1 | [112] | |
| Circ-ITSN2 | MiR-218-5p | LMO7 | Promotes myoblast proliferation and differentiation | Ross 308 and White Longhorn | Pectoral and leg muscles | E10, E13, E16, E19 | [113] | |
| Sponging miRNAs act on satellite cells | CircTMTC1 | MiR-128-3p | MSTN | Inhibition of satellite cell differentiation | Ross 308 and White Longhorn | Pectoral muscles | E10, E13, E16,E19 | [114] |
| Circ-PPP1R13B | MiR-9-5p | IGF2BP3 | Promotes satellite cell proliferation and differentiation | Ross 308 and White Longhorn | Pectoral muscles | [115] | ||
| Circ-FNDC3AL | MiR-204 | BCL9 | Promotes satellite cell proliferation and differentiation | Ross 308 | Pectoral muscles | E10, E13, E16, E19 | [116] | |
| Translating directly into protein | Circ-FAM188B | CircFAM188B-103aa | Promotes satellite cell proliferation and inhibits differentiation | Ross 308 | Pectoral and leg muscles | D1, D3, D5, D7, D14, D21, D28, D35 | [117] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, H.; He, Y.; Li, X.; Du, Y.; Zhao, J.; Ge, C. Regulation of Non-Coding RNA in the Growth and Development of Skeletal Muscle in Domestic Chickens. Genes 2022, 13, 1033. https://doi.org/10.3390/genes13061033
Shi H, He Y, Li X, Du Y, Zhao J, Ge C. Regulation of Non-Coding RNA in the Growth and Development of Skeletal Muscle in Domestic Chickens. Genes. 2022; 13(6):1033. https://doi.org/10.3390/genes13061033
Chicago/Turabian StyleShi, Hongmei, Yang He, Xuzhen Li, Yanli Du, Jinbo Zhao, and Changrong Ge. 2022. "Regulation of Non-Coding RNA in the Growth and Development of Skeletal Muscle in Domestic Chickens" Genes 13, no. 6: 1033. https://doi.org/10.3390/genes13061033
APA StyleShi, H., He, Y., Li, X., Du, Y., Zhao, J., & Ge, C. (2022). Regulation of Non-Coding RNA in the Growth and Development of Skeletal Muscle in Domestic Chickens. Genes, 13(6), 1033. https://doi.org/10.3390/genes13061033






